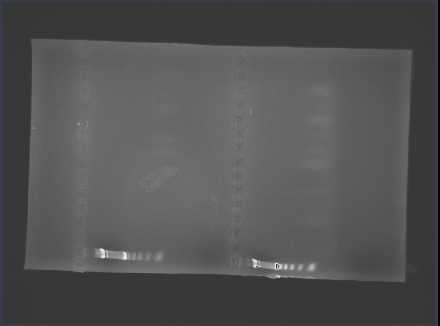
https://spreadsheets.google.com/ccc?key=tdo5r3XmdttbbOaODGrGU3Q&hl=en#gid=0
Project Proposal Development
https://docs.google.com/document/edit?id=1pGGEKz-BPRqF9deokhxhbu-S1JytLyJicm7wVWkgSLQ#
~July 21, 2010
We sampled Armina californica collected by the FHL Neurobiology class (Jim Murray). The animals had been collected several weeks prior and were held in a single tank at FHL. Some showed signs of dorsal erosion and slight blistering. Samples were excised of the dorsal lesions and sent to Cornell by Drew Harvell for histology. We observed RLO colonies in what appears to be digestive gland:
Figure 1. Armina digestive gland with RLO colonies 20x magnification.
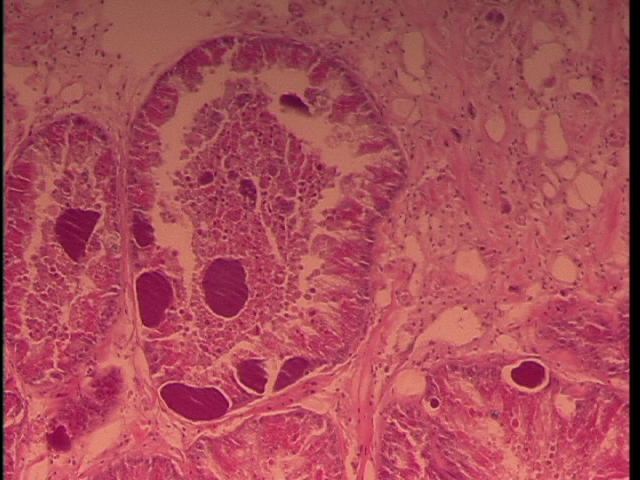
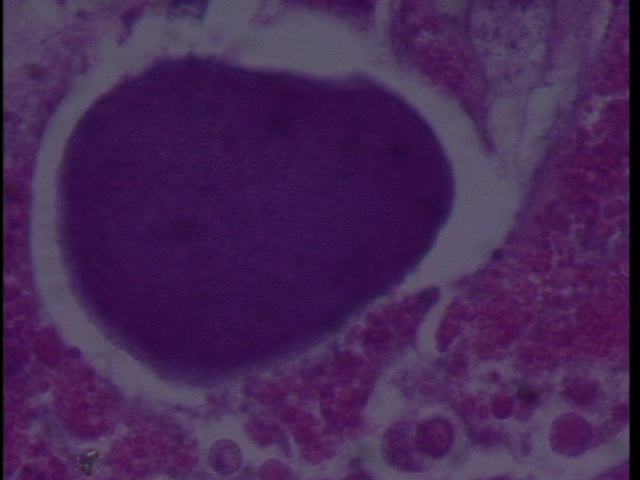
Figure 2. RLO colony 100x magnification in Armina.
July 29, 2010
Jim Murray's class collected more Armina californica from: south of Seattle, near Tacoma?
One very moribund (may be dead) and two alive with initial lesions (white discoloration and erosion on dorsal epithelium). Jim excised brains from two live and then we took cross sections and placed in 4% formalin for 4 d (oops). Parallel samples were placed in 100% ethanol for PCR analyses.
A. early lesion from new collection (2 cassettes)
B. Had two lesions on back
C. Dead Armina
Sent for histology 8-3-10.
Aug 2, 2010
Sampled two more Armina:
D: One from old group with white lesions on dorsal surface
E. One from the new group collected July 28th, 2010 - this animal has a dark, bumpy lesion with black spots, which is different from the typical white eroded lesions seen previously.
Aug 3, 2010
Sampled two Armina-assumed both from new group (Jul 28), will need to double check with neuro
8:30p F. dead, was sacrificed by neuro. around 1:30p
11:30p G. lesion dead, was sacrificed by neuro. around 10:30p, neuro. reported this one had a few back lesions
Future plans:
1. Extract DNA from samples
2. Amplify with WSRLO primers and universal eubacterial primers
3. Clone and sequence
4. make PCR assay and ISH assay
5. Publish in Science (then wake up and publish in Diseases of Aquatic Organisms or Journal of Aquatic Animal Health)
cPCR and QPCR recipes on pg 12&13 of the follow doc: WS in Abalone.pdf
Wednesday 8/4/2010
will run PCR on RLP+ DNA from abalone with withering syndrome Rickettsia using 2 types of master mix
1) “generic” with Go Taq (following recipe used with sybr green in lab)
per tube (ul)
BSA 1.5
Go Taq 12.5
F primer 0.5
R primer 0.5
H20 8
template 2
2) from Andre et al. 2000.
refer to: http://bio533.wikispaces.com/file/view/WS+in+Abalone.pdf
per tube (ul)
5x buffer 4
MgCl2 1.2
BSA 0.8
H2O 11.08
dNTPs 0.4
F primer 0.1
R primer 0.1
Taq 0.32
template 2
-made master mixes x5 for each recipe
-set up 2 negative blanks and 2 template wells for each of the master mixes
-pipetted out almost all of Andre master mix, also initially pipetted incorrect amounts of Andre master mix and so transferred into new strip tubes
The following was programmed in the PCR machine in the wet lab and named “C”
cPCR Thermal Cycler Program
Time Temp (°C)
Step 1 3 min 95
Step 2 1 min 95
Step 3 30 sec 62
Step 4 30 sec 72
Repeat steps 2-4, 40 times
Step 5 10 min 72
5p histology cassettes from yesterday into 70% EtOH, now sitting in fume hood
Thursday 8/5/2010
9:30a shared agarose PCR gel with Matt (0.5g agar, 50ml 1xTBE gel, 5ul EtBr-added after microwaved), 115V 20min
-noted the first A+ mixture had less volume than the second before I filled the well
-filled each well with 15ul: ladder, A-, A-, G-, G-, A+, A+, G+, G+
-saw bands only for G+, G+
6:30p DNA extraction on sample D (0.07g cross-sec incl. epidermis)
-step 5 centrifuged in cold centrifuge
-took break at step 3 for ice cream social
-stopped process for digestive tract sample between steps 9-10
-step 13 had to wait a few extra minutes for block to heat up
11p cut up slug H, put cassette in fixative
for all DNA extractions, using Stool Kit with incl. protocol, whole tablet, spinning at 8000 rpm right after adding buffer AW1, at end elute in 100ul buffer AE and incubate at room temp for 5 min before spinning down
Friday 8/6/2010
11a briefed Lisa C. on DNA extraction, had to throw out digestive tract sample that was stopped previous evening between steps 9-10
11:45a Matt joined team Armina
-cut up and fixed slugs I & J, TOD 8:30a, no lesions observed on either slug
2p Carolyn gave a demonstration on making histology slides:
-Prepare a 40 C water bath
-Place the histology block in the microtome and align it with the blade
-Place several drops of 50% ethanol on a clean microscope slide
-Cut a thin section from the histology block, using probes, place the section over the ethanol
-Place the section into the water bath to let the paraffin relax
-Dip a positively charged microscope slide into the water bath, place the section on the slide and remove the section out of the water bath
-Place the slide on a slide warmer to dry
Saturday August 7, 2010
1:30p extracted DNA with Matt from samples A (0.07g - Matt) & B (0.07g - Lisa), cut cross-sec incl. epidermis for both
-made one negative control without water as a blank
-control was started 10 min behind samples, caught up by end of process
-froze samples in left freezer when done
4pm put cassettes I & J fixed yesterday into EtOH
Monday August 9, 2010
Indep Project: Extracted DNA from Armina C, E, F,G samples.
Weight of sample C: 0.08g
Weight of sample E: 0.08g
C & E (Matt),
F (0.08g with some residual EtOH) & G (0.05g) (Lisa)
one blank extraction (Lisa)
We made a master mix following the "generic" recipe posted on 8/4 with universal bacteria primer, EUB-A and EUB-B.
We prepared for 13 reactions, leaving a little extra for pipetting errors:
19.5µL BSA
6.5µL EUB-A
6.5µL EUB-B
104µL PCR H2O
162.5µL GO TAQ
-Samples AàG
-Two blanks
-Negative control of PCR water
-Two positive controls of RickettsiaPlaced 2µL of template in appropriate wells with master mix.
Our plate was hand spun to force everything to the bottom of the wells, but our plate cracked in the middle. It did not appear to affect any wells.
PCR settings: (MJ Research Inc PTC-100)
Temp Time #Cycles
95 180sec 1 cycle
----------------------------------------------
95 45sec
55 30sec (35 cycles)
72 90sec
----------------------------------------------
72 300sec 1 cycle
*From now on, when we run PCR we will end with a temp drop to low temp (4-25 degrees) and hold for 30 minutes or so.
We removed the plate after PCR and placed it in the freezer until we can run a gel the next day.
Tuesday, August 10, 2010
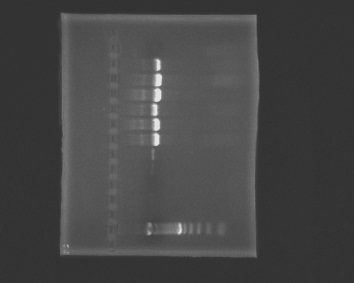
Prepare a 1% agarose gel (0.5g of agarose, 50mL of TBE buffer), load wells and run the gel.
1. ladder, 2. negative, 3. blank1, 4. blank2, 5. positive (Rickettsia), 6. positive (Rickettsia), 7. A, 8. B, 9. C, 10. D, 11. E, 12. F, 13. G
 |
| armina_8-10-2010.jpg |
Our negative controls were negative and we got a band in our positive controls. Samples A-F had large bands.
No bands appeared in Sample G.
Extracted DNA from Armina samples H (0.05g), I (0.05g), J(0.07g)
Made a master mix with EHR 16 S primer and ran a gel.
1. neg c, 2. pos c, 3. A, 4. B, 5. C, 6. D, 7. E, 8. F, 9. G, 10. H, 11. I, 12. J
| armina_8-10.JPG |
August 11, 2010
Prepared master mix with RA primers and IMMO instead of GO TAQ.
Neg, pos, samples A-->J
Prepared master mix with EUB primers.
Neg, samples B, H, I, J
Prepared a 1% TBE gel. For product using IMMO: placed 1uL bead of loading dye on paraffin. The mixed 9uL of PCR product in a bead of loading dye before placing in wells.
First row: 1. Neg, 2. Pos, 3. A, 4. B, 5. C, 6. D, 7. E, 8. F, 9. G, 10. H, 11. I, 12. J
Second row: 1. Neg, 2. B, 3. H, 4. I, 5. J
The bands were cut out, placed in labeled individual 1.5mL microcentrifuge tubes and stored in the freezer.
Thursday 8/12/2010
1p set up PCR with two master mixes:
12 units of “generic” using glydo_hydro_70 forward and reverse primers, IMMO mix with ~2ul loading dye per well
12 units of “generic” using SP forward and reverse primers, IMMO mix with ~2 ul loading dye per well
-gel was run with one negative control, RA+, A-J samples for each set of primers
-no bands anywhere
-both sets of primers were from the Roberts collection. Steven said they were degraded (made from degraded samples? or primers were degraded?)