lisafong@ucla.edu
Monday 8/9/2010
10a extracted DNA from samples C & E (Matt), F (0.08g with some residual EtOH) & G (0.05g) (Lisa), plus one blank extraction (Lisa)
3p ran PCR using universal eubacterial primers:
1 negative control (PCR H2O + Master Mix)
1 blank extraction from 8/6
1 blank extraction from 8/9
1 each of DNA extracted from individuals A-G
2 RA + control
Master Mix:
per tube (ul)
BSA 1.5
Go Taq 12.5
F primer 0.5
R primer 0.5
H20 8
template 2
Temperature Cycle (saved as ARLO1 in the thermo-cycler upstairs in lab 10):
step 1 3min 95C
step 2 1 min 95C
step 3 45 sec 55C
step 4 90 sec 72C
step 5 repeat steps 2-4 35x
step 6 5 min 72C
Lisa’s notes:
-didn’t vortex between 700ul additions in step 3, vortexed 2 min after all ASL added
-inhibit tab incubated in samples F, G, and the blank
-observed precipitate in blank after step 12
-step 17 centrifuged at full speed
Saturday 8/6/2010
1:30p extracted DNA with Matt from samples A (0.05g) & B (mass?), cut cross-sec incl. epidermis for both
-made one negative control without water as a blank
-control was started 10 min behind samples, caught up by end of process
-froze samples in left freezer when done
4pm put cassettes I & J fixed yesterday into EtOH
Friday 8/6/2010
9:45a Went to look for sea stars with Drew at Eagle Cove. Tide not super low, water a little rough. No seastars observed, few Mopalia, no mussel beds.
11a briefed Lisa C. on DNA extraction, had to throw out digestive tract sample that was stopped previous evening between steps 9-10
11:45a Matt joined team Armina
-cut up and fixed slugs I & J, TOD 8:30a, no lesions observed on either slug
2p Carolyn histology cutting/slides/tubes
Thursday 8/5/2010
9:30a shared agarose PCR gel with Matt (0.5g agar, 50ml 1xTBE gel, 5ul EtBr-added after microwaved), 115V 20min
-noted the first A+ mixture had less volume than the second before I filled the well
-filled each well with 15ul: ladder, A-, A-, G-, G-, A+, A+, G+, G+
-saw bands only for G+, G+
6:30p DNA extraction on sample D (0.07g cross-sec incl. epidermis)
-step 5 centrifuged in cold centrifuge
-took break at step 3 for ice cream social
-stopped process for digestive tract sample between steps 9-10
-step 13 had to wait a few extra minutes for block to heat up
11p cut up slug H, put cassette in fixative
for all DNA extractions, using Stool Kit with incl. protocol, whole tablet, spinning at 8000 rpm right after adding buffer AW1, at end elute in 100ul buffer AE and incubate at room temp for 5 min before spinning down
Wednesday 8/4/2010
9:30a helped Emma move setup to Moose’s lab, learned about the acidification setup there
will run PCR on RLP+ DNA from abalone with withering syndrome Rickettsia using 2 types of master mix
1) “generic” with Go Taq (following recipe used with sybr green in lab)
per tube (ul)
BSA 1.5
Go Taq 12.5
F primer 0.5
R primer 0.5
H20 8
template 2
2) from Andre et al. 2000.
refer to: http://bio533.wikispaces.com/file/view/WS+in+Abalone.pdf
per tube (ul)
5x buffer 4
MgCl2 1.2
BSA 0.8
H2O 11.08
dNTPs 0.4
F primer 0.1
R primer 0.1
Taq 0.32
template 2
-made master mixes x5 for each recipe
-set up 2 negative blanks and 2 template wells for each of the master mixes
-pipetted out almost all of Andre master mix, also initially pipetted incorrect amounts of Andre master mix and so transferred into new strip tubes
The following was programmed in the PCR machine in the wet lab and named “C”
cPCR Thermal Cycler Program
Time Temp (°C)
Step 1 3 min 95
Step 2 1 min 95
Step 3 30 sec 62
Step 4 30 sec 72
Repeat steps 2-4, 40 times
Step 5 10 min 72
5p histology cassettes from yesterday into 70% EtOH, now sitting in fume hood
Tuesday 8/3/2010
-change of plan re: host response portion of group project
will look at differential response to CO2 and Vt stress in Emma’s oyster larvae
-came in this morning to see one of the clean Armina laying eggs
10:45a Talked to neuro people about getting Armina, arranged to receive corpses with brain and rhinophores extracted. Also received Jim Murray’s blessing to take Armina.
10:50a Started “Test for protease” exercise using 1mL of Vt cultured in marine broth
http://bio533.wikispaces.com/Lab_OA
11:30a put histology cassettes from yesterday in 70% EtOH
11:45a bagged up cassettes wrapped in a paper towel, added a 70% EtOH to wet, and sent out
4p helped change water/set up counting/larvae in RNA later for end of C.gigas larvae/Vt/acidification preliminary study
4:30p part of “right group” for sea fan differential display
Morgan made master mix
arbitrary primer 19
Sampled two Armina-assumed both from new group (Jul 28), will need to double check with neuro
8:30p F. dead, was sacrificed by neuro. around 1:30p, from Dash Pt Jul 28 batch
11:30p G. lesion dead, was sacrificed by neuro. around 10:30p, neuro. reported this one had a few back lesions, from Dash Pt Jul 28 Batch
Monday 8/2/2010
-removed new Armina agar plates from incubator at 10am, only saw growth in plate from clean dorsal, questioning my plating method, would like to try using cotton swabs and toothpicks, when using metal loop should have put warm loop in corner of agar to bring temperature of loop to agar temp
-discussed Armina group project with Sarah & Carolyn: ID bacteria, histology, differential host response to stress [lesions] (hsp 70? qPCR?)
-got in loop regarding OA Vt class project:
Goals/Questions: what is the influence of ocean acidification and Vt on oyster larvae health?
8 soda bottles:
2 control (no Vt)
2 with Vt diluted to 10^-1; 10^8 colony forming units
2 with Vt diluted to 10^-3; 10^6 cfus
2 with Vt diluted to 10^-5; 10^4 cfus
Sun 8/1
1) grew up Vt in marine broth over night
2) diluted to c = 10^-1 = 150mL infected marine broth
3) RNA sampling: pipette ~1000 indiv. into tube with RNAlater
Mon 8/2
4) red dye indicates live larvae b/c they take it up, whereas dead larvae should show up clear. Problem: there are some pink ones, and not all the live ones take up the dye…so decided not to use dye
5) RNA sampling
6) change sea water every day
7) plate count bacteria (30-300 method); each plate wal 100ul spread around
10^-3 TNTC (too numerous to count)
10^-4 TNTC
10^-5 (1000, 1104) avg. 1052
10^-6 (167, 144) avg. 155
10^-7 (24, 28) avg. 21
10^-8 (2, 2) avg. 2
colony forming units/ml = (# colonies)(dilution factor)(plate dilution) = 155 x 10^6 x 10 = 1.55 x 10^9 cfu/ml
*will get complete mortality around 10^5 cfu
-gram stained smears from old and new Armina lesions
-fixed portions of new lesion and old lesion Armina for histology
-put tissue sample in 200 proof EtOH to preserve for DNA studies
-did serum agglutination exercise following directions at http://bio533.wikispaces.com/Lab_OA
-Sarah plated out Armina new and old lesion swabs and non-lesioned area swabs, sent to incubator 6pm 37C
7p got 2 lesioned, 3 non-lesioned Armina from neuro lab, put in tanks: 2 clean in their own tank, the other 3 with the 2 already old lesioned individuals in hope that we will have a spread of lesions
Saturday 7/31/2010
-Steven talked about using the computer program
-checked plates around 10:30am from chitons only saw some growth in quadrant 1 of healthy-infected girdle
-found new Armina in Neuro lab with one big bumpy lesion; lesion looked different from other Armina lesions observed; lesion had dark stuff in between bumps
-plated lesion and non-lesion cultures of new Armina dorsal side using only metal loop, incubated at 3pm at 37C
-looked at scrapes from lesion and non-lesion areas under compound scope, noted no noticeable difference between the two
-took Chiton plates out of incubator 7pm, put down in lab 5
-helped change larvae water
~July 21, 2010
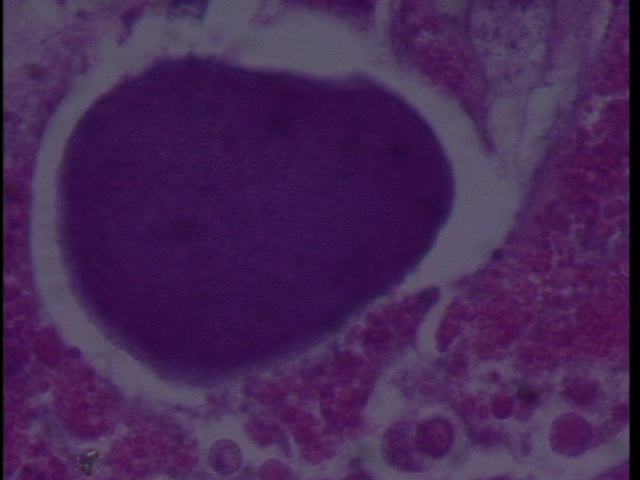
We sampled Armina californica collected by the FHL Neurobiology class (Jim Murray). The animals had been collected several weeks prior and were held in a single tank at FHL. Some showed signs of dorsal erosion and slight blistering. Samples were excised of the dorsal lesions and sent to Cornell by Drew Harvell for histology. We observed RLO colonies in what appears to be digestive gland:
Figure 1. Armina digestive gland with RLO colonies 20x magnification.
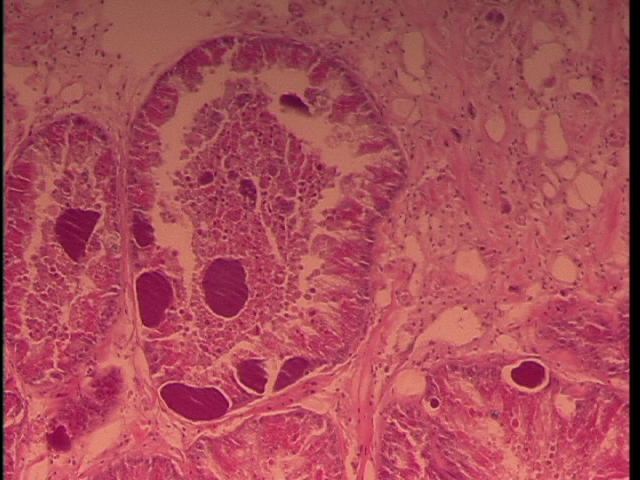
Figure 2. RLO colony 100x magnification in Armina.
July 29, 2010
Jim Murray's class collected more Armina californica from: south of Seattle, near Tacoma?
One very moribund (may be dead) and two alive with initial lesions (white discoloration and erosion on dorsal epithelium). Jim excised brains from two live and then we took cross sections and placed in 4% formalin for 4 d (oops). Parallel samples were placed in 100% ethanol for PCR analyses.
A. early lesion from new collection (2 cassettes)
B. Had two lesions on back
C. Dead Armina
Sent for histology 8-3-10.
Aug 2, 2010
Sampled two more Armina:
D: One from old group with white lesions on dorsal surface
E. One from the new group collected July 28th, 2010 - this animal has a dark, bumpy lesion with black spots, which is different from the typical white eroded lesions seen previously.
Future plans:
1. Extract DNA from samples
2. Amplify with WSRLO primers and universal eubacterial primers
3. Clone and sequence
4. make PCR assay and ISH assay
5. Publish in Science (then wake up and publish in Diseases of Aquatic Organisms or Journal of Aquatic Animal Health)
Friday 7/30/2010
Western Blot
-our western blot didn’t look too successful. Maybe the HSproteins weren’t in sufficiently high concentrations because we mashed up miscellaneous portions of the bodies. Apparently the HSps are found in the tentacles.
Chitons
-picked up chitons (Mopalia ciliata) at Argyle Pt because heard reports of some flaky behavior
-found one that had bare patches on girdle, 3 acorn barnacles growing on it, and a little anemone
-collected assortment of “healthy” (held fast to rocks) and “sick” (not holding fast to rock)
-all other chitons, regardless of our classification gross examination looked ok and were behaving similarly and as expected in bag (balling up, clinging to each other, walking on sides of bag
-plated smears from 3 chitons: sicko, a healthy, and a healthy looking one from the sick bag on marine agar and TCBS
-for healthy-healthy and healthy-sick, made 2 types of plate each: foot, girdle
-for sick-sick, made plates for lesioned girdle, healthy spot on girdle, foot, messed up plate area by barnacles, healthy looking spot from plates
Thursday 7/29/2010
Ran another gel
-class ran gel electrophoresis with 30ul of sample and buffer to prep for western blot
Western Blot
-used gel for western blot following proteomics protocol
http://bio533.wikispaces.com/Lab_Proteomic
Friday 7/30/2010
Western Blot
-our western blot didn’t look too successful. Maybe the HSproteins weren’t in sufficiently high concentrations because we mashed up miscellaneous portions of the bodies. Apparently the HSps are found in the tentacles.
Chitons
-picked up chitons (Mopalia ciliata) at Argyle Pt because heard reports of some flaky behavior
-found one that had bare patches on girdle, 3 acorn barnacles growing on it, and a little anemone
-collected assortment of “healthy” (held fast to rocks) and “sick” (not holding fast to rock)
-all other chitons, regardless of our classification gross examination looked ok and were behaving similarly and as expected in bag (balling up, clinging to each other, walking on sides of bag
-plated smears from 3 chitons: sicko, a healthy, and a healthy looking one from the sick bag on marine agar and TCBS
-for healthy-healthy and healthy-sick, made 2 types of plate each: foot, girdle
-for sick-sick, made plates for lesioned girdle, healthy spot on girdle, foot, messed up plate area by barnacles, healthy looking spot from plates
Thursday 7/29/2010
Ran another gel
-class ran gel electrophoresis with 30ul of sample and buffer to prep for western blot
Western Blot
-used gel for western blot following proteomics protocol
http://bio533.wikispaces.com/Lab_Proteomic
Bonamia in Oysters
-cut open European flat oysters
-used Giemsa stain; stained blots of heart, hemolymph droplets and smear
-looked for Bonamia infection in stained slides
-tried to get the hang of identifying hemocytes
Zostera
-looked at the leading edge margin of a Zostera blade supposedly infected with Labrynthula under the compound scope
-compared to website of lab group with photographed examples of Labrynthula in culture and in vivo
http://www.unf.edu/coas/biology/SeagrassImages/SeagrassPageRoss08.dwt
-couldn’t tell whether there were Labrynthula in the cells…suggestions: slice thinner piece of grass blade for more light to shine through, get fresher blades
-cut open European flat oysters
-used Giemsa stain; stained blots of heart, hemolymph droplets and smear
-looked for Bonamia infection in stained slides
-tried to get the hang of identifying hemocytes
Zostera
-looked at the leading edge margin of a Zostera blade supposedly infected with Labrynthula under the compound scope
-compared to website of lab group with photographed examples of Labrynthula in culture and in vivo
http://www.unf.edu/coas/biology/SeagrassImages/SeagrassPageRoss08.dwt
-couldn’t tell whether there were Labrynthula in the cells…suggestions: slice thinner piece of grass blade for more light to shine through, get fresher blades
Wednesday 7/28/2010
We extracted the protein following the ‘Proteomics’ protocol, but did not quantify it. Our class had 6 anemones: 3 were heat stressed at ~28 C, and 3 were left in the water table (control). Morgan and I were responsible for 2 of the control anemones, but noted that our anemone #1 (Cont. 1 LM) was introverted, suggesting it might have been slightly stressed at the time we sacrificed it.
http://bio533.wikispaces.com/Lab_Proteomic
-did not run the Bradford Assay nor quantification
-ran 30, 50, and 10 ul in gel electrophoresis.
-30 uL looked pretty even across groups, with the exception of one group, so we decided to run 30ul samples for western blot, adjusting the proportion of the first group with the darker column to account for their high protein concentration (reduced or diluted their sample)
-ISH didn’t work light up classic and new Rickettsia as it was supposed to
-saw the various colonies of Rickettsia in H&E slides
-if we had a slide with the probe attached to the target DNA, we could compare that with the H&E slides by matching anatomical regions between the slides
Tuesday 7/27/2010
-continued ISH protocol
Monday 7/26/2010
ISH protocol
http://bio533.wikispaces.com/Lab_Withering
-using ISH probe designed for classic withering to see if binds to other type
-slides of matching cuts of abalone in paraffin provided
-our group (Morgan, Sarah, me) ran two pairs of slides
-ISH probe attaches to DNA of targeted Rickettsia, which in our case was the classic/new type. The other existing type in the abalone is stippled, and not associated will abalone illness.
-thought that classic and new types are the same, but now thought that new is just classic with a bunch of phages, which cause it to appear to stain differently from the typical classic type
-2 slides to have one of them negative control. If there is stuff on the control, it indicates contamination.
-purpose of multiple washes in protocol is to have fresh xylene/functional_equivalent strip solution
-probe is a sequence with dye, try to get to anneal to denatured Rickettsia DNA, stains rickettsia tissue still on slide so it stands out from other tissues under visual examination
Saturday 7/24/2010
Learning to analyze QPCR data
goal: see relative gene abundance
| Well |
Well Type |
Threshold |
Ct |
expression in gene |
| H5 |
Unkown |
2155.942 |
no ct |
|
| H6 |
Unkown |
2155.942 |
39.79 |
0.281353291 |
| H7 |
Unkown |
2155.942 |
no ct |
|
| H8 |
Unkown |
2155.942 |
no ct |
|
| H9 |
Unkown |
2155.942 |
no ct |
|
| H10 |
Unkown |
2155.942 |
no ct |
|
| H11 |
Unkown |
2155.942 |
no ct |
|
| H12 |
Unkown |
2155.942 |
no ct |
Where
H5-6 is actn negative control
H7-8 is actn
H9-10 is c jun negative control
H11-12 is c jun
-the lack of expression was likely due to the frying of the cDNA sample I submitted for QPCR
To find the expression value, used equation given on Littorina exercise page:
Arbitrary expression value =10^(-(0.3012*Ct)+11.434)
To normalize data, divide c-jun expression value by actin expression value in each row (i.e. separately for each sample). The comparison is based on relative proportions, not specific units. Pooled class results showed a notable, but not significant difference between infected (n = 3) and uninfected (n = 3) groups when the normalized data was placed in a bar chart
If the negative control is not clean (ours were not), options for trouble shooting include (in order of preference):
1) re-do PCR
2) check master mix for contamination
3) go back and re-extract tissue
-to check for gDNA, run PCR on RNA (QPCR if that’s the type of date you’re already working with). If there’s product, it’s coming from gDNA because RNA wouldn’t yield anything. Then can use DNAse to clean up the sample.
Friday 7/23/2010
10am
1.Zostera plates by Kathy and Morgan
-saw growth coming from the infected looking pieces of Zostera plated on agar (marine and TCDS)
-some spreading on the TCDS have discreet boundaries, suggesting the coloration may be due to something other than bacteria
-did not note much growth from non-infected samples on agar
2.cDNA
-produced cDNA from RNA using methods:
1) 17.75 uL RNA
2) heat 70C 5min
3) transfer to ice > 10 min (I put my sample on ice for ~3 min)
4) add 7.25 uL of master mix to RNA
5) 42C 60 min
6) 95C 3 min
where master mix recipe is:
5 uL - 5XMMLV buffer
1.25 uL – MMdNTPs
0.5 uL – MMLV RTase
0.5 uL – oligo dT primers
-left my sample (KM4) in the last heating phase for >>>3 minutes
2:45pm
3. QPCR
-created two master mixes, one for c jun kinase gene, and one for actin gene
-the general master mix recipe used:
12.5 uL – 2XSYBRMM
1.5 uL – BSA
0.5 uL – F primer
0.5 uL – R primer
8 uL – SH2O
where forward and reverse primers varied depending on which gene was being targeted
-each master mix was prepared in 5x the quantity required for one sample
-2 uL portions of either cDNA (test) or SH2O (negative control) were added to tubes in the QPCR plate, along with 23 uL of a master mix
-in total had 2 tubes of each test/negative control and master mix combination:
H5-6 was actn negative control
H7-8 was actn
H9-10 was c jun negative control
H11-12 was c jun
note: SYBR master mix contains: MgCl3, 10x buffer, DNA taq polymerase, DNTP, SYBR fluorescent dye
note: actn is reference gene, cjun is the gene under examination
4. lesioned Tritonia sacrificed by neuro lab
-C.F. put chunks of skin to culture bacteria in a six-walled liquid culture of QPX media
-C.F. also prepped some samples to be sent for histology
Thursday 7/22/2010
A lot went down today:
8:30am
1. Field trips to Picnic Bay, False Bay
-I went with Sandy Wyllie-Echeverria and Co. to Picnic Bay, where we examined the Z. marina, looking for Labrinthula
-blades of healthy and "infected" grass were collected for further investigation
-"infected" blades had de-pigmented (dead?) portions of the blade, with brown leading edges and no evidence of herbivory
-the brown leading edges were the supposed Labrinthula, but they also could have been caused by alternative items
-we also collected several dungeness crabs, a few cockles
1:30pm
2. Check bacterial plates from Armina
-we looked at the plates streaked on 7/21 with swabs from the sea slugs
-our marine agar plates (from heal-heal 1) yielded some bacteria, but we couldn't delineate separate colonies
-our TCBS (thiosulfate citrate bile salts sucrose) plates yielded no evidence of Vibrio from our individual
-other groups (with lesioned slugs) had evidence of Vibrio. Yellow on the TCBS plates indicated sucrose digesting strains, whereas green spots showed an absence of sucrose digestion
3. RNA extraction
-Steven led the class through the steps of RNA extraction, given in the "Day 3" section of the Littorina exercise (http://bio533.wikispaces.com/Lab_Littorina)
-in step 27, I resuspended my pellet in 50uL H2O. I lost a little bit of pellet when I jammed a pipet tip in the bottom of the tube containing the extract, but there was a visible amount remaining. I changed the pipet tip and resuspended the remaining amount.
4. gel running
-Lisa oversaw the electrophoresis of yesterday's DNA samples in lab 10
5. Hard agar
-Morgan and Kathy put pieces of 'infected' and healthy Zostera on hard agar plates to see whether bacterial colonies would expand from them
6. wet isolation of Labrynthula-associated bacteria
-Tiffany and Sarah placed bits of 'infected' and healthy Zostera in wet culture dishes with solution
-the samples went on a shaker table overnight to provide uniform access to nutrients within each sample
-tomorrow the samples will be centrifuged to see whether bacteria grew
-there was variation in their tool cleaning methods between different samples
7. hemolymph extraction from crabs
-Carolyn showed some of us how to extract hemolymph from crabs by inserting a hypodermic needle into their hemolymph sinuses
-hemolymph was dropped onto slides and topped with slipcovers
-ciliates were noted immediately in the sick crab hemolymph, ~30 min later noted ciliates in healthy crab h.l., gone from sick crab slide
10pm
8. Dungeness crab dissection (dead > 5 hrs
-dissected dead crab from Picnic Bay with Carrie, James, and Lisa
-made squash mounts of digestive tract, gonad?, gill tissue
-noted no cysts, saw two types of ciliates (the ‘pear’ and the ‘cucumber’), a ton of ciliates in gill tissue
Wednesday 7/21/2010
10am Armina swabs
-Armina sent from Tacoma have been developing lesions along their backs, then dying within a few days
-we wanted to take a look at the bacteria from their body surfaces by plating out swabs
-Morgan and I swabbed a slug without lesions (slug was labeled heal-heal 1)
-we sectioned the plates into quadrants and used a streaking method, diluting the streak in each consecutive quadrant
-I streaked 3 TCBS plates
-Morgan streaked 3 marine agar plates
-we both used plastic loops, which we cleaned 10% in bleach solution between each smear
-we also set up a squash mount of cells scraped off the back of our slug, and saw no ciliates on the slide
-squash mounts from the lesioned areas of sick slugs contained ciliates
1:30pm DNA extraction
-followed method for Qiagen Stool Kit given to us in lab
-my tissue weighed <0.01g
-Macerated tissue as best I could using a razor blade
-in step 4, only initially added 700 uL Buffer ASL before heating (step 5). To rectify, added additional 700 uL after heating, vortexed, then heated again. Sample was not fully homogenized because tissue was not cut into sufficiently small pieces for that result.
-canceled step 5 given in handout
-ran steps 12-15, but the amount of liquid in the tube didn't look right, so I threw that run out and re-did it with a different micropipette (later learned bench was missing appropriate tips for 20-200 uL pipette, which made quantities using that size pipette incorrect)
-in step 22 realized tips didn't match 20-200 pipette after attempting to use it for step 22 and seeing very little (< 100uL) AE buffer in spin column. To rectify, spun out the amount in the spin column without giving prescribed incubation period. Used small micropipette to roughly measure the amount of supernatant in the bottom of the collection tube (~25 uL), piped that amount back into the spin column, then added the difference (75 uL) to total 100 uL. Then proceeded with final methods.
-Carrie prepped a DNA test mix for the class to add their samples to
3:30 pm Gram Stain Demo
-Carolyn demo-ed gram stain preparation of bacterial samples on slides
-instructions of ‘day 2’ of Littorina lab methods page
Tuesday 7/20/2010
1:30pm
Cattle Point, Histology, Littorina dissections and sample fixing for DNA and RNA extractions
Cattle Point:
-at site ~8am
-collected littorines from tidepools in upper ITZ near areas with a lot of bird guano
-L. sitkana was frequently found higher up away from the tide pools (~0-2 ft), whereas L. scutulata was more frequently found right at the water edge (generally >0.5 ft from edge, underwater in tidepool)
-each of us collected ~6 of each species, as well as whatever large Nucella looked good to us
Histology:
-examined C. gigas (Pacific oyster) hematocyte slide, couldn't find hematocytes
tip for finding hematocytes: focus on edge of slide or label, then scan slide for cells (it still didn't work in this case)
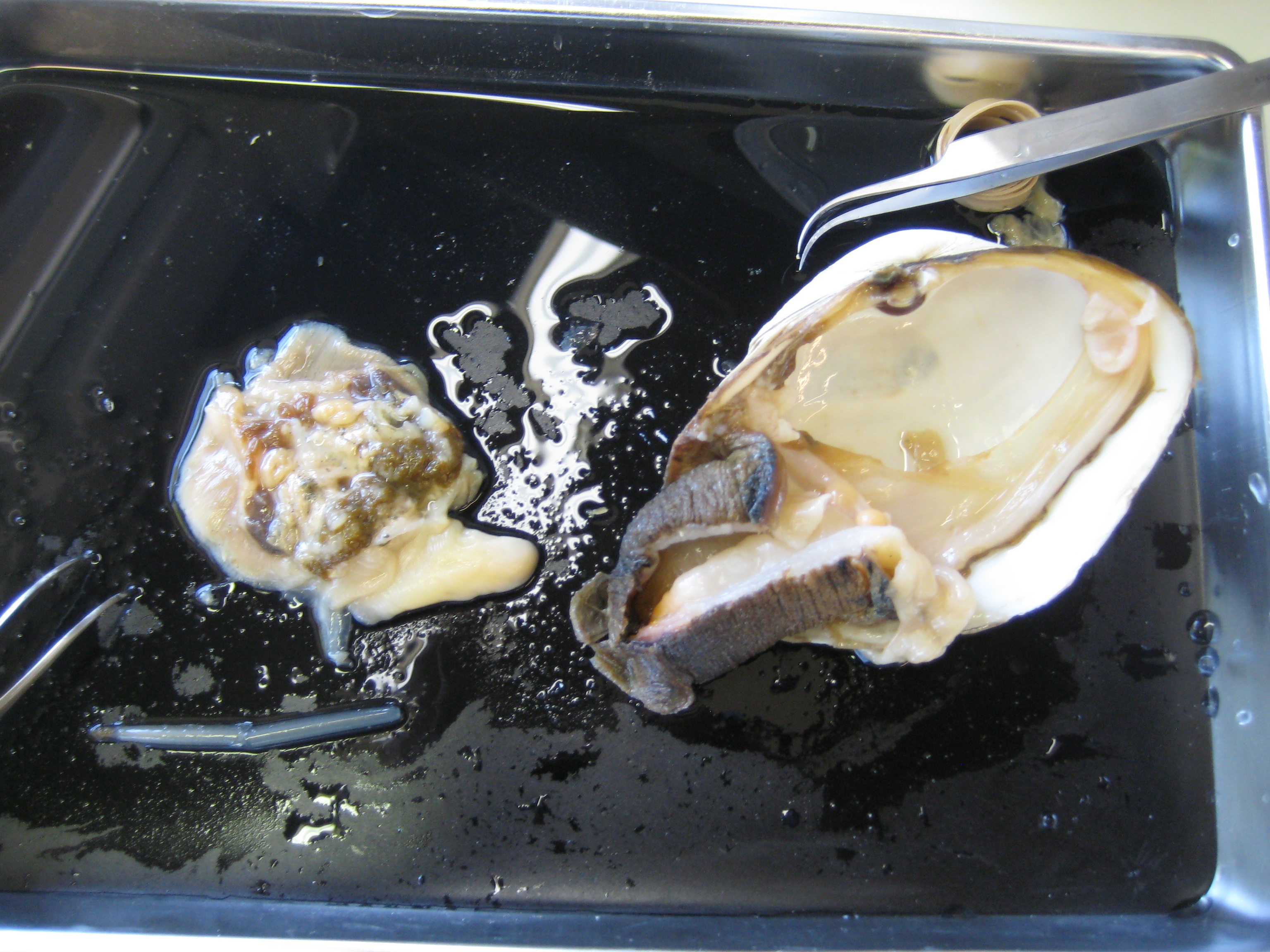
-examined healthy H. rufescens (red abalone) slide, IDed: hypobranchial gland, oocytes, digestive gland, mantle, posterior esophagus epithelium, intestine, gill (labeled photos in binders helpful for tissue ID)
Littorina:
-cracked shells of live snails using a (lock?) wrench
-separated shell from body
-cut off digestive system parts at posterior, head (because of pigments contained there), and operculum
-added sea water to check for parasites (not found in any specimens I dissected)
-cut off a roughly 1 x 2 mm portion of the foot, labelled "LF[#]", and put on dry ice in freezer for later RNA extraction
-put remaining tissue in freezer in vials with corresponding labels for each snail for later DNA extraction
-contributed tissue 4 L. sitkana for molecular tests to class total
-additionally opened up ~2 L. sitkana and ~2 L. scutulata (or possibly similar L. plena) to search for trematodes--did not save tissue
-saw trematodes from infected snails dissected by classmates--learned that rediae and sporocyst stages of trematode development are not very mobile relative to cercaria stage
Monday 7/19/2010**
2:30pm
Invertebrate Dissections
Sea cucumber (sp?):
-Partnered with Morgan, who cut
-followed cutting directions given for Cucmaria frondosa in Invertebrate Anatomy Online
http://webs.lander.edu/rsfox/invertebrates/cucumaria.html
-cucumber slowly eviscerated intestine and respiratory trees in response to cutting attempts
-found we initially only cut through the epidermis, and so had to re-cut through the dermis to open up specimen
-didn't find reproductive organs in cucumber
-internal parts IDed: calcareous ring, esophagus, gut, respiratory trees, longitudinal muscle bundles, buccal tentacle, poladian vesicle
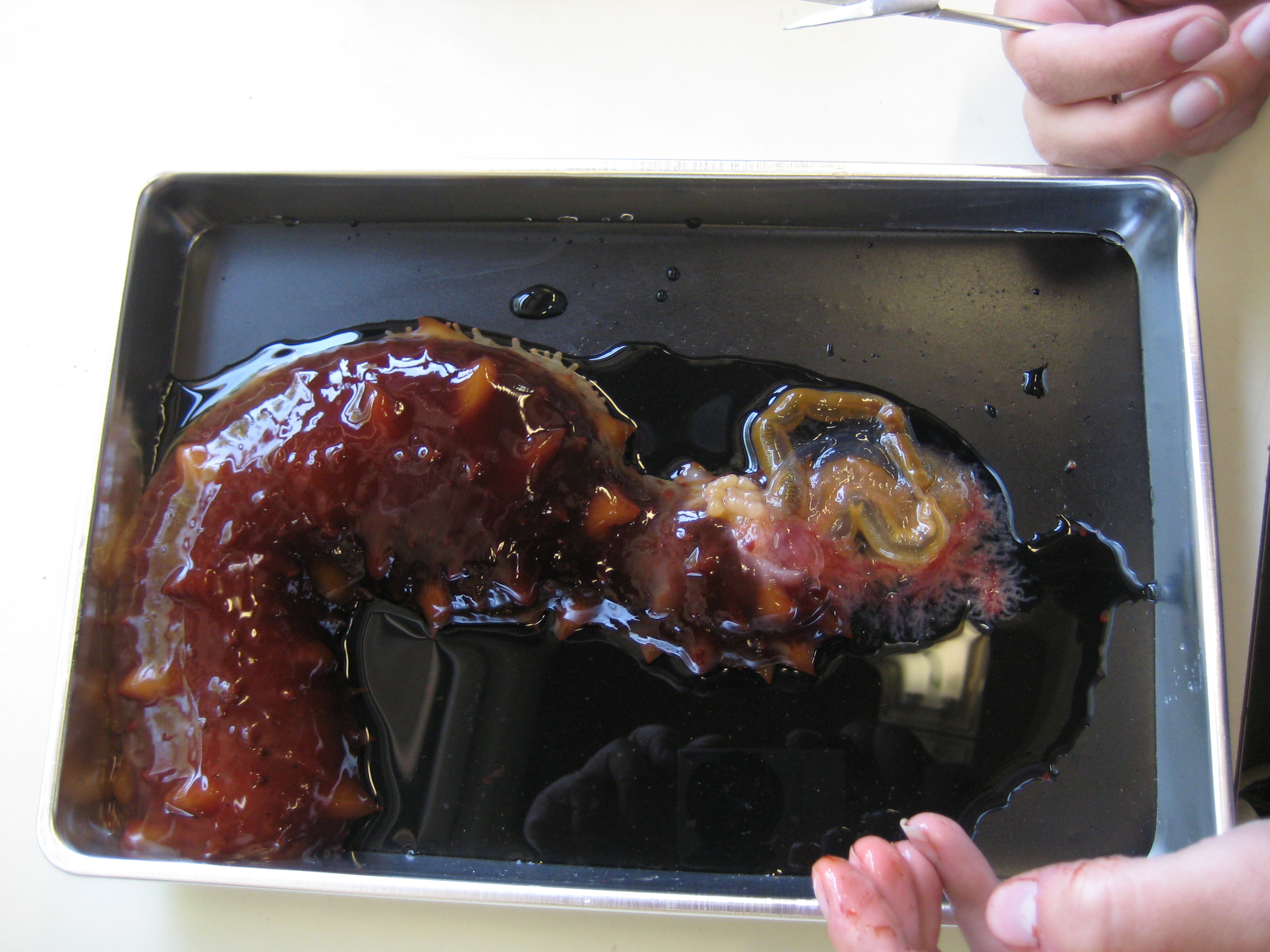 |
| sea cucmber evisceration |
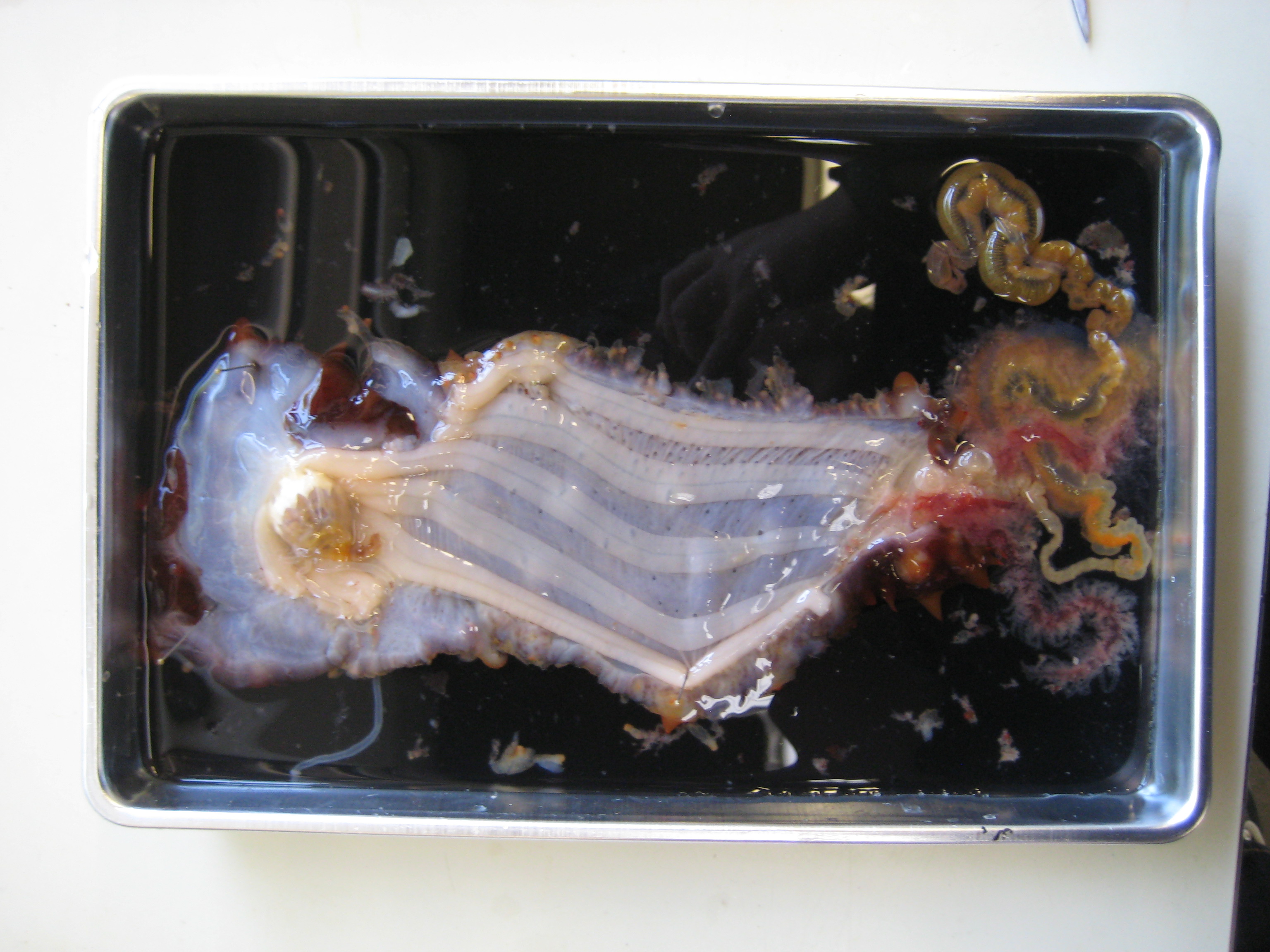 |
| dissected cucumber |
Sea star (Pisaster brevispinus?):
-cut of tips off all arms, then cut up along arms towards central disk. clipped fused ossicles (ambulacral???) giving structure to center of animal so body could be separated by top and bottom
-noted the cardiac stomach was the blob adjacent to the mouth, and the pyloric stomach was the pentagonal organ in the ring canal
-I thought P. brevispinus is generally more pink, whereas this individual was a muted purple
 |
| dissected sea star |
Other noteworthy items:
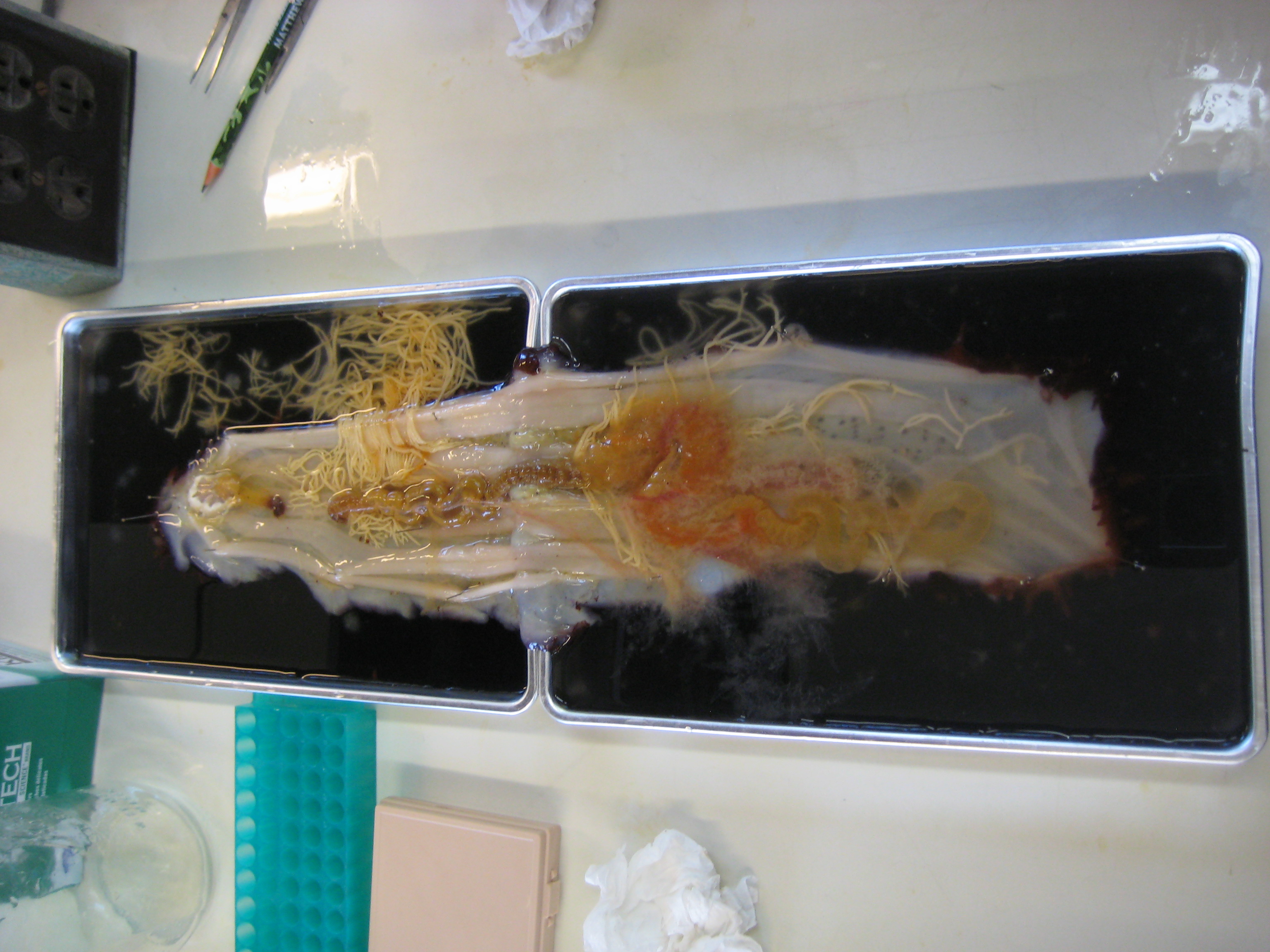
-Lani and Matt opened up a huge sea cucmber with a lot of gonads. Its poladian vesicles were much larger than the small cucumber's.
-saw the crystalline style of the geoduck used in digestion
-Morgan induced nematocyst firing of medtridium tissue
-saw the eye and tentacle of a snail Sarah worked on under a dissection scope. For the eye structure, I could see the small dot that is exposed to the outside stimuli, and the larger portion that is contained in the head.
 |
| big cucumber |
 |
| geoduck |