Slides and associated material associated with the presenation given at the Friday Harbor Laboratories on August 13th, 2014 -
_"Does DNA methylation facilitate phenotypic plasticity in marine invertebrates?"_.
##Slides
coming soon
##Papers Highlighted
Olson CE and Roberts SB. (**2014**). [Genome-wide profiling of DNA methylation and gene expression in _Crassostrea gigas_ male gametes](http://journal.frontiersin.org/Journal/10.3389/fphys.2014.00224/abstract) Frontiers in Physiology. 5:224. doi: 10.3389/fphys.2014.00224
Gavery MR and Roberts SB. (**2014**) [A context specific role for DNA methylation in bivalves](http://bfg.oxfordjournals.org/content/early/2014/01/06/bfgp.elt054.short) Briefings in Functional Genomics. doi:10.1093/bfgp/elt054 ([pdf](http://eagle.fish.washington.edu/cnidarian/Briefings%20in%20Functional%20Genomics-2014-Gavery-217-22.pdf))
Gavery MR and Roberts SB. (**2013**) [Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc](https://peerj.com/articles/215/) PeerJ 1:e215. doi:10.7717/peerj.215
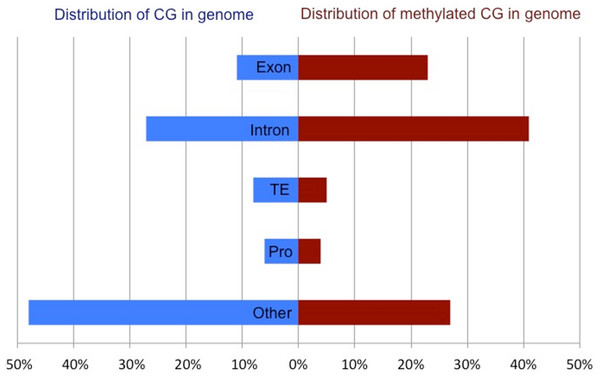 Roberts SB and Gavery MR (**2012**) [Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates?](http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249382/?tool=pubmed) Frontiers in Physiology 2:116. doi:10.3389/fphys.2011.00116
Roberts SB and Gavery MR (**2012**) [Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates?](http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249382/?tool=pubmed) Frontiers in Physiology 2:116. doi:10.3389/fphys.2011.00116
 Gavery MR and Roberts SB. (**2012**) [Characterizing short read sequencing for gene discovery and RNA-Seq analysis in Crassostrea gigas](http://www.sciencedirect.com/science/article/pii/S1744117X11001018) Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 7:2 94-99. doi:10.1016/j.cbd.2011.12.003
Gavery MR and Roberts SB. (**2010**) [DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (_Crassostrea gigas_)](http://www.biomedcentral.com/1471-2164/11/483) BMC Genomics 11:483. doi:10.1186/1471-2164-11-483
##Data
###_Crassostrea gigas_ high-throughput bisulfite sequencing (gill tissue)
Citation: Gavery, Mackenzie; Roberts, Steven (2013): Crassostrea gigas high-throughput bisulfite sequencing (gill tissue). figshare.
This fileset contains genomic feature tracks from **methylation-enriched** high-throughput bisulfite sequencing and RNA-seq analysis for Pacific oyster (Crassostrea gigas) gill tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012). All data and instructions are also available at .
File descriptions:
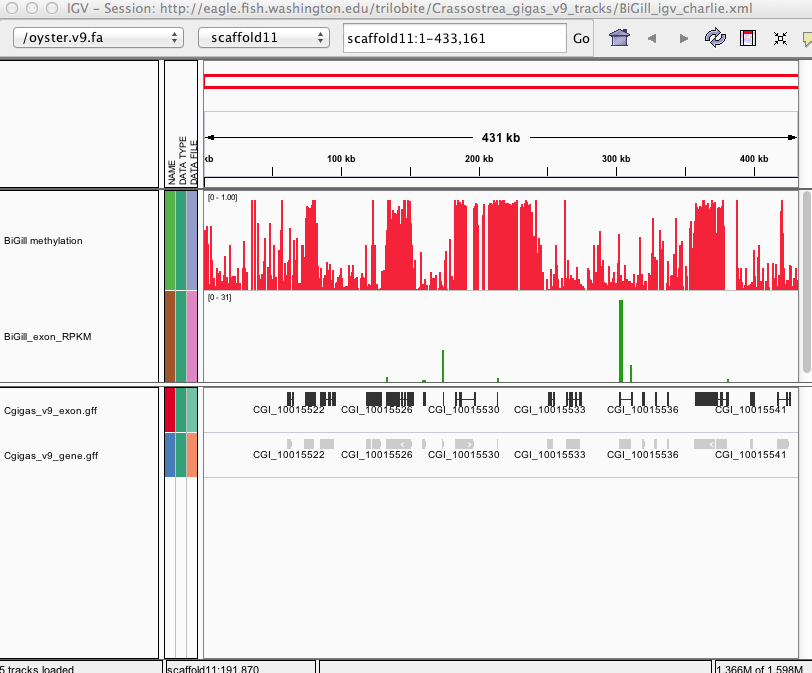
[BiGill_CpG_methylation.igv](http://files.figshare.com/1252773/BiGill_CpG_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGill_exon_clc_rpkm.igv](http://files.figshare.com/1252772/BiGill_exon_clc_rpkm.igv) - Exon-specific gene expression values (RPKM) from RNA-seq analysis.
[BiGill_igv_charlie.xml](http://files.figshare.com/1252770/BiGill_igv_charlie.xml) - A session file, which loads methylation and RNA-seq feature tracks as well as the location of C.gigas genome features.
[Query to derive_CG_AllData_IGV.txt](http://files.figshare.com/1252771/Query_to_derive_CG_AllData_IGV.txt) - Query (SQLShare) used to derive the methylation feature track from the original methratio output (http://goo.gl/5LGq9Q)
Screenshot
***
###_Crassostrea gigas_ high-throughput bisulfite sequencing (male gamete)
This file contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) male gonad tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012).
File descriptions:
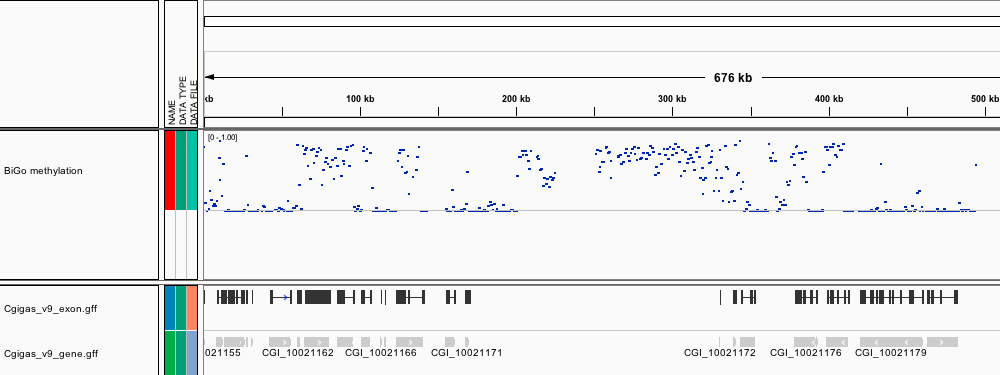
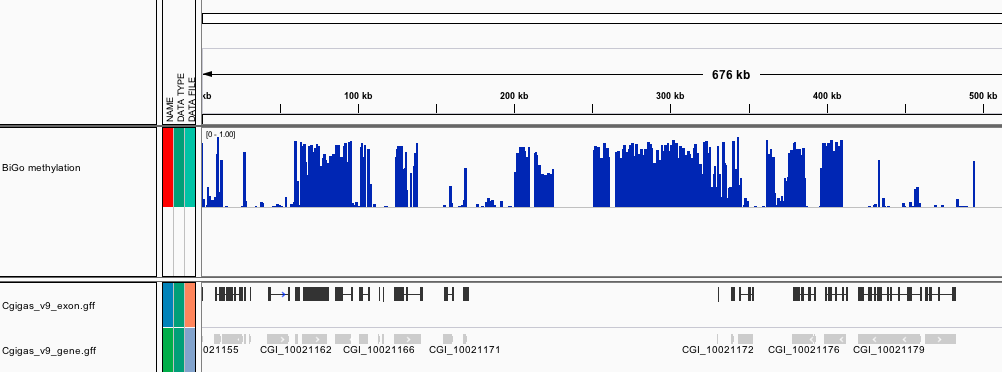
[BiGo_methylation.igv](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGo_igv.xml](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_igv.xml) - A session file, which loads methylation and location of C.gigas genome features.
Screenshots
***
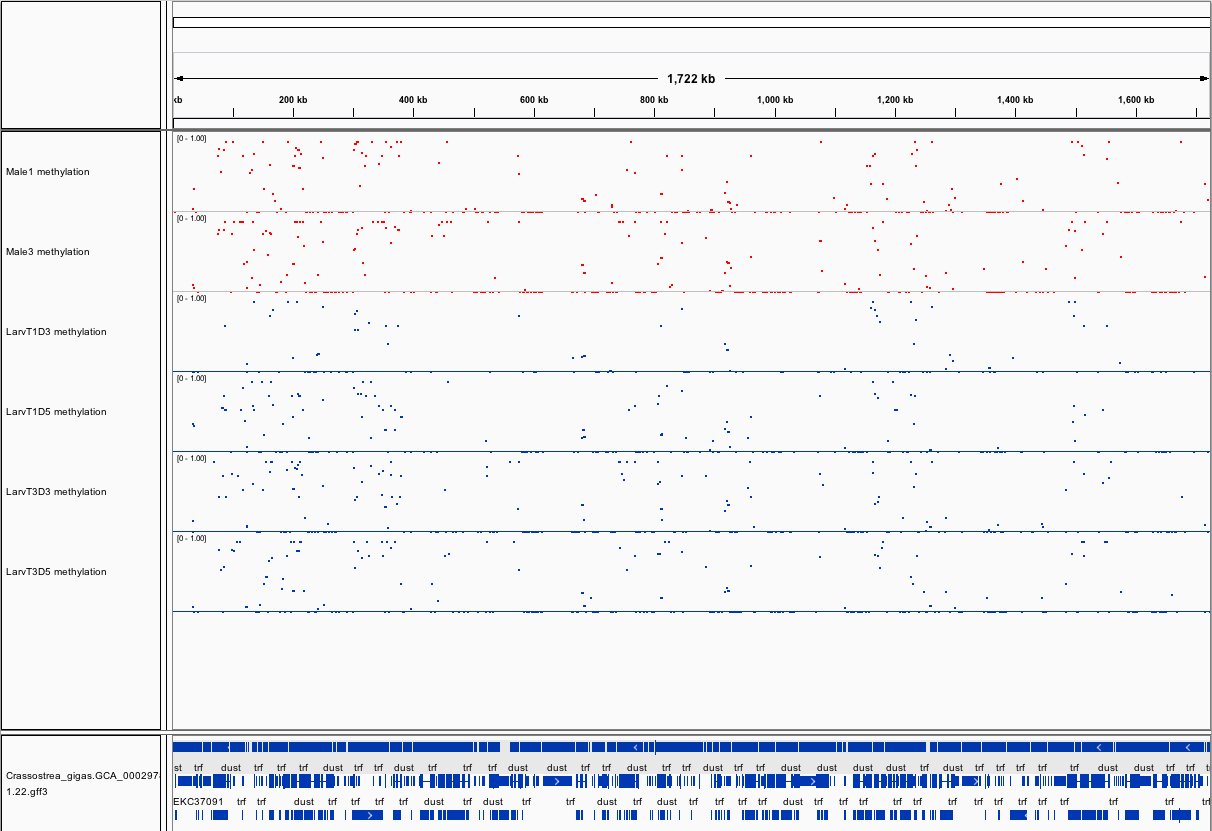
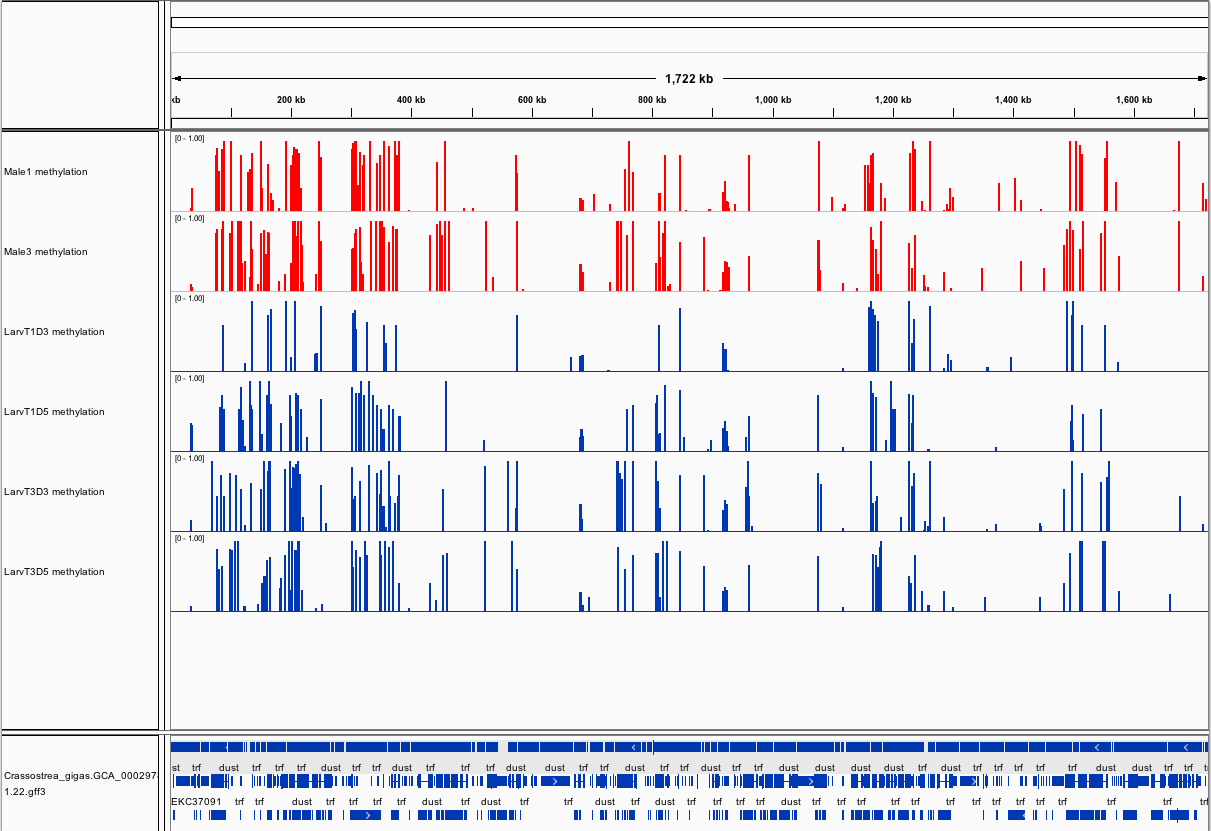
###_Crassostrea gigas_ high-throughput bisulfite sequencing (sperm and larvae)
Citation: Olson, Claire; Roberts, Steven (2014): Crassostrea gigas high-throughput bisulfite sequencing (larvae and sperm tissues). figshare.
This fileset contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) spermatozoa and larvae samples. Samples were taken from two males (Male 1 and Male 3) and larvae taken from these males at Day 3 (D3) and Day 5 (D5) post-fertilization. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Ensembl).
File descriptions:
[methratio_out_CG5x_IGV_M1.igv](http://files.figshare.com/1538436/methratio_out_CG5x_IGV_M1.igv) - Male 1 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_M3.igv](http://files.figshare.com/1538437/methratio_out_CG5x_IGV_M3.igv) - Male 3 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D3.igv](http://files.figshare.com/1538440/methratio_out_CG5x_IGV_T1D3.igv) - Larvae from male 1 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D5.igv](http://files.figshare.com/1538441/methratio_out_CG5x_IGV_T1D5.igv) - Larvae from male 1 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D3.igv](http://files.figshare.com/1538443/methratio_out_CG5x_IGV_T3D3.igv) - Larvae from male 3 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D5.igv](http://files.figshare.com/1538444/methratio_out_CG5x_IGV_T3D5.igv) - Larvae from male 3 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[LarvaeSperm_igv.xml](http://files.figshare.com/1538478/LarvaeSperm_igv.xml) - A session file, which loads sample methylation tracks and location of C.gigas genome features.
Screenshots
##Related Presentations
##People doing the science
###[Mackenzie Gavery](http://students.washington.edu/mgavery/Welcome.html)
Gavery MR and Roberts SB. (**2012**) [Characterizing short read sequencing for gene discovery and RNA-Seq analysis in Crassostrea gigas](http://www.sciencedirect.com/science/article/pii/S1744117X11001018) Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 7:2 94-99. doi:10.1016/j.cbd.2011.12.003
Gavery MR and Roberts SB. (**2010**) [DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (_Crassostrea gigas_)](http://www.biomedcentral.com/1471-2164/11/483) BMC Genomics 11:483. doi:10.1186/1471-2164-11-483
##Data
###_Crassostrea gigas_ high-throughput bisulfite sequencing (gill tissue)
Citation: Gavery, Mackenzie; Roberts, Steven (2013): Crassostrea gigas high-throughput bisulfite sequencing (gill tissue). figshare.
This fileset contains genomic feature tracks from **methylation-enriched** high-throughput bisulfite sequencing and RNA-seq analysis for Pacific oyster (Crassostrea gigas) gill tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012). All data and instructions are also available at .
File descriptions:
[BiGill_CpG_methylation.igv](http://files.figshare.com/1252773/BiGill_CpG_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGill_exon_clc_rpkm.igv](http://files.figshare.com/1252772/BiGill_exon_clc_rpkm.igv) - Exon-specific gene expression values (RPKM) from RNA-seq analysis.
[BiGill_igv_charlie.xml](http://files.figshare.com/1252770/BiGill_igv_charlie.xml) - A session file, which loads methylation and RNA-seq feature tracks as well as the location of C.gigas genome features.
[Query to derive_CG_AllData_IGV.txt](http://files.figshare.com/1252771/Query_to_derive_CG_AllData_IGV.txt) - Query (SQLShare) used to derive the methylation feature track from the original methratio output (http://goo.gl/5LGq9Q)
Screenshot

***
###_Crassostrea gigas_ high-throughput bisulfite sequencing (male gamete)
This file contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) male gonad tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012).
File descriptions:
[BiGo_methylation.igv](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGo_igv.xml](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_igv.xml) - A session file, which loads methylation and location of C.gigas genome features.
Screenshots


***
###_Crassostrea gigas_ high-throughput bisulfite sequencing (sperm and larvae)
Citation: Olson, Claire; Roberts, Steven (2014): Crassostrea gigas high-throughput bisulfite sequencing (larvae and sperm tissues). figshare.
This fileset contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) spermatozoa and larvae samples. Samples were taken from two males (Male 1 and Male 3) and larvae taken from these males at Day 3 (D3) and Day 5 (D5) post-fertilization. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Ensembl).
File descriptions:
[methratio_out_CG5x_IGV_M1.igv](http://files.figshare.com/1538436/methratio_out_CG5x_IGV_M1.igv) - Male 1 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_M3.igv](http://files.figshare.com/1538437/methratio_out_CG5x_IGV_M3.igv) - Male 3 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D3.igv](http://files.figshare.com/1538440/methratio_out_CG5x_IGV_T1D3.igv) - Larvae from male 1 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D5.igv](http://files.figshare.com/1538441/methratio_out_CG5x_IGV_T1D5.igv) - Larvae from male 1 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D3.igv](http://files.figshare.com/1538443/methratio_out_CG5x_IGV_T3D3.igv) - Larvae from male 3 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D5.igv](http://files.figshare.com/1538444/methratio_out_CG5x_IGV_T3D5.igv) - Larvae from male 3 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[LarvaeSperm_igv.xml](http://files.figshare.com/1538478/LarvaeSperm_igv.xml) - A session file, which loads sample methylation tracks and location of C.gigas genome features.
Screenshots


##Related Presentations
##People doing the science
###[Mackenzie Gavery](http://students.washington.edu/mgavery/Welcome.html)
 My research aims to improve our understanding of the physiological responses of marine invertebrates to natural and human-driven environmental change. Specifically, I am interested the role of epigenetics, processes that can alter gene activity without changes to the underlying DNA sequence, in mediating environmental responses in shellfish. I am using the Pacific oyster (Crassostrea gigas) as a model to examine the role of DNA methylation in regulating interactions between genes and the environment. Through this work I hope to better understand the pathways through which environmental contaminants, such as endocrine disruptors, induce physiological changes and to determine whether these induced changes can be passed on to future generations
---
###[Claire Olson](http://students.washington.edu/che625/)
My research aims to improve our understanding of the physiological responses of marine invertebrates to natural and human-driven environmental change. Specifically, I am interested the role of epigenetics, processes that can alter gene activity without changes to the underlying DNA sequence, in mediating environmental responses in shellfish. I am using the Pacific oyster (Crassostrea gigas) as a model to examine the role of DNA methylation in regulating interactions between genes and the environment. Through this work I hope to better understand the pathways through which environmental contaminants, such as endocrine disruptors, induce physiological changes and to determine whether these induced changes can be passed on to future generations
---
###[Claire Olson](http://students.washington.edu/che625/)
 I am currently a masters student in Steven Roberts’ lab at the University of Washington in the School of Aquatic and Fishery Sciences. My general research interests are studying aquatic ecosystem health as a function of environmental change, with an emphasis on the study of epigenetics.
I am currently a masters student in Steven Roberts’ lab at the University of Washington in the School of Aquatic and Fishery Sciences. My general research interests are studying aquatic ecosystem health as a function of environmental change, with an emphasis on the study of epigenetics.
 My research aims to improve our understanding of the physiological responses of marine invertebrates to natural and human-driven environmental change. Specifically, I am interested the role of epigenetics, processes that can alter gene activity without changes to the underlying DNA sequence, in mediating environmental responses in shellfish. I am using the Pacific oyster (Crassostrea gigas) as a model to examine the role of DNA methylation in regulating interactions between genes and the environment. Through this work I hope to better understand the pathways through which environmental contaminants, such as endocrine disruptors, induce physiological changes and to determine whether these induced changes can be passed on to future generations
---
###[Claire Olson](http://students.washington.edu/che625/)
My research aims to improve our understanding of the physiological responses of marine invertebrates to natural and human-driven environmental change. Specifically, I am interested the role of epigenetics, processes that can alter gene activity without changes to the underlying DNA sequence, in mediating environmental responses in shellfish. I am using the Pacific oyster (Crassostrea gigas) as a model to examine the role of DNA methylation in regulating interactions between genes and the environment. Through this work I hope to better understand the pathways through which environmental contaminants, such as endocrine disruptors, induce physiological changes and to determine whether these induced changes can be passed on to future generations
---
###[Claire Olson](http://students.washington.edu/che625/)
 I am currently a masters student in Steven Roberts’ lab at the University of Washington in the School of Aquatic and Fishery Sciences. My general research interests are studying aquatic ecosystem health as a function of environmental change, with an emphasis on the study of epigenetics.
I am currently a masters student in Steven Roberts’ lab at the University of Washington in the School of Aquatic and Fishery Sciences. My general research interests are studying aquatic ecosystem health as a function of environmental change, with an emphasis on the study of epigenetics.
 Roberts SB and Gavery MR (**2012**) [Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates?](http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249382/?tool=pubmed) Frontiers in Physiology 2:116. doi:10.3389/fphys.2011.00116
Roberts SB and Gavery MR (**2012**) [Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates?](http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249382/?tool=pubmed) Frontiers in Physiology 2:116. doi:10.3389/fphys.2011.00116
 Gavery MR and Roberts SB. (**2012**) [Characterizing short read sequencing for gene discovery and RNA-Seq analysis in Crassostrea gigas](http://www.sciencedirect.com/science/article/pii/S1744117X11001018) Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 7:2 94-99. doi:10.1016/j.cbd.2011.12.003
Gavery MR and Roberts SB. (**2010**) [DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (_Crassostrea gigas_)](http://www.biomedcentral.com/1471-2164/11/483) BMC Genomics 11:483. doi:10.1186/1471-2164-11-483
##Data
###_Crassostrea gigas_ high-throughput bisulfite sequencing (gill tissue)
Citation: Gavery, Mackenzie; Roberts, Steven (2013): Crassostrea gigas high-throughput bisulfite sequencing (gill tissue). figshare.
Gavery MR and Roberts SB. (**2012**) [Characterizing short read sequencing for gene discovery and RNA-Seq analysis in Crassostrea gigas](http://www.sciencedirect.com/science/article/pii/S1744117X11001018) Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 7:2 94-99. doi:10.1016/j.cbd.2011.12.003
Gavery MR and Roberts SB. (**2010**) [DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (_Crassostrea gigas_)](http://www.biomedcentral.com/1471-2164/11/483) BMC Genomics 11:483. doi:10.1186/1471-2164-11-483
##Data
###_Crassostrea gigas_ high-throughput bisulfite sequencing (gill tissue)
Citation: Gavery, Mackenzie; Roberts, Steven (2013): Crassostrea gigas high-throughput bisulfite sequencing (gill tissue). figshare.