###Canonical Feature Tracks (Ensembl)
Ensemble provides a feature tracks that are updated on a regular basis.
They can be directly accessed at
Note this will ensure you have the most current version.
_Version 25_
GTF
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.gtf
GFF3
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.gff3
Screenshot
List of gff feature (v25)
```
5 EnsemblGenomes RNA
2530 EnsemblGenomes exon
13 EnsemblGenomes gene
28 EnsemblGenomes miRNA
28 EnsemblGenomes miRNA_gene
1410 EnsemblGenomes pseudogenic_tRNA
13 EnsemblGenomes rRNA
13 EnsemblGenomes rRNA_gene
47 EnsemblGenomes snRNA
47 EnsemblGenomes snRNA_gene
20 EnsemblGenomes snoRNA
20 EnsemblGenomes snoRNA_gene
994 EnsemblGenomes tRNA_gene
2422 EnsemblGenomes transcript
186890 GigaDB CDS
186938 GigaDB exon
26101 GigaDB gene
26101 GigaDB transcript
650376 dust repeat_region
224899 trf repeat_region
```
---
###Canonical Feature Tracks (version 9)
Gene
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff
Exons
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff
Intron
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_intron.gff
Promoter (= 1kbp 5' of genes)
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff
Transposable Elements
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff
Complement to Gene, Promoter, and TE tracks
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_COMP_gene_prom_TE.bed
All CGs
http://eagle.fish.washington.edu/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff
Screenshot:
_Details regarding the development of these tracks can be found in [this IPython Notebook](http://nbviewer.ipython.org/github/sr320/ipython_nb/blob/master/TJGR_OysterGenome_IGV.ipynb) as well as in this [methods section](https://peerj.com/articles/215/#p-7)._
quicklook
```
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff <==
C16582 flankbed promoter 386 395 . - . ID=CGI_10000001;
C17212 flankbed promoter 1 30 . + . ID=CGI_10000002;
C17316 flankbed promoter 1 29 . + . ID=CGI_10000003;
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff <==
scaffold38980 fuzznuc nucleotide_motif 63420 63421 2 + . ID=scaffold38980.741;note=*pat pattern:CG
scaffold38980 fuzznuc nucleotide_motif 63670 63671 2 + . ID=scaffold38980.742;note=*pat pattern:CG
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff <==
scaffold1479 WUBlastX LTR_Gypsy 2608 4209 104 + . .
C33730 WUBlastX LTR_Pao 1960 2589 652 - . .
C33730 WUBlastX LTR_Pao 3358 5868 1471 - . .
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE.gff <==
C21242 TRF Tandem_Repeat 38 100 72 + . .
C21306 TRF Tandem_Repeat 35 143 112 + . .
C21306 TRF Tandem_Repeat 574 947 208 + . .
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TEx.gff <==
scaffold1479 WUBlastX LTR_Gypsy 2608 4209 104 + . .
C33730 WUBlastX LTR_Pao 1960 2589 652 - . .
C33730 WUBlastX LTR_Pao 3358 5868 1471 - . .
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff <==
C16582 GLEAN CDS 35 385 . - 0 Parent=CGI_10000001;
C17212 GLEAN CDS 31 363 . + 0 Parent=CGI_10000002;
C17316 GLEAN CDS 30 257 . + 0 Parent=CGI_10000003;
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff <==
C16582 GLEAN mRNA 35 385 0.555898 - . ID=CGI_10000001;
C17212 GLEAN mRNA 31 363 0.999572 + . ID=CGI_10000002;
C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003;
==> /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_intron.gff <==
C17476 subtractBed intrn 75 103 . - . Parent=CGI_10000004;
C19392 subtractBed intrn 184 451 . + . Parent=CGI_10000015;
C20262 subtractBed intrn 539 641 . - . Parent=CGI_10000025;
```
***
##Experimental Feature Tracks
###_Crassostrea gigas_ high-throughput bisulfite sequencing (gill tissue)
Citation: Gavery, Mackenzie; Roberts, Steven (2013): Crassostrea gigas high-throughput bisulfite sequencing (gill tissue). figshare.
This fileset contains genomic feature tracks from **methylation-enriched** high-throughput bisulfite sequencing and RNA-seq analysis for Pacific oyster (Crassostrea gigas) gill tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012). All data and instructions are also available at .
Associated Publicaiton: XXXXXXX
File descriptions:
[BiGill_CpG_methylation.igv](http://files.figshare.com/1252773/BiGill_CpG_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGill_exon_clc_rpkm.igv](http://files.figshare.com/1252772/BiGill_exon_clc_rpkm.igv) - Exon-specific gene expression values (RPKM) from RNA-seq analysis.
[BiGill_igv_charlie.xml](http://files.figshare.com/1252770/BiGill_igv_charlie.xml) - A session file, which loads methylation and RNA-seq feature tracks as well as the location of C.gigas genome features.
[Query to derive_CG_AllData_IGV.txt](http://files.figshare.com/1252771/Query_to_derive_CG_AllData_IGV.txt) - Query (SQLShare) used to derive the methylation feature track from the original methratio output (http://goo.gl/5LGq9Q)
Screenshot
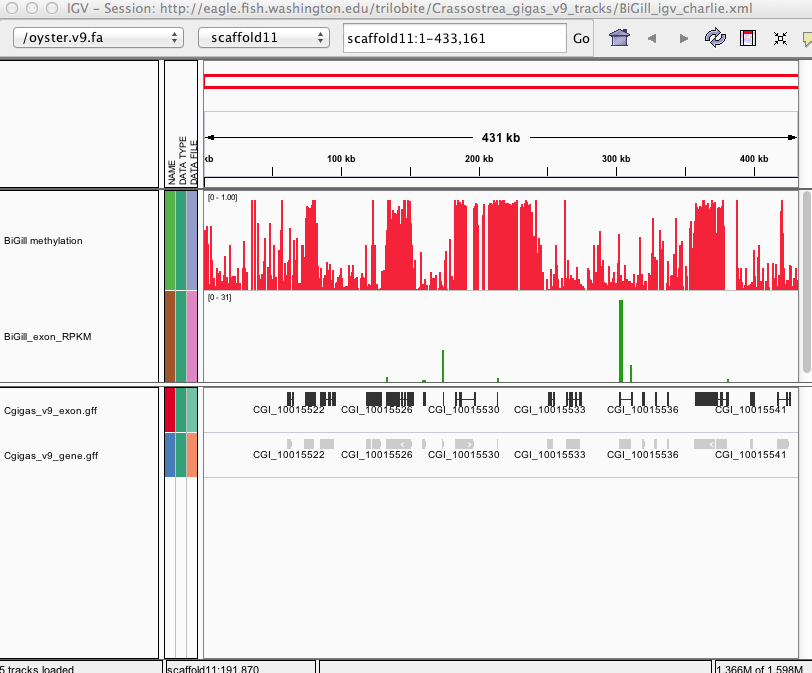
***
###_Crassostrea gigas_ high-throughput bisulfite sequencing (male gamete)
This file contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) male gonad tissue. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Fang et al. 2012).
Associated Publicaiton: XXXXXXX
File descriptions:
[BiGo_methylation.igv](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_methylation.igv) - Location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[BiGo_igv.xml](http://eagle.fish.washington.edu/Mollusk/174gm_analysis/BiGo_igv.xml) - A session file, which loads methylation and location of C.gigas genome features.
Screenshots
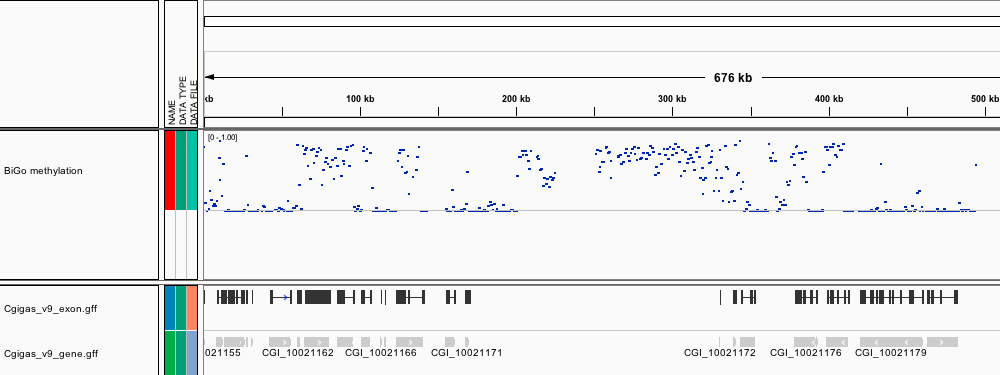
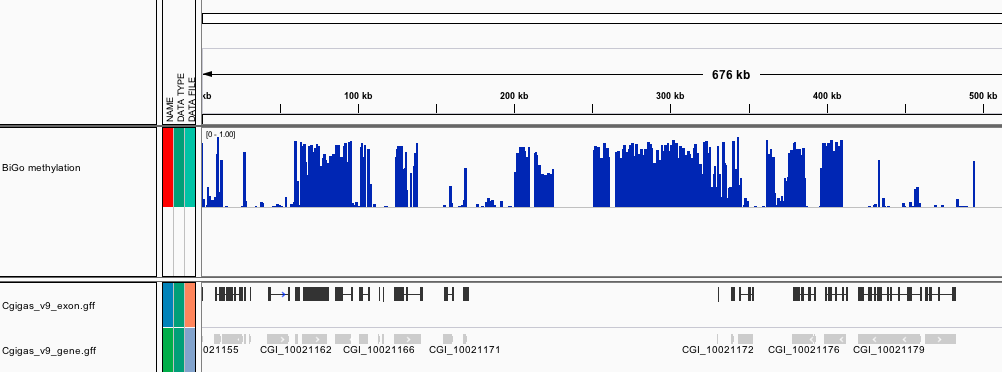
***
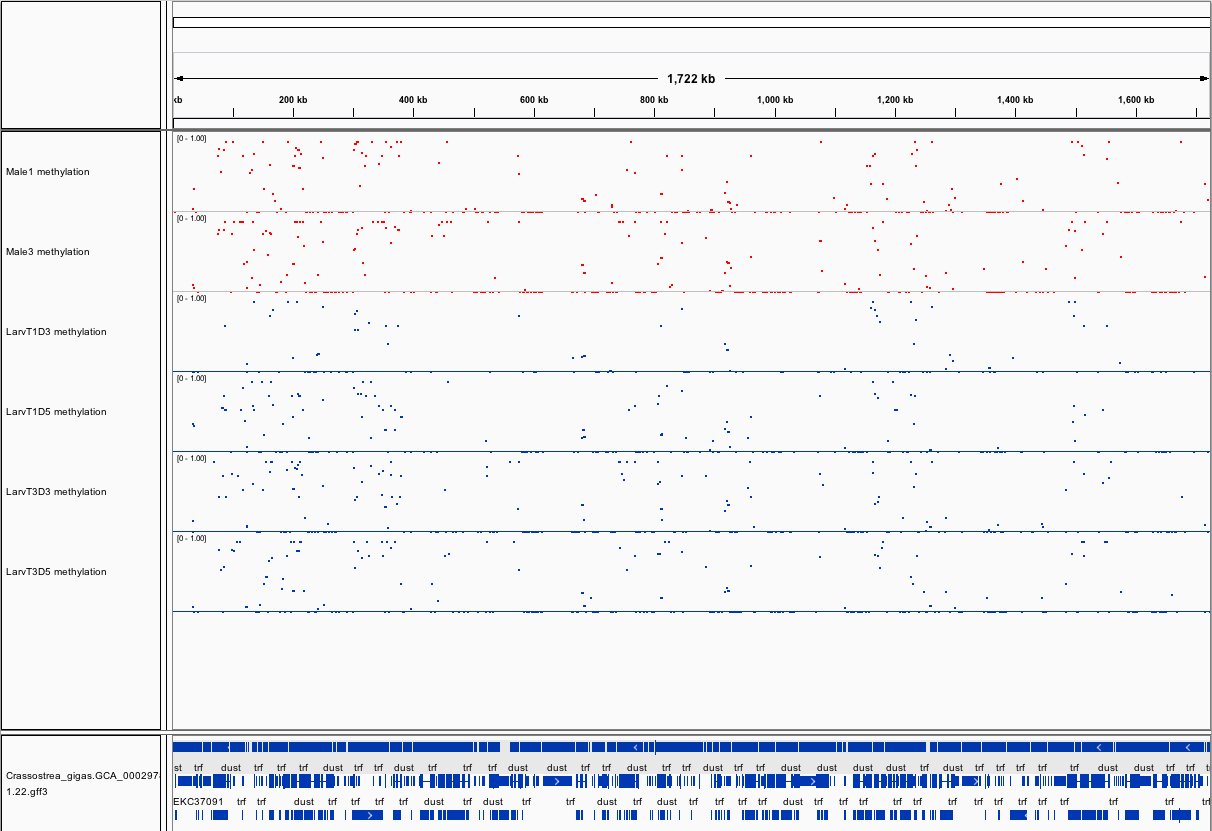
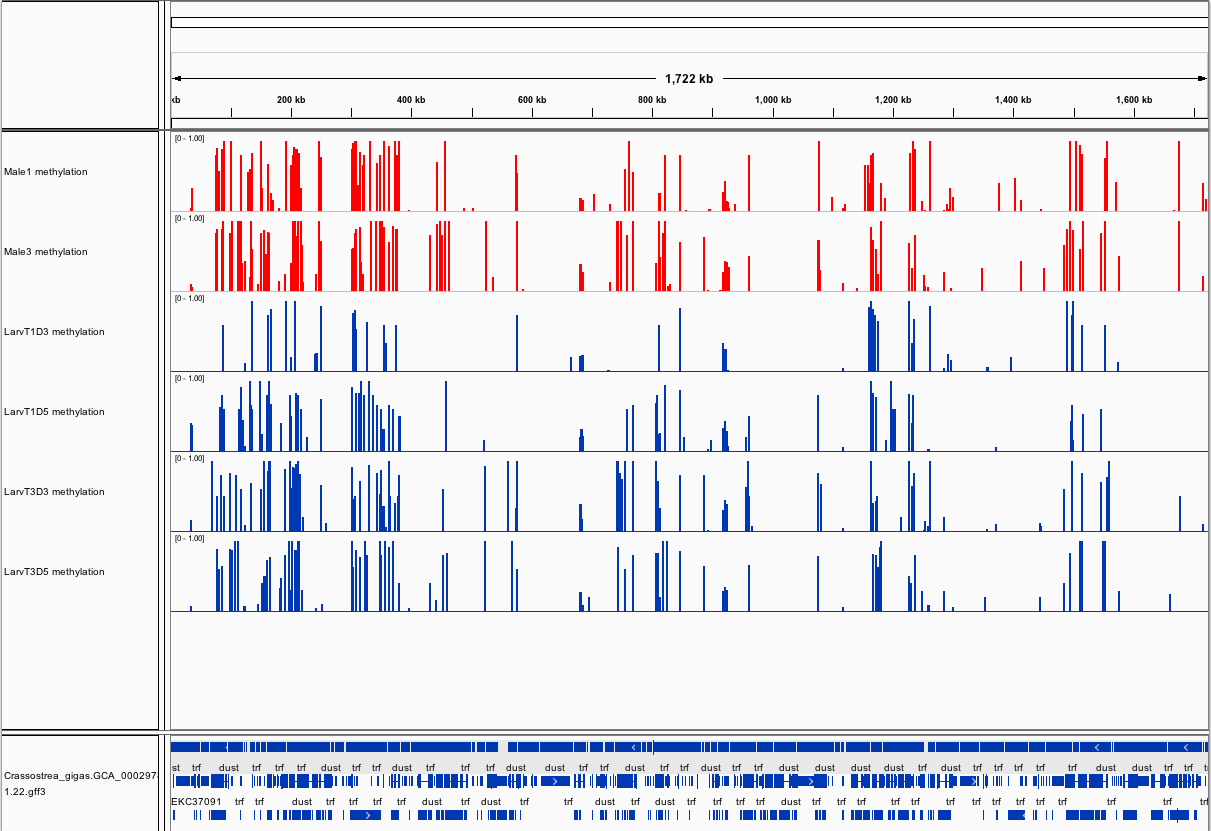
###_Crassostrea gigas_ high-throughput bisulfite sequencing (sperm and larvae)
Citation: Olson, Claire; Roberts, Steven (2014): Crassostrea gigas high-throughput bisulfite sequencing (larvae and sperm tissues). figshare.
This fileset contains genomic feature tracks from high-throughput bisulfite sequencing for Pacific oyster (Crassostrea gigas) spermatozoa and larvae samples. Samples were taken from two males (Male 1 and Male 3) and larvae taken from these males at Day 3 (D3) and Day 5 (D5) post-fertilization. Feature tracks were developed to be viewed with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) in conjunction with the C. gigas genome (Ensembl).
Associated Publicaiton: XXXXXXX
File descriptions:
[methratio_out_CG5x_IGV_M1.igv](http://files.figshare.com/1538436/methratio_out_CG5x_IGV_M1.igv) - Male 1 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_M3.igv](http://files.figshare.com/1538437/methratio_out_CG5x_IGV_M3.igv) - Male 3 location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D3.igv](http://files.figshare.com/1538440/methratio_out_CG5x_IGV_T1D3.igv) - Larvae from male 1 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T1D5.igv](http://files.figshare.com/1538441/methratio_out_CG5x_IGV_T1D5.igv) - Larvae from male 1 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D3.igv](http://files.figshare.com/1538443/methratio_out_CG5x_IGV_T3D3.igv) - Larvae from male 3 day 3 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[methratio_out_CG5x_IGV_T3D5.igv](http://files.figshare.com/1538444/methratio_out_CG5x_IGV_T3D5.igv) - Larvae from male 3 day 5 post-fertilization location and proportion of methylation for all analyzed CpG dinucleotides with greater than 5x coverage.
[LarvaeSperm_igv.xml](http://files.figshare.com/1538478/LarvaeSperm_igv.xml) - A session file, which loads sample methylation tracks and location of C.gigas genome features.
Screenshots