![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 20150521_trimmed_2212_lane2_1000ppm_CTTGTA.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 57867827 |
| Filtered Sequences | 0 |
| Sequence length | 1-51 |
| %GC | 42 |
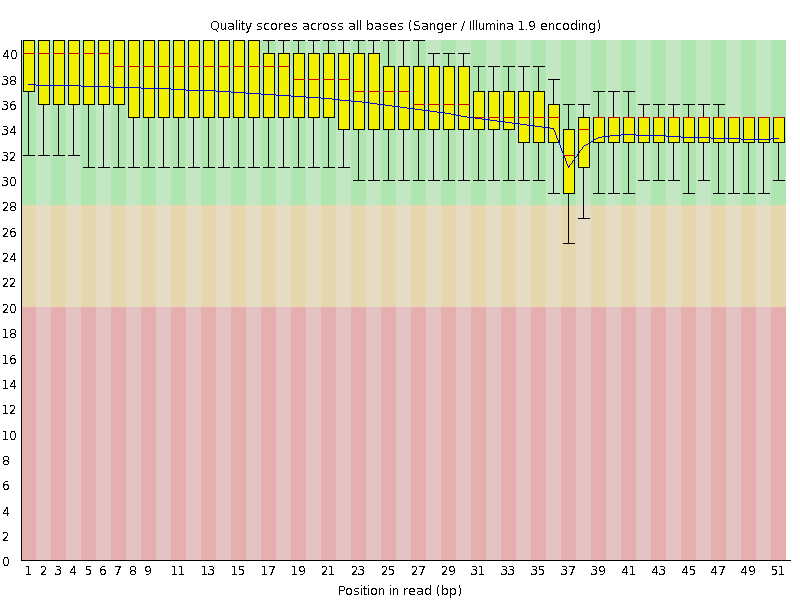
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
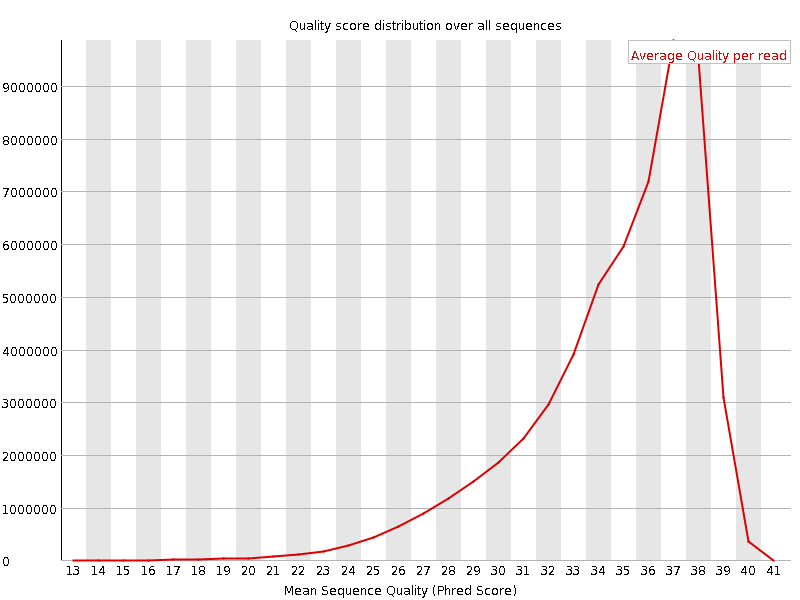
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[OK]](Icons/tick.png) Per base GC content
Per base GC content

![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

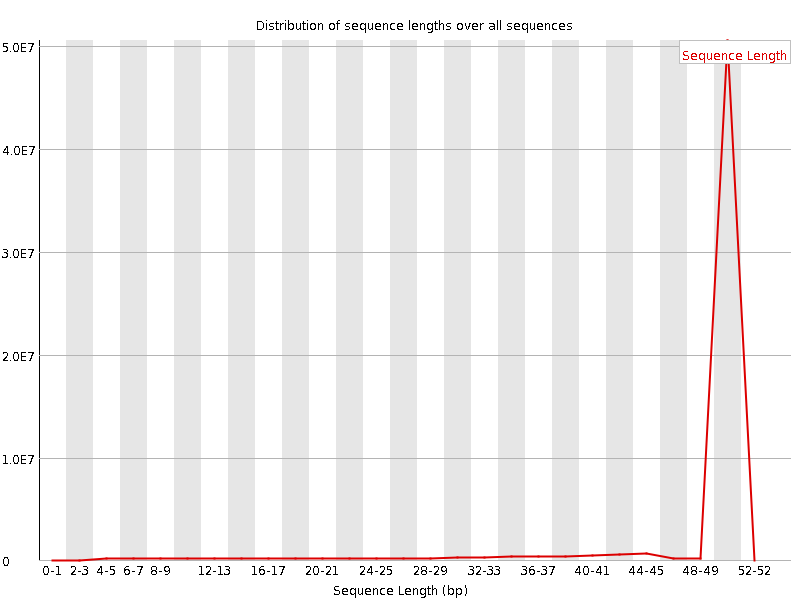
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
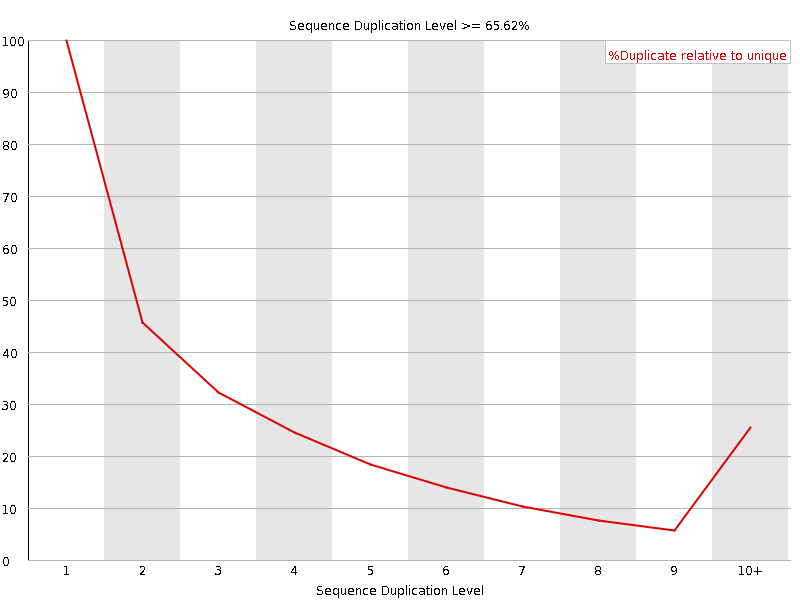
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
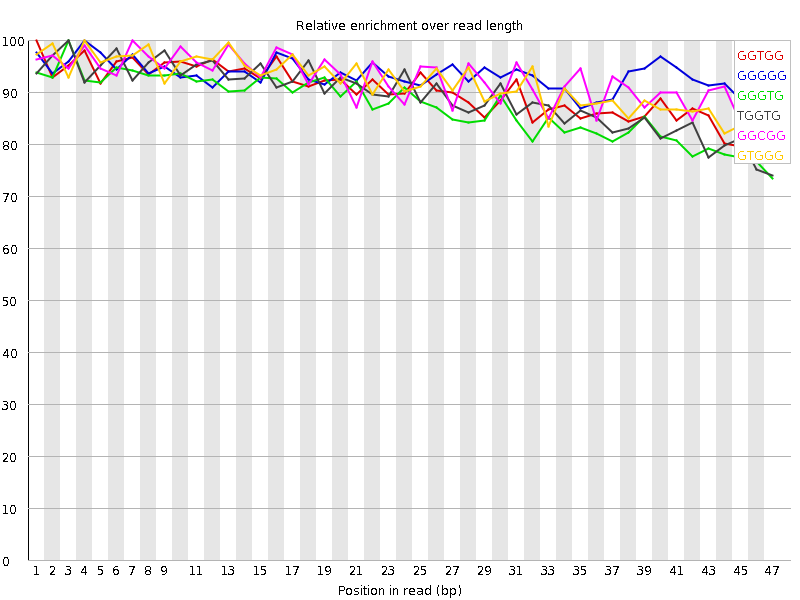
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGTGG | 1667150 | 28.072369 | 31.050274 | 1 |
| GGGGG | 666900 | 24.236992 | 25.984175 | 4 |
| GGGTG | 1195380 | 20.128452 | 22.972113 | 3 |
| TGGTG | 2448040 | 19.098934 | 21.339375 | 3 |
| GGCGG | 1534530 | 17.711245 | 19.078068 | 7 |
| GTGGG | 1035720 | 17.440008 | 18.960598 | 4 |
| GTGTG | 2140275 | 16.697836 | 22.899574 | 1 |
| TGGGG | 879005 | 14.801157 | 16.091248 | 9 |
| GTGGT | 1888035 | 14.729929 | 16.799067 | 2 |
| GGAGG | 1377430 | 14.090473 | 14.916147 | 7 |
| GGTTG | 1779730 | 13.884965 | 15.394164 | 3 |
| GTTGG | 1676655 | 13.080802 | 14.476382 | 4 |
| GGGGT | 768950 | 12.947993 | 14.6253195 | 2 |
| GGTGT | 1637035 | 12.771698 | 14.230278 | 1 |
| TGTTG | 3527480 | 12.75092 | 13.72501 | 42 |
| GGGCG | 1095080 | 12.639198 | 13.704639 | 25 |
| TGTGG | 1566785 | 12.223626 | 13.460924 | 4 |
| GCGGG | 999035 | 11.530664 | 12.489025 | 5 |
| GGGTT | 1391155 | 10.85341 | 14.885671 | 3 |
| GAGGG | 1044350 | 10.683217 | 11.617266 | 1 |
| GGGAG | 927430 | 9.487181 | 10.014297 | 10 |
| TGGGT | 1203920 | 9.3926525 | 10.455533 | 2 |
| TTGGG | 1153325 | 8.997925 | 10.024115 | 5 |
| TGTGT | 2407615 | 8.702901 | 11.5789 | 6 |
| GTTGT | 2396925 | 8.664259 | 9.62965 | 8 |
| GTTTG | 2314230 | 8.365337 | 9.637457 | 46 |
| TGGCG | 1476105 | 7.8936377 | 8.498713 | 3 |
| TTGGT | 2160495 | 7.8096266 | 8.640214 | 2 |
| GATGG | 1645935 | 7.8010845 | 8.635516 | 4 |
| GGTGA | 1625200 | 7.70281 | 8.442287 | 10 |
| GGGGA | 749975 | 7.671898 | 8.225767 | 28 |
| CGGGG | 659975 | 7.617301 | 8.2287445 | 16 |
| CGGCG | 2047910 | 7.5065446 | 8.090735 | 1 |
| GCTGG | 1349370 | 7.215908 | 7.983678 | 1 |
| GGGGC | 624625 | 7.209298 | 7.800057 | 6 |
| TTGTG | 1990835 | 7.196349 | 8.053194 | 2 |
| GTGTT | 1959610 | 7.0834785 | 7.8065934 | 5 |
| TGGTT | 1949110 | 7.0455236 | 7.813073 | 5 |
| GCGGC | 1907900 | 6.993343 | 7.615674 | 11 |
| GCGCG | 1889510 | 6.925935 | 7.3258753 | 3 |
| AGGGG | 662025 | 6.772209 | 7.3490887 | 2 |
| GGATG | 1381690 | 6.548668 | 7.0793486 | 15 |
| TTTGG | 1783960 | 6.44855 | 7.015011 | 7 |
| GGTCG | 1147720 | 6.137562 | 6.7811213 | 22 |
| CGGTG | 1131850 | 6.0526953 | 6.634908 | 10 |
| GAGGT | 1232430 | 5.8412337 | 6.5080233 | 40 |
| GCGGT | 1084430 | 5.799111 | 6.2560315 | 17 |
| GGCGC | 1562955 | 5.728959 | 6.1959977 | 1 |
| TGAGG | 1203835 | 5.7057047 | 6.103026 | 37 |
| GGTTT | 1550890 | 5.6060634 | 6.1355915 | 6 |
| TGCGG | 1043650 | 5.581036 | 5.8590565 | 1 |
| GTCGG | 1032925 | 5.5236826 | 6.223159 | 2 |
| GGCTG | 1004385 | 5.371062 | 5.83316 | 12 |
| CGCGG | 1461475 | 5.356987 | 5.954775 | 1 |
| AGGTG | 1125590 | 5.334854 | 5.797502 | 9 |
| TTGTT | 3176980 | 5.32081 | 5.522712 | 41 |
| GGCGT | 994040 | 5.3157406 | 5.7872734 | 14 |
| TGATG | 2415365 | 5.304096 | 5.8814836 | 3 |
| AGGGT | 1117600 | 5.2969847 | 7.756057 | 2 |
| GGTGC | 969565 | 5.184858 | 5.5383496 | 11 |
| GTGAG | 1072955 | 5.085385 | 5.809968 | 17 |
| GTGGA | 1059955 | 5.02377 | 5.5172873 | 5 |
| GTGGC | 924145 | 4.941969 | 5.5348463 | 5 |
| TGGAG | 1023480 | 4.8508925 | 5.1700873 | 3 |
| GGCGA | 1487390 | 4.8321066 | 5.244955 | 20 |
| GCGTG | 900940 | 4.8178782 | 5.3152766 | 3 |
| GAGTG | 1005515 | 4.7657456 | 5.7542214 | 2 |
| GGCCG | 1209240 | 4.4324284 | 4.944248 | 1 |
| TGTTT | 2625675 | 4.397484 | 4.7751555 | 42 |
| AGTGG | 903935 | 4.2842965 | 5.199709 | 3 |
| TCGGG | 795715 | 4.2551756 | 4.7321577 | 15 |
| TTTGT | 2531360 | 4.239525 | 4.654876 | 43 |
| CGTGG | 784425 | 4.194801 | 4.732473 | 19 |
| GCCGG | 1133100 | 4.1533394 | 4.5293984 | 17 |
| GTGCG | 769315 | 4.1139984 | 4.448177 | 2 |
| TGGGC | 768660 | 4.1104956 | 4.4715014 | 9 |
| GGGAT | 856555 | 4.0597343 | 4.403179 | 8 |
| ATGGG | 846730 | 4.0131674 | 4.4074516 | 8 |
| CGGGC | 1094140 | 4.010533 | 4.339549 | 6 |
| GTTGA | 1794710 | 3.9411497 | 4.300632 | 4 |
| TAGGG | 830935 | 3.9383051 | 6.0264206 | 1 |
| TTTTG | 2326170 | 3.8958724 | 4.2645407 | 6 |
| GTAGG | 817170 | 3.8730645 | 4.3046064 | 43 |
| GTGAT | 1720205 | 3.7775378 | 4.185009 | 2 |
| TGGGA | 784635 | 3.7188613 | 3.989138 | 12 |
| GAGGA | 1272910 | 3.6651495 | 4.0351663 | 2 |
| TGCTG | 1477470 | 3.6607065 | 3.9363317 | 6 |
| GAAGG | 1265315 | 3.6432807 | 3.7871382 | 17 |
| GGTAG | 749800 | 3.5537574 | 3.7370894 | 16 |
| GCAGG | 1085790 | 3.5274222 | 3.754496 | 9 |
| GAGAG | 1222665 | 3.5204768 | 4.176428 | 1 |
| GTTTT | 2095775 | 3.5100064 | 3.8549201 | 7 |
| GATGT | 1596695 | 3.5063121 | 3.8599572 | 1 |
| GATTG | 1570175 | 3.4480748 | 3.7874599 | 9 |
| GTTGC | 1375365 | 3.4077225 | 5.0079646 | 47 |
| GGTAT | 1549800 | 3.403332 | 4.407991 | 40 |
| GACGG | 1034930 | 3.3621924 | 3.6105459 | 2 |
| CGGGT | 625015 | 3.3423376 | 3.6333523 | 2 |
| GTATG | 1503420 | 3.301482 | 4.3111067 | 41 |
| GGAGT | 695655 | 3.2971315 | 3.649847 | 3 |
| CTGGG | 610855 | 3.2666159 | 3.5386045 | 8 |
| CGAGG | 990515 | 3.2179008 | 3.6222596 | 1 |
| GGAAG | 1114740 | 3.2097235 | 3.535488 | 46 |
| AGGCG | 977080 | 3.1742544 | 3.4454813 | 15 |
| GGCAG | 975525 | 3.169203 | 3.4078052 | 44 |
| ATGGT | 1430650 | 3.1416805 | 3.4556289 | 2 |
| AGGAG | 1078245 | 3.1046414 | 3.339159 | 45 |
| GCGGA | 955575 | 3.1043909 | 3.3817565 | 14 |
| GGGCT | 576600 | 3.0834334 | 3.3617783 | 4 |
| AGCGG | 946610 | 3.075266 | 3.2658 | 4 |
| GGGTC | 565370 | 3.0233798 | 3.2825084 | 3 |
| AGATC | 1378065 | 0.58385587 | 6.9355297 | 47 |