What is the relation between transposable elements and methylation?
For purposes of proposal and reports, I have gone back to look at a small project done in collaboration with scientist at IFREMER looking at pesticide exposure on oyster larvae methylation.
---
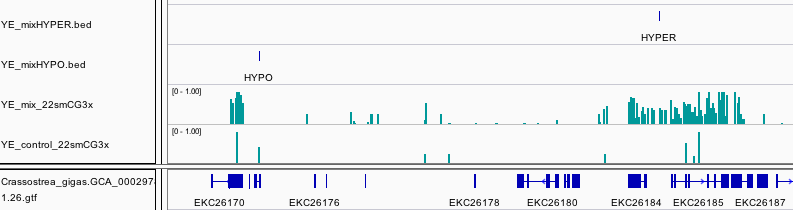
The control library had limited yield so the number of loci with data from the treated and the control library was restricted. However using a liberal 3x coverage for both, we found a total of 823 DMRs (544 hypermethylated and 279 hypomethylated).
---
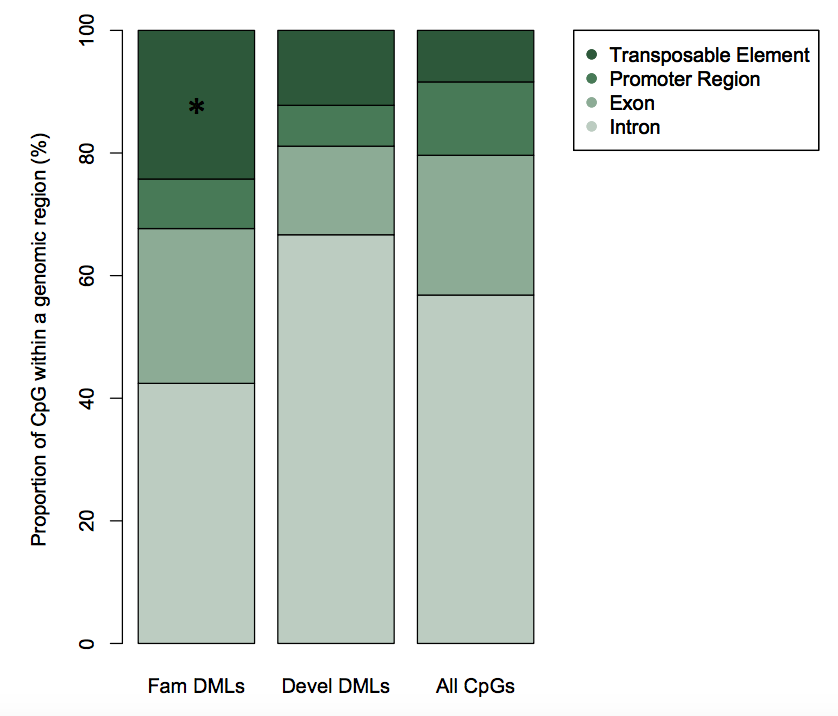
Intriguingly, when one accounts for all CGs in the genome, these DMRs are predominantly in transposable elements.
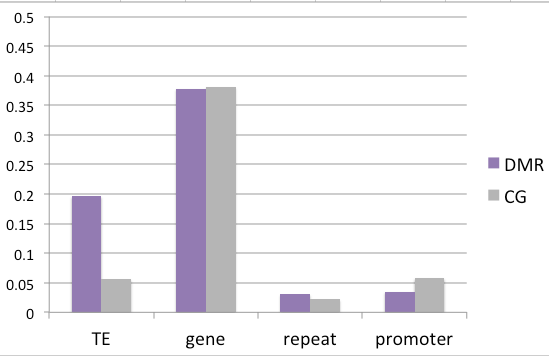 Reminiscent of ...
Reminiscent of ...
 ---
The analysis is not pretty, but here is what I have to offer.
https://github.com/sr320/nb-2015/blob/master/Cg/YE-pesticide/YE-pesticide-summary.ipynb
---
The analysis is not pretty, but here is what I have to offer.
https://github.com/sr320/nb-2015/blob/master/Cg/YE-pesticide/YE-pesticide-summary.ipynb Reminiscent of ...
Reminiscent of ...
 ---
The analysis is not pretty, but here is what I have to offer.
https://github.com/sr320/nb-2015/blob/master/Cg/YE-pesticide/YE-pesticide-summary.ipynb
---
The analysis is not pretty, but here is what I have to offer.
https://github.com/sr320/nb-2015/blob/master/Cg/YE-pesticide/YE-pesticide-summary.ipynb