Epigenetics Morning lecture
Definition: heritable changes in a trait or phenotype, caused by a mechanism other than mutation to the DNA sequence
ex) why do cells have different types with same DNA? expression
Epigenome provides regulatory aspects of genome
Marks: chromatin packaging
histone modifications: acetylation or methylation, DNA methylation, non-coding RNAs ex, microRNA ~21 bp complimentary base pair to bind to mRNA and prevent expression
DNA methylation: Cytosine next to a Guanine in the sequence in animals - 5' CG 3'
in a promoter can inhibit transcription by blocking access to transcription factors
functions: tissue differentiation, imprinting & X-inactivation (only one copy of gene gets expressed), development (orchestrated event with genes on and off), genome stability (transposable element silencing)
Epigenetic inheritance:
1)mitotic inheritance
2)intergenerational inheritance
by DNA methyltransferases (DNMT's) family of enzymes which function to methylate DNA
3)transgenerational inheritance - inherit epigenetic marks from parents
DNA methylation and the environment
factors shown to affect them: in identical mice, toxins/diet cause different expression of the Agouti coat color and obese gene. Exposed pregnant mothers to BPA- more babies with obese phenotype, but soy counteracted.
-sex determination temperature promotion ex) sea bass aromatase promoter to convert androgens to estrogens = more males
-Plants have more methylation and demethylation, ex)salinity, drought, temperature, frost
-behavior of mother rat (licking, grooming) affects methylation status of glucocorticoid receptor
-aging- twins look more different when older because different environments affect methylation
-disease- virus/host interactions, cancer, diabetes, asthma
What could be negative effects of methylation?
-bad passed on- after stress is removed, information still passed on ex)famine grandchildren still had disease
-positive- adaptive more quickly
Variations between taxa- not all this serves all taxa
1)we work on non-models, primarily studied in mammals
2) variation in eukaryotes: density, distribution, context
3) methods: limited genomic information, rely on sequences but molecular pathways may not be clear
-Invertebrates- only a handful of species lack DNA methylation, most 30-60% methylation, primarily in exons, bees good study organism b/c worker vs drone
Epigenetics Afternoon lecture:
characterization of DNA methylation in oysters
Ecologically important filter-feeders and bioindicators, economically important to have healthy populations and hatcheries, facing environmental stresses
What does the oyster methylome look like? more like a fruit fly or a bee? which genes are methylated? where is methylation, exons, introns, repeats? what is the role of methylation in oysters???
Approaches: methylation specific restriction digest, bioinformatics, genome-wide methylation (MBD-seq, Bisulfite sequencing)
Methylation specific restriction digests
-Restriction enzymes recognize a sequence and cut DNA
-HpaII and MspI recognize same CCGG site, but Hpa can't cut the site while Msp can always cut it.
-ex) mouse, sea star, fruit fly. Split up dna into two parts, half digested with HpaII and the other have with MspI, when run on a gel will look very short if heavily methylated (lots of big chunks) in HpaII, but spread out in MspI becuase of many cuts.
-mussel and oyster get light protection
-does your organism have expressed sequence tags for methylation enzymes?
Bioinformatics: in silico approach
-methylated cytosines are highly mutable, switch C>T, so methylated regions get depleted from CpG to TpG. CpG O/E = observed/expected
-can tell which are low or high by function: predict housekeeping functions to be most methylated, inducible genes in stress response to be less methylated
Genome -wide methylation analysis useing MBD-seq (Methyl-binding domain isolated - genome sequencing) shows which are enriched in the methylated fraction
experiment to confirm in silico results
can we confirm with high throughput sequencing?
1) reduce genome and only sequence a portion, use same methyl-binding protein as before
2)illumina library, bisulfite conversion (gold standard) converts cytosine to uracil. Doesn't convert methylated cytosines, so any remaining cytosines are methylated.
3) seuqence then
4) BSMAP software
5) reference: de novo contigs, characterized
visualize results with galaxy
map back reads , count percent methylation for methylatable sites, find most heavily methylated regions, blast to see use or importance
ex) hexokinase
company: Nanostring: nCounter(R)
screen for methylation at different tissues or life stages to see differential methylation in different tissues or just do PCR.
combining methods into Targeted Bisulfate sequencing
So it's characterized. But so what is the role in oysters?
Functional Role: MERV
theory: housekeeping genes methylated to keep transcription factors from laying down on "spurious transcription sites" and make efficient
less methylated: environmental response genes to get greater variation so you can have different isoforms, alternative splicing, different versions. Absence could promote SNPs with sequence variation. Transient methylation? "inducible"
Methylation and environment: endocrine disrupters, plastics, etc., cause changes in DNA methylation patterns, can disregulate negatively and have consequences, like feminized male fish.
experiment: expose pacific oysters to synthetic estrogen, look at male:female ratio, gonad devt, gene expression and dna methylation
Hypothesis: ECD exposure will skew sex ratios toward more females, alter expression patterns for genes involved in reproduction stress and detox
, and methylation patterns will be altered upon exposure to EDC's in oysters.
Summary: probably important. how will epigenetics be affected by the environment? ongoing research to test these mechanisms.
8/22/12- Touch ups on the presentation today, then presented in the evening!
also killed sand dollars, cleaned sea tables, made and autoclaved broth for morning inoculation of sand dollar bacteria.
8/21/12- Working on the presentation today- building our slides, doing the stats. Took last measurement on sand dollar transmission today.
8/20/12-Counting the total counts for the oysters post bleaching. Some wells have absurd numbers of oysters- like over 300. Also, data analysis getting broken down.
figuring out the data and background information. Spent hours at Carolyn's figuring out why our LD-50 and pH data are so, so very weird. And then we figured it out later that night!
8/19/12-collecting the 48 hour data for oysters. water chemistry is FRUSTRATING.
8/18/12 - Why Share?
Don't reinvent the wheel, you don't know everything
Alaska salmon program - group data entry, freely available
everyone can work on the same data analysis to complement yours
what you've done is based on what others have done- share the love, share the science.
Lab: open notebooks allow you to know what everyone in your lab is doing without keeping the same hours
obligation by being paid by the taxpayer to share with the public
right now, there's limited risk
Emma: save oysters from OA scifund (crowdsourcing)
Blogger
Twitter - post journal articles, ask and answer questions
Lab webpage, project blogs, wordpress
links
sharing unfunded grant proposals, datasets, supplemental materials to papers
FigShare - citable
Imaging:
Google Fusion
Wordle
prezi
ManyEyes
8/17/12-
OAysters- set up and going!
3 strains x 2 ages x 2 acidities x 3 replicates = 12 12-well plates of oyster larvae.
Lecture Notes (Steven)
Why study Genetic Variation?
-different responses to stress- resilience
-population level/metagenomics
-evolutionary metagenomics
-conservation biology
-predict response to variation in environment
-origin of pathogen- epidemiology (source of patient zero)
-absence of genetic variation may allow phenotypic variation
How to assess Genetic Variation?
-SNP's - single nucleotide polymorphisms - change in single base pair - high throughput data useful
-RFLP's - restriction fragment length polymorphism - digest DNA, enzymes break up at specific places, different in each species, dif. banding patterns
-Microsatellites - short repeated segments
-sequence genome or single gene -
-old fashioned - allozymes at protein level
Technique: RAD- looking at SNPs
SNPs
-variations in genome through mutations, SNPs are substitutions, occur >1% of population
-0.1 % unique base pairs different between people
-can be neutral or harmful depending on where found
-most found in non-coding regions
-synonymous does not change amino acid composition - third codon position least likely to change
Herring
- Non-recovering populations- Exxon-Valdez, pathogens, etc. What was the change in the structure?
-CLC- de novo assembly - contigs - finds SNPs
-Output: consensus sequence and allelic variation
-transcriptome data- DNA that has been turned into RNA -gives genes to study something like virulence, host immune response, susceptibility
-Expt: 454, SOLiD, 4 individuals within 4 populations
-pooled individuals to generate 2 libraries (liver- toxins, testes-affected and clean easy to get DNA)
-Sequencing output- reads 1.9 mil, 272 bp, matched 1.7 mil, average contig length 728
-targeting genes in the xenobiotic/stress response, see SNPs within those gene sets
-Synonymous vs. Non-Synonymous - how do you know? WORKFLOW
>Consensus sequences > EMBOSS getorf (from EBI website) orf=open read frames=start and stop minimum 200bp > reference assembly > SNP detection > Annotate SNP positive features > Filter based on e-value
found 7000 snps, 2,500 putatively nonsynonymous > what does GO slim say they do?
used rarefaction curve to economically sequence to hit all the contigs possible economically
local adaptation- how to measure across populations? use PCR to cheaply assess SNPs across many individuals, dual labeled Taqman lay on sequence
high resolution melt temperature analyses- sequence amplified, double chained DNA- heat very slowly and see different H-bonds
What else can you do with this data?
-metagenomics? look for the virus! find it in the liver tissue
New ways of assessing SNPs
-RAD sequencing > digest DNA with restriction enzymes > each fragment from different individuals > form fragments that are random, but specific, forms stacks > discovery and genotyping in one step (!!!) > much cheaper, more efficient, no genome needed (!!!) BUT they're not genes. you get no function or indication of intron or exon.
8/16/12-
trying to analyze the abalone transcription data: metagenomics!
Many Eyes is a cool website for graphics, which I will have to play with later when I have flash player.
Got our oyster larvae today! set them up in bins on the benches.
learned about abalone, Rickettsia and phages.
Setting up for the OAysters project tomorrow- autoclaved tubes,
8/15/12 -
OAysters:
Met with Carolyn over lunch to plan experiment for this week.
THE PLAN (bold is today)
Revive Bacteria strains (RE 22, RE98 and RE101)
-pass onto plate
-inoculate media and grow overnight
-Next day: inoculate into 0.25% tryp SW and tryp 2000 SW and allow to grow under those conditions.
Prepare seawater (from OA lab)
-clean 4 large bottles and autoclave
-Make 1L 0.25% tryptone in SW and 1L 0.25% tryptone in 2000ppm SW
-Filter 1L SW, 1L 2000 SW, 1L tryp SW, 1L tryp 2000 SW through 0.2 micron filter
-Find 0.2 micron syringe filters and set up bubblers
Oysters
-acclimate for 24 hours upon arrival
-AM: put in wells and add half 2000 SW (gradual introduction to acidified water) if treatment
-PM: add bacteria and start challenge
AM: Using limited short-read data
C. gigas polluted vs. non polluted sites, how does gene expression vary between the two sites?
get reads and do de novo assembly, reference assembly from EST's for C gigas (compare two approaches, both useful, comparable)
annotated with GO slim compare differentially expressed genes to see if there's an over or under representation of GO terms
arrange based on function
result: integrins cadherins etc. expressed higher in polluted area
Ostrea lurida- did not show difference on growth or survival
DEGs = differentially expressed genes
larvae turn on genes to compensate for acidification or they're negatively impacted
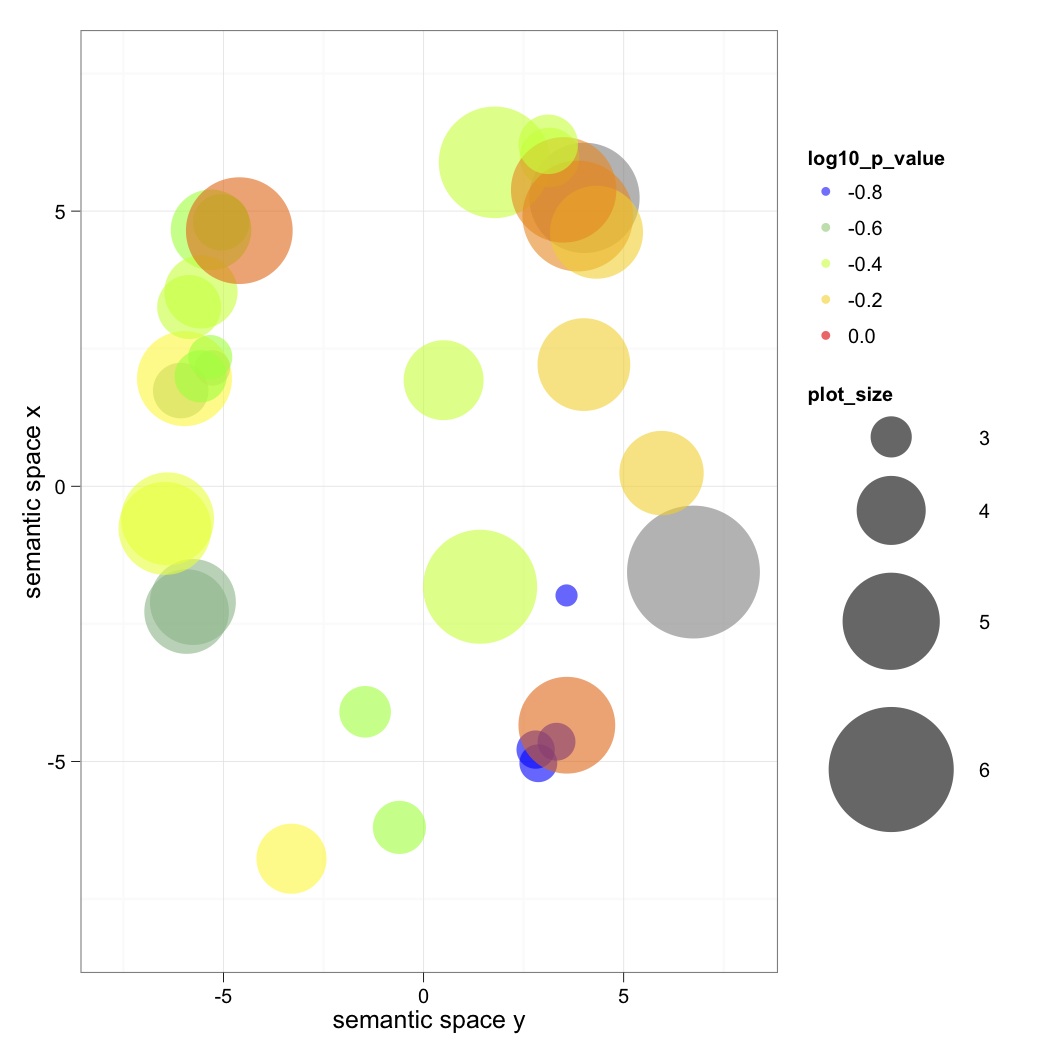
making pretty graphs:
Outburst: "what are these graphs? what do they mean?
WHYYYYY. I am desperate to know what the size of these circles is supposed to mean. "
second half- DAVID analysis for microarrays
we're using it for next gen, this same analysis can be done in CLC
We're giving it our two lists of the two text files. One must be designated the background, and both must be put through the DAVID conversion tool to convert from UNIPROT-ID to UNIPROT Accession #. Then we went to the functional annotation tool and could expand certain analyses, and download the text files.
Finally, we used REVIGO to input our GO# and P-value and made pretty pretty scatterplots. Export to R and it looks cool!
PM:
WHOLE OF BIOINFORMATICS SO FAR:
Illumina sequences in two libraries
trimmed (got rid of poor quality reads) and assembled contigs in CLC (Steven did this) to make de novo assembly
| Annotation steps: >>BLAST sequences against Swiss-Prot (annotation step) in Terminal >>>into Galaxy to join tables- add together our BLAST and Gene Association Tables and making pie charts |
DEG steps: >>RNA Seq to see (# reads/library =fold difference) which genes were differentially expressed (DEGs), came from CLC |
Into DAVID for enrichment analysis (compare DEG to entire transcriptome)
Chose terms to examine (GO FAT BP) and downloaded
Look at output in REVIGO
8/14/12 - QPX data analysis
Workflow:
reads come back from Illumina
Use CLC to form de novo assembly
map back to library in CLC
export into a tab delimited text file
RPKM- reads per kilobase per million mapped reads (normalizes for library size)
Do the other library!
map back to file that compares the two (QPX expt), high negative fold change numbers indicate large differences between the two libraries.
Use terminal to BLAST the results
1) TEXT WRANGLER to open up transcriptome fasta file and change spaces to underscores
>QPX_transcriptome_v1_Contig_1
2) start up blast
./blastx -query /Users/Shared/EIMD_blast/query/QPXtranscriptome.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot -out /Users/Shared/EIMD_blast/query/QPXtranscriptomeOUT -outfmt 6 -evalue 1E-20 -max_target_seqs 1 -num_threads 2
query looks for the input file
db establishes which database to use
out sends the output file somewhere
outfmt ->6 is fasta
evalue is the threshold that we want
max_target_seqs is the number of hits per contig
num_threads is how much CPU space to use
PM LAB:
Go to GALAXY (penn state)
gaining a full understanding of the transcriptome.
Get file into Galaxy:
1) Get Data > Upload File > check convert to tab delimited >run
view file by clicking eyeball
Separate ID number:
1)Text Manipulation > convert delimiters to tab > select pipes
View Stevens databases from the wiki and import to our table- swisspro gene id codes and descriptions
1) click eyeball to view
2)green plus sign to import
Join the descriptions to our data
1) Join, Subtract and group > join two datasets > select datasets and columns > change all options to yes > execute
JOIN ALL THE TABLES.
Export to Excel, delete unnecessary columns, play with in pivot table!
8/13/12 - AM: Exam
PM:
8/11/12-
8/10/12 - Immunology lecture in afternoon
-48 hour oyster counts
| Strain |
Hrs |
Well |
# |
Dead |
Total |
Live |
Proportion Survival |
| X00123 |
48 |
Control |
1 |
||||
| X00123 |
48 |
Control |
2 |
||||
| X00123 |
48 |
Control |
3 |
||||
| X00123 |
48 |
10^4 |
1 |
||||
| X00123 |
48 |
10^4 |
2 |
||||
| X00123 |
48 |
10^4 |
3 |
||||
| X00123 |
48 |
10^5 |
4 |
||||
| X00123 |
48 |
10^5 |
5 |
||||
| X00123 |
48 |
10^5 |
6 |
||||
| X00123 |
48 |
10^6 |
7 |
||||
| X00123 |
48 |
10^6 |
8 |
||||
| X00123 |
48 |
10^6 |
9 |
||||
| X00123 |
48 |
Vt10^5/Am10^6 |
10 |
||||
| X00123 |
48 |
Vt10^6/Am10^6 |
11 |
||||
| X00123 |
48 |
Am only |
12 |
Sand Dollar transmission expt health assessment
8/9/12- Immunology lecture and Vibrio day 3
-Talk on shellfish immunology by Carolyn
-Assessed living/dead oyster larvae in all treatments
| Strain |
Hrs |
Well |
# |
Dead |
Total |
Live |
Proportion survival |
| X00123 |
24 |
Control |
1 |
0 |
|||
| X00123 |
24 |
Control |
2 |
0 |
|||
| X00123 |
24 |
Control |
3 |
1 |
|||
| X00123 |
24 |
10^4 |
1 |
2 |
|||
| X00123 |
24 |
10^4 |
2 |
0 |
|||
| X00123 |
24 |
10^4 |
3 |
0 |
|||
| X00123 |
24 |
10^5 |
4 |
5 |
|||
| X00123 |
24 |
10^5 |
5 |
2 |
|||
| X00123 |
24 |
10^5 |
6 |
1 |
|||
| X00123 |
24 |
10^6 |
7 |
2 |
|||
| X00123 |
24 |
10^6 |
8 |
2 |
|||
| X00123 |
24 |
10^6 |
9 |
3 |
|||
| X00123 |
24 |
Vt10^5/Am10^6 |
10 |
2 |
|||
| X00123 |
24 |
Vt10^6/Am10^6 |
11 |
1 |
|||
| X00123 |
24 |
Am only |
12 |
4 |
RESULTS of analysis and bacterial idenfication steps on Vt- X00123
2. Gram stain - GRAM NEGATIVE
-had trouble focusing, but culture was overwhelmingly red.
3. Serum agglutination test - POSITIVE
-bubbles make trouble.
4. Azocasein protease assay - POSITIVE
8/9/12- More oyster vibrio!
-Another talk by Dr. Hershberger
-Counted out 35-60 oyster larvae into each well, inoculated with Vibrio and Aeromonas.
-Plated vibrio and aeromonas to test concentrations.
-Afternoon seminars by Steve and Drew on their research- lots of fun to hear about methylation and coral diseases! Looking forward to hearing more about the new data Drew just got in :)
8/8/12- Talk about fish disease management
-I have a definite interest in management and government work. It's nice to work in theory and tackle the basic questions of ecology and such, but it also seems like it'd be rewarding to address specific epidemics and manage them. What's the point of knowing about disease unless we try to use the information to fight it? I really enjoyed Dr. Hershbergers talk about the influence of the environment on fish diseases.
-Learned how to test spectrophotometric pH! Follow the protocol. Yay!
8/7/12- Projects in progress:
1. Oyster Vibrio:
-Plating on agar cultures to see if Aeromonas bacteria has probiotic effects against different strains of Vibrio tubiashi.
-Plating on TCBS to see which strains of Vibrio tubiashi grow better and have the ability to metabolize sucrose.
2. Sand Dollars:
-Observations from yesterday: colonies growing on TCBS plates on the interior sand dollar surface for all the unhealthy ones and one healthy.
-Transmission expt: some discoloration on a purportedly healthy sand dollar, healthy control boxes all dollars are stacked, in transmission ones, they were spread. TODAY however, same behavior not observed.
3. Laby:
-other people are plating the autoclaved eelgrass into laby cultures.
-Sammi and I are talking about looking for hits on the Lacuna and Phyllaplysia DNA extractions again. For Phyllaplysia we'll try to isolate where in the organism the Laby is carried (or not carried), ie, digestive gland, foot, head, etc. In Lacuna that dissection would be difficult, so we'll try testing Lacuna from different substrates (kelp, rock, eelgrass) and see whether we get any that don't carry Laby. Looking for a vector of transmission! Exciting!
8/6/12-
8/5/12- Checking on our plates: Looks like we've got some VIBRIO!
8/4/12- Bleaching and lesioned sand dollars from East Sound- protocol
I. Histology
-Two bleached, one healthy small sand dollars put in 3.5% formalin, to be transferred to EtOH, then to be decalcified and sectioned and put in cassettes.
-One healthy (Accession #: SB-1-H), one bleached (SB-3-B) large sand dollars internal organs (gonads, digestive tract) put in cassettes and into Davidson's fixative, to be transferred to EtOH.
->Observations- large bleached sand dollar was male- looked at sperm under microscope. Sex unknown for bleached.
->Gross morphology from dissection- bleached were easier to crack in half, somewhat more brittle test. Healthy specimens interior was clear and dark, diseased interior was cloudy and pale (fluid contained sperm after injuries to organs)
II. Cultures
| Accession # |
Bleaching |
Lesions |
Histo? |
Plates |
Swab |
| SB-1-H (large) |
no |
no |
organs |
Marine Agar & TCBS |
Interior and Exterior |
| SB-6-H (small) |
no |
no |
whole individual |
Marine Agar & TCBS |
Exterior |
| SB-2-B |
yes |
no |
no |
Marine Agar & TCBS |
Interior and Exterior |
| SB-3-B |
yes |
no |
organs |
Marine Agar & TCBS |
Interior and Exterior |
| SB-4-BL |
yes |
yes |
no |
2x Marine Agar & TCBS lesion and bleaching |
Bleaching Interior and Exterior, Lesion Interior and Exterior |
| SB-5-BL |
yes |
yes |
no |
Marine Agar & TCBS |
Lesion Interior and Exterior |
III. Transmission Experiment
-mesh sided boxes in common sea table (same water in all boxes)
-individuals placed touching in boxes at start
-new mud placed in boxes immediately prior to placement
-length is measured along the axis of symmetry of the sand dollar
-Will check on health of sand dollars daily.
-Some healthy sand dollars had evidence of previous injuries or predation, but all injuries were completely healed.
-All extra sand dollars (all are bleached or lesioned) are being kept in separate sea table.
| Box 3 2 Healthy -51 mm -66 mm 1 Bleached -77 mm |
Box 6- Control 3 Healthy -78 mm -75 mm -58 mm (previous predation) |
Box 2- Healthy baby holding box -22 mm (has lightening to underside) -31 mm -30 mm |
| Box 8 - Control 3 Healthy -48 mm -71 mm (previous predation) -74 mm (previous predation) |
Box 13 2 Healthy - 72 mm -67 mm 1 Bleached -66 mm |
Box 4- Bleached baby box -29 mm -30 mm -26 mm |
| Box 16 2 Healthy -46 mm -68 mm 1 Bleached -89 mm |
Box 14 3 Healthy -72 mm -49 mm -54 mm |
empty |
8/3/12- Electrophoresis, brief introduction to next-gen sequencing, sand dollar collecting at East Sound (Orcas Isl.), lecture on modeling, and discussing of future directions of research for the laby project.
Our electrophoresis looked so much better than last time!

8/2/12-Field Trip to False Bay, lectures, data entry (and PCR)
8/1/12- Field trip to Picnic Cove, lectures, DNA extraction.
Sammi had set up a neat experiment with
7/31/12- Field trip to Beachcomber Cove on Orcas Island today. Much more successful sampling of eelgrass- the beds were healthier, although the presumptive laby lesions were still pretty prevalent. We even saw them in the eelgrass mesocosms outside our lab that have been maintained for several years. We've got the protocol we followed here: Eel Grass Protocol. I noticed while entering the data this afternoon that some people tended to mark lesions as many small short ones, while other people would group it into one large 30 cm lesion. Perhaps in analysis we should sum the lengths of the lesions? I'm concerned that we're not accounting for different number of blades, and I'm really curious how the blades grow so we know which ones are the newest or oldest and how those get infected. That kind of life history information doesn't exist though, according to Sandy. We see the infections more commonly on the longer blades, but lesions also make blades more liable to break off, and the oldest blades seem to be the ones on the exterior of the plant, like the lettuce leaves you'd peel off and not eat. I feel like those basic life history traits are really essential to understanding the disease.
It is fun to be able to direct our project a bit more now- everyone has good ideas for what to test and I'm excited to see how the eelgrass mesocosms work, although I don't think I'll wind up specializing in this project.
Our lecture today was about coral diseases. My impression is that there are a couple of symptoms that are caused by many agents. Talking about the different factors made me want to try to model it- plug in some factors regarding transmissibility, virulence, environmental stressors, presence and abundance of potential pathogens in the environment, and if you're lucky you might get a prediction for how likely an epidemic is and how far it might spread if there already is one. I'd be afraid to come up with results like Roger Bradbury's, but if they turned out to be useful for predicting outbreaks (like the temperature anomaly map was earlier in lectures) then it would be worthwhile. I'm still working out in my own head how to incorporate my interests in modeling and community dynamics- we're learning so much on the organismal level and smaller, and I'd love to go into the impacts that disease can have on the ecosystem as a whole, how long recovery takes. I've particularly enjoyed talking about the microbiome and holobiont for that reason.
7/30/12- First field trip! Trip to False Bay today. The tide came in faster than we expected, so we were only able to survey the shallow transect.
Afternoon lecture by Steve, and Laby culture samples prepared. I had some trouble following Steve's lecture- when I got the big picture it was very interesting, but since I have essentially no molecular or cellular background, I found myself wishing for a lot more background information and simplification. I'll ask more questions in the future, but I found myself Googling every other word to try to understand the context of the information that was on the screen.
7/29/12- No class today, but went tidepooling with Ana and Jamie at Lime Kiln State Park. Saw abundant limpets, snails, chitons (Katharina, Tonicella, and Cryptochiton), Cucumaria miniata, Urticina, Hemigrapsus nudus and H. oregonensis, some kind of boring clam (rubbery edged siphon, about 2 cm diameter), Rostanga, sea stars, green urchins, lots of Saccharina sessile. Site was a steep drop, with few pools and protected areas. -1.8 ft tide, foggy all around and cool, but sunny where we were. Two harbor seals on the rocks.
We found one abnormal Strongylocentrotus droebachiensis. It appeared to have patchy dark lesions all over its body- areas where spines had fallen off and were replaced with goopy purple epidermis or potentially microbial colonies. There was a white margin to the presumptively infected patches, with darker purple or red centers. The urchin was still alive, and we collected him in a coffee cup and put him in a dish in the lab. I'll put him in a sea table tomorrow morning. His tube feet are lethargic. We may do epidermal scrapes or cultures to try to isolate the pathogen, but will likely have to mix up some new broth or agar plates (my hypothesis is that it's bacterial, based on a hypothesis that it's bald urchin disease). I'm curious how histology might work, given the crustyness of a calcareous skeleton.
I've found urchin tests with purple lesions before, and am curious as to whether this is a similar pathology.
7/28/12
7/27/12
7/26/12-
AM Lecture- introduction to infectious disease, AM lab- DNA extraction, PM Lecture- Bacterial diseases of Molluscs, PM lab- PCR
My reintroduction to lab benchwork was a little rough today. You'd think that following the recipe and just keeping it sterile would be easy, but it's really very challenging to keep up with the steps and maintain sterile technique. Haven't done that in a while! At the end we did wind up with two tubes of Armina DNA
7/25/12- Diagnostic Methods and Good microbes
Summary of morning lecture: Today we covered the primary methods of identifying and confirming disease and the situations in which they would be used. In a surveillance setting, such as monitoring a population, or a certification for a stock or facility, there are optimal numbers of samples to take in order to detect pathogens or parasites of a certain prevalence. For example, if you expect that a pathogen affects 5% of the population, you need to examine 60 specimens (regardless of the population size) to be 95% confident that if it was there, you saw it. Diagnostic sensitivity (type I error) and specificity (type II error) were discussed in the context of proofing a test, such as a new PCR test. In the context of DNA, analytical sensitivity refers to how dilute a sample can be and still be accurate, and specificity is how well it distinguishes between the target DNA and similiar sequences.
Methods discussed included:
Gross examination- does it look funny? Lesions, exterior scraping, gill squash, tissue squashes
Histology
Transmission Electron Microscopy (TEM) - similar process to histology, but better magnification.
Antigen based tests- fluorescence shows where the pathogen is, or ELISA method can mark the epitope
PCR- identifies whether gene sequences from pathogens are present, or qPCR can estimate quantity, but doesn't tell whether the pathogen is viable or virulent, just that it's present.
In-Situ Hybridization- places a fluorescent marker/probe on a target DNA sequence
SNP's- identify variations between strains with PCR
and a few other DNA based tests.
We dissected out Armina californica digestive glands and took samples for histology and for DNA extraction, which we'll do tomorrow. It was cool to see the many species of nudibranchs in the neuroethology lab where they had taken out the brains before we could use them. There were many species there- I hadn't seen Tritonia before, and some of the Triopha catalinae didn't look so good either.
Our PM lecture covered the microbes of a holobiont- the beneficial microbial community that lives on the surface (or within the tissues) of the organism. In corals and squid, these can produce antimicrobial compounds and interact competitively with harmful microbes. Really blurs the line between individual and community!
7/24/12- Introduction to invertebrate anatomy, histology, and histopathology
We were never going to recognize a diseased marine organism if we didn't know what a healthy one looked like, so today we got to know them inside and out. In the morning Carolyn lectured on the basic anatomy and biology of molluscs, echinoderms, and crustaceans, then we proceeded to the lab to see for ourselves. We pulled up our sleeves, pulled on our gloves, and proceeded to (scientifically) dismantle oysters, sea cucumbers, shrimp, sea stars, an urchin, and a snail. Highlights: a Parastichopus who just poured out his own back end, the sea urchin that wouldn't die, makin' some baby oysters, and breaking into a Nucella using the vice in the shop. Also, first injury of the class- I pinched myself in the shop with the vice and have a tiny blood blister. So much for the safety talk. All the filamentous stuff inside the cucumber was particularly mystifying, but we narrowed the white stuff down to the spermatophores at least, though the orange was still unknown.
The afternoon was spent on histology, both in lecture and lab. Carolyn gave us a slideshow on histological techniques and major tissues of oysters, clams, abalone, and shrimp and their pathologies. I found it interesting that types of pathogens cluster in certain hosts, though I will have to ask what that correlation is. I found that my memory of histology from human anatomy in undergrad had me familiar enough with what vessels, muscles, blood cells and nuclei generally look like in stained sections. Hopefully with some more time and looking at a lot more slides I'll be able to recognize the differences between the different species' tissues. Some disease, like WS in abalone, mycosis in bivalve shells, and certain lesions and cysts were quite obvious in section, while others required some significant microscopic power and knowledge of the nuances of the color of a nucleus to identify.
Fun Fact: Shrimp eyes contain PCR inhibitors!
7/23/2012- First Day of Class and Orientation (written on 7/24)
Orientation! We had tours of the computer lab, library, stockroom, shop, our lab, and the docks (for checking out rowboats) and I think that thanks to the great coffee in the cafeteria, most of the information stuck. We also had two lectures from Drew Harvell- an introduction to the context of the Ecology of Infectious Marine Disease, and a briefing on what we know about how climate change will affect diseases in the ocean so far, particularly in corals.
It's so exciting to be taking this class and to see just how wide open the field is. We've observed a fair number of marine diseases, but how many go unnoticed or undiagnosed? The most dramatic examples are ones which rapidly decimated whole populations, like the Diadema urchin die-offs in the early 1980's, situations in which the epidemic came and went so quickly that all we know is that it happened, not who or how or why. It seems that every species must have diseases, and for how many species there are under the water that we're not watching, it amazes me that everything isn't already dead. Through ecological cascades, something as small as a virus can completely shift the dominant species in an ecosystem.
The other thing that never fails to shock me is the magnitude of the effects we're already seeing from climate change. Drew's afternoon lecture brought us up to date with how diseases are changing due to climate change. Pushing species out of their thermal and pH optimums is increasing diseases (although decreasing in some cases) by altering host immunity and susceptibility, pathogen development rates, and potentially by changing transmission. Reduced overwintering is also a factor- imagine that a hypothetical pathogen dies below 10 degrees C and is extra virulent above 15 degrees C. In a normal year, all winter it would be too cold for it to survive, perhaps, and so the host would only have to fight it off for a couple months of the year, and it would only have to worry about the extra virulence on weird hot days. With a rise in a couple degrees all year round, you have less time the pathogen spends being dead, and more times it probably pops into that extra virulent temperature range. Knowing how coral diseases are correlated with temperature anomalies and strange weather events helps us find populations that are hit and study them, not to mention all the other diseases.
The final reason that I'm so excited is that this class is a piece of the Marine Infectious Disease Research Coordination Network- a concerted effort of disease scientists around the world to study infectious marine diseases. We're learning what the forefront of knowledge in the field is, and the hands-on techniques to take it further in the future. It's going to be a fun couple of weeks!