Too meta?
Having just given a talk on reproducibility, I am in the midst of responding to reviewer comments about what we did (12 months ago!) and boy can I say every minute of putting [this notebook](https://github.com/che625/olson-ms-nb/blob/master/BiGo_dev.ipynb) was worth it. I even found [where we ran the entire notebook, so all result files are easily accessible](http://eagle.fish.washington.edu/cnidarian/index.php?dir=olson-ms-nb-master_12_1714%2Fwd%2F). Beyond praising Claire, I will document my follow up analysis here.
Essentially the want more quantitative information on differential methylation beyond ..
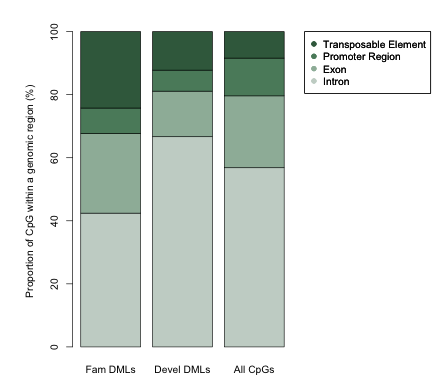 Makes sense.
Here is what was originally done.
Makes sense.
Here is what was originally done.
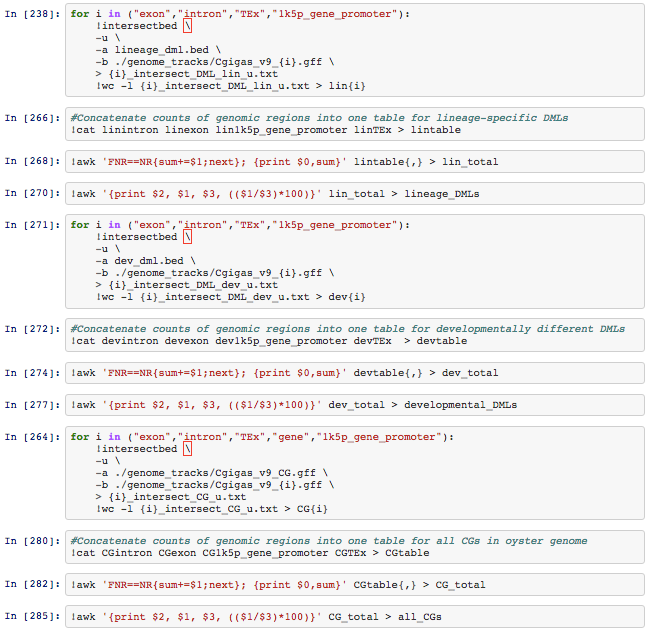 For example the file named _linexon_ contained `16 exon_intersect_DML_lin_u.txt`. The 4 files were concatenated to produce `lintable` ....
For example the file named _linexon_ contained `16 exon_intersect_DML_lin_u.txt`. The 4 files were concatenated to produce `lintable` ....
 and a little awk
```
awk 'FNR==NR{sum+=$1;next}; {print $0,sum}' lintable{,} > lin_total
awk '{print $2, $1, $3, (($1/$3)*100)}' lin_total > lineage_DMLs
```
to create `lineage_DMLs`...
and a little awk
```
awk 'FNR==NR{sum+=$1;next}; {print $0,sum}' lintable{,} > lin_total
awk '{print $2, $1, $3, (($1/$3)*100)}' lin_total > lineage_DMLs
```
to create `lineage_DMLs`...

 Makes sense.
Here is what was originally done.
Makes sense.
Here is what was originally done.
 For example the file named _linexon_ contained `16 exon_intersect_DML_lin_u.txt`. The 4 files were concatenated to produce `lintable` ....
For example the file named _linexon_ contained `16 exon_intersect_DML_lin_u.txt`. The 4 files were concatenated to produce `lintable` ....
 and a little awk
```
awk 'FNR==NR{sum+=$1;next}; {print $0,sum}' lintable{,} > lin_total
awk '{print $2, $1, $3, (($1/$3)*100)}' lin_total > lineage_DMLs
```
to create `lineage_DMLs`...
and a little awk
```
awk 'FNR==NR{sum+=$1;next}; {print $0,sum}' lintable{,} > lin_total
awk '{print $2, $1, $3, (($1/$3)*100)}' lin_total > lineage_DMLs
```
to create `lineage_DMLs`...
