 August 18, 2010
August 18, 2010
http://www.phdcomics.com/comics/archive.php?comicid=1325
August 16, 2010
http://www.phdcomics.com/comics/archive.php?comicid=1325
August 13, 2010
http://www.phdcomics.com/comics/archive.php?comicid=1325
August 12, 2010
Prepared two master mixes, GH and SP primers, for 13 rxn. Used PCR water instead of BSA.
123.5µL PCR H2O
6.5µL F Primer
6.5 µL R Primer
162.5µL IMMO
Loaded 23µL of master mix and 2µL of template (Neg, Pos, Samples A à J) into wells and ran PCR
*well plate tipped over, sample J with SP primer master mix was low compared to others.
August 11, 2010
Indep Project:
Prepared master mix with RA primers and IMMO instead of GO TAQ.
Neg, pos, samples A-->J
Prepared master mic with EUB primers.
Neg, samples B, H, I, J
Prepared a 1% TBE gel. For product using IMMO: placed 1uL bead of loading dye on paraffin. The mixed 9uL of PCR product in a bead of loading dye before placing in wells.
First row: 1. Neg, 2. Pos, 3. A, 4. B, 5. C, 6. D, 7. E, 8. F, 9. G, 10. H, 11. I, 12. J
Second row: 1. Neg, 2. B, 3. H, 4. I, 5. J
| armina_8-11.JPG |
August 10, 2010
Indep Project:
Prepare a 1% agarose gel (0.5g of agarose, 50mL of TBE buffer), load wells and run the gel.
1. ladder, 2. negative, 3. blank1, 4. blank2, 5. positive (Rickettsia), 6. positive (Rickettsia), 7. A, 8. B, 9. C, 10. D, 11. E, 12. F, 13. G
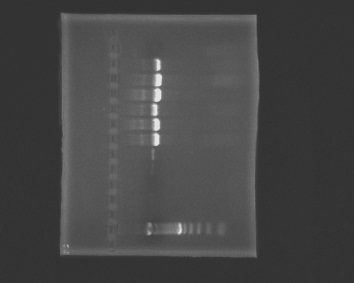 |
| armina_8-10-2010.jpg |
Extracted DNA from Armina samples H, I, J
Made a master mix with EHR 16 S primer and ran a gel.
1. neg c, 2. pos c, 3. A, 4. B, 5. C, 6. D, 7. E, 8. F, 9. G, 10. H, 11. I, 12. J
| armina_8-10.JPG |
August 9, 2010
Indep Project: Extracted DNA from Armina C and E samples.
Weight of sample C: 0.08g
Weight of sample E: 0.08g
We made a master mix with universal bacteria primer, EUB-A and EUB-B.
We prepared for 13 reactions, leaving a little extra for pipetting errors:
19.5µL BSA
6.5µL EUB-A
6.5µL EUB-B
104µL PCR H2O
162.5µL GO TAQ
-Samples A à G
-Two blanks
-Negative control of PCR water
-Two positive controls of Rickettsia Placed 2µL of template in appropriate wells with master mix.
Our plate was hand spun to force everything to the bottom of the wells, but our plate cracked in the middle. It did not appear to affect any wells.
PCR settings: (MJ Research Inc PTC-100)
Temp Time #Cycles
95 180sec 1 cycle
95 45sec
55 30sec (35 cycles)
72 90sec
72 300sec 1 cycle
*From now on, when we run PCR we will end with a temp drop to low temp (4-25 degrees) and hold for 30 minutes or so.
We removed the plate after PCR and placed it in the freezer until we can run a gel the next day.
August 7, 2010
Indep Project: Extracted DNA from Armina A sample
Weight of sample A: 0.07g The extracted DNA was placed in the freezer.
August 6, 2010
Indep Project: Carolyn gave a demonstration on making histology slides
-Prepare a 40 C water bath
-Place the histology block in the microtome and align it with the blade
-Place several drops of 50% ethanol on a clean microscope slide
-Cut a thin section from the histology block, using probes, place the section over the ethanol
-Place the section into the water bath to let the paraffin relax
-Dip a positively charged microscope slide into the water bath, place the section on the slide and remove the section out of the water bath
-Place the slide on a slide warmer to dry
August 5, 2010
Differential Display on Sea Fans Part2:
Made a 1% agarose gel.
-placed 0.5g of agarose powder and 50mL of 1x TBE buffer solution into an erlenmeyer flask
-microwaved the solution for 45 seconds
-mixed in 5µL of ethidium bromide
-poured the gel into the gel box and placed the comb
-let it set for 15 minutes to solidify
-removed the comb, placed the gel in the rig, and added 1x TBE buffer solution until the wells are covered
I loaded 7µL of 100bp ladder into well 1. Then I loaded 15µL into each well the following:
[With Primer A5] (A) (L) (C) (-) (-) [With Primer A17] (A) (L) (C) (-) (-)
 |
| dd_seafan_8.3.10b.jpg |
Indep Project: No growth on the mucus plates. Inoculated the plates with bacteria colonies grown from Cattle Point anemone.
August 4, 2010
Indep Project: Suspended a large anemone on the 1mm x 1mm fine mesh over a custard glass for 5 minutes. Scraped the underside of the mesh and collected 0.25mL of mucus. 50µL of the mucus was pipetted onto each marine agar and TCBS plate. A metal loop was bent into an L, dipped in ethanol, flamed and used to spread the mucus over each plate. Then the plates and lids were exposed to UV light to sterilize them.
Differential Display on Sea Fans:
I chose the arbitrary primers A5 and A17. I prepared a master mix containing:
12µL arbitrary primer
6µL ACP2
36µL H2O
60µL 2x MX
I loaded 19µL of master mix into each well. Then added 1µL of cDNA from tubes A, L, C. I have two negative control wells for
each primer.
August 3, 2010
Testing for protease:
1. Placed 1.5mL of marine broth with Vibrio tubiashii into a microcentrifuge tube
2. Centrifuged for 10 minutes
3. Placed 100µL of supernatant and 400µL of Azocasien into a new microcentrifuge tube
4. Incubated at 37 C for 30 minutes
5. Added 600µL of 10% trichloric acetic acid (TCA)
6. Placed tube on ice for 30 minutes
7. Centrifuged at 13,000 rpm for 5 minutes
8. Added 200 μl of 1.8 N NaOH to a new microcentrifuge tube
9. Added 800 μl of the supernatant to the tube with the NaOH
Result: The supernatant turned orange indicating a positive result for protease.
Indep. project: Stressed anemone in distilled water and collected mucus. Method found in paper:
EFFECTS OF A TOXIN FROM THE MUCUS OF THE
CARIBBEAN SEA ANEMONE (BUNODOSOMA
GRANULIFERA) ON THE IONIC CURRENTS OF
SINGLE VENTRICULAR MAMMALIAN
CARDIOMYOCYTES
EDUARDO M. SALINAS,’ JORGE CEBADA,’ ALBERT0 VALD&*
ANOLAND GARATEIX,’ ABEL ANEIROS* and JULIO L. ALVAREZ3*
Toxicon,
Vol. 35, No. 12, pp. 169!%1709, 1997
Will centrifuge to see if I can get a pellet of mucus.
Centrifuged two tubes at 3000rpm for 10 minutes. A dark pellet formed but I wasn't able to isolate mucus from the pellet or water.
I tried hanging an anemone in the air for a few minutes on 1mm x 1mm fine nylon mesh but did not see much mucus. Method found in paper:
Do anemonefishes use molecular mimicry to avoid being stung by host anemones?
J. K. Elliott*, R.N. Mariscal, K. H. Roux
J. Exp. Mar. Biol. Ecol. 179 (1994) 99-113
Differential Display on Sea Fans: (Group: Sarah, Loni, Marie and Matthew)
Our group received the A14 arbitrary primer. Sarah prepared a master mix containing:
14µL arbitrary primer
7µL ACP2
42µL H2O
70µL 2x MX
1µL of cDNA samples A, L, C were placed in the wells. There were two negative controls.
August 2, 2010
Indepedent Group Project: Katy and Morgan made a master mix with GoTaq Green Master Mix, BSA, sterile water, and universal bacteria primers. The master mix and A nthopleura elegantissima template were added to the strip wells in the thermocycler. We will run a test PCR to see if we can detect the presence of bacteria with the universal bacteria primers. We will not be able to identify the bacteria with this test.
Observed slides of Serum Agglutination Test with Vibrio tubiashii. On the control slide, the bacteria were spread out and no clumps were present. On the treated slide, the bacteria clumped together.
Class project:
Goal/Question: Examine the influence of ocean acidification (OA) and Vibrio tubiashii (Vt) on larval oyster health
Timeframe: 3 days
Setup: Static system, only air is flowing in but not water. The water is refreshed everyday.
The oysters were dyed with Neutral Red. Living tissue takes up the dye and appears red. Pink larva can be dying or dead.
Vt was grown in broth overnight.
The larval oysters were inoculated on 8/1 and fed. The serial dilutions to determine amount of bacteria to add was not done before inoculation. The larval oysters were sampled on 8/1 and 8/2 at 5pm. The larval oysters were strained using 70 micron mesh to collect a sample size ~1000.
Calculating the serial dilutions CFU/mL = (# colonies)(dilution factor)(plate dilution)
10^-6= 15^5 x 10^6 x 10
1.55 x 10^9 CFUs which means the amount of bacteria added to the larval oyster tanks were 10^4, 10^6, and 10^8. Lisa said that Vt will kill all the larva at 10^3.
10^-3: TNTC
10^-4: TNTC
10^-5: 1000; 1104 Ave: 1052
10^-6: 162; 144 Ave: 155
10^-7: 24;18 Ave: 21
10^-8: 2; 2 Ave: 2
July 30, 2010
Primer design tutorial:
Reviewed quiz 1 and discussed different strategies and tools.
Steven prepared a new gel with class anemone samples, ladder and oyster samples.
Western blotting results: The antibody bound to the oyster but not to the sea anemone.
July 29, 2010
We checked our gels to see how the color bands would appear in the gel, decide what amount to load. Our M+S HS sample bands appeared dark blue indicating a high amount of protein. So we diluted 25µL of our supernatant with 75µL of CelLytic MT and placed into a new tube. Then we pipetted 30µL of the diluted solution and 30µL of the 2X Reducing Sample Buffer into a screwcap tube. Our sample was centrifuged for 10 seconds, boiled for 5 minutes and centrifuged for 1 minute. We prepared a new protein gel and loaded 30µL of our sample for Western blotting and 20µL for staining and visual analysis.
I measured out Marine Agar powder to make more agar plates.
Oyster analysis for Bohemia sp . protists:
I cracked open an oyster. Removed some hemolymph with a syringe and placed a few drops on a slide. The I removed the ventrical of the heart and swabbed a small area on the slide. I placed the slides in a methanol bath for a few minutes and in a Giemsa Stain bath for several minutes. I looked at my slide under 100X oil immersion but I did not identify any hemocytes on my slides.
July 28, 2010
We looked over our ISH slides and compared them to the H&E slide.
H&E: I can see the Rickettsia in the digestive tissue . The inclusion bodies are large (about 26µm), round and dark purple. Each inclusion body contains hundreds of Rickettsia. The H&E also stains the nucleus (around 5µm) of the cells purple. The hemocytes are 11µm.
08:1-12 (+): The probe did not bind to the Rickettsia.
08:1-12 (-): The control slide does not have the probe so the Rickettsia are not colored.
Protein extractions from A nthopleura elegantissima
Sarah and I worked with a heat shocked anemone. Sample tube labeled M+S HS
Method:
-Blotted dry and weighed, 0.82 grams
-Cut and weighed an 1/8th section of the anemone, 0.06 grams
-Minced with a razor blade and placed in a microfuge tube with 800µL of CelLytic MT
-Spun the tube at maximum speed in a refrigerated microfuge for 10 minutes
-Transfered 500µL of supernatant to a new labeled tube and store on ice
-Vortex supernatant
-Place 50uL of supernatant and 50uL of 2X Reducing Sample Buffer in a screwcap tube
-Centrifuge for 10 seconds
-Boil tube for 5 minutes
-Centrifuge for 1 minute
-Prepare a protein gel, wash out wells with buffer solution
-Loaded wells with 10uL, 30uL and 50uL
-Run for 45 minutes at 150 volts
July 27, 2010
We continued with the ISH protocol (steps are in the lab manual).
5. Stringency washes
6. Detection
The slides will be ready to view tomorrow. The probes would attach to classic WS-RLP, turning them brown for easier identification.
We discussed potential group projects during lunch:
- acidification and larval disease
- acidification, stress on Anthopleura elegantissima
- microbial community on Anthopleura elegantissima
- wasting disease on P isaster ochraceus
- Rickettsia and nudibranchs
June 26, 2010
We went through the protocol for In-situ hybridization (ISH) to probe for WS-RLP, an abalone pathogen. The probes came from PCR DIG Probe Synthesis kit and prepared the previous day. I worked in a team of three with Marie and Loni. We had two histology slides, the sections were in paraffin. One slide will get the probes, the other slide will be our control and no probes.
We went through the steps from the lab manual for:
1. Tissue deparaffinization – we used Safe Clear instead of Xylene.
2. Permeabilization of tissues
3. Prehybridization – we used the alternative buffer
4. Hybridization
I'm reviewing what other students have done for the quiz. It looks good to go through literature to avoid reinventing the wheel if someone has done similar work and published. Then doing searches on NCBI taxonomy looking for a genome of the organism or closely related organisms, genes and then running BLAST. I'm playing with NCBI: checking out the taxonomy section to find "tremetoda." Looking at what the genome looks like.
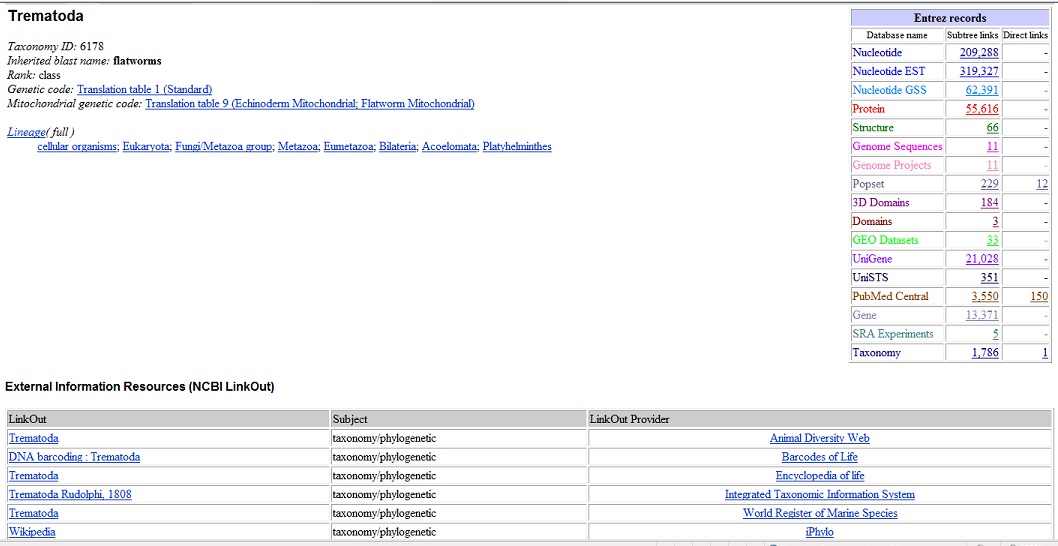 |
| taxonomy.jpg |
June 25, 2010
I worked on the quiz question. I wanted to learn more about the tools Steven covered in his lecture, NCBI, BLAST, Entrez Gene, and others. It was my first time trying to look for genes and figuring out primers. It took a lot of trial and error but I learned a lot. I made sure I was able to repeat my steps and find my primers again.
I found a long list of mRNA sequences when I search NCBI for “trematode.” I wasn’t sure what any of the results meant. I had to Google certain things trying to figure out what a particular gene was and if it was in trematodes.
If money and time was unlimited, it would interesting to run PCR of Littorina (same species) that were infected with various trematodes. Then look at the genes that are not in the Littorina , find gene(s) shared by all the trematodes, and make a primer based on that.
June 24, 2010
We reviewed and discussed the Littorina Q-PCR results. It was interesting to go through and see all the different results and hypothesize what might have happened:
If the negative control was not a flat line, but instead peaked at around 80o C, there could have been contamination at some point or in the nanopure water.
If a gene was not expressed but lab technique and cDNA were good and no contamination occurred, it could be possible the gene was not expressed.
I did not get a clear result from both my cjunk samples, just two squiggly lines that descended and no peak. Possible reason is poor lab technique. For actin, the graph looked proper although the two lines were not on top of each other. There was only one peak at 85o C which was consistent with other samples where actin was expressed. My actin Ct were 31.54 and 33.80. I used this formula: Arbitrary expression value =10^(-(0.3012*Ct)+11.434) and my arbitrary values were 94.69346 and 17.92421 with a mean 56.30884. I could not normalize my values because I did not have Ct numbers for cjunk.
It was interesting to see the chart of gene expression: the unhealthy Littorina had higher mean gene expression than the healthy ones. Although various factors had to be ignored like small sample size, if they are all the same species or not, gene carryover, etc.
I checked my nudibranch streak plate. The bacteria from NL2 grew very fast and fermented sucrose turning the surrounding agar yellow. The bacteria from NL3 were green round colonies and also grew very fast.
June 23, 2010
Nudibranch Streak Plate results
No lesions #1 (NL1) Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 “ “, 4 colonies
Q3 “ “, >5 colonies
Q4 none
NL2 Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 “ “, >20 colonies
Q3 none
Q4 none
NL3 Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 “ “, >20 colonies
Q3 none
Q4 none
NL1 TCBS
Q1 none
Q2 none
Q3 none
Q4 none
NL2 TCBS
Q1 2 yellow shiny round colonies grew larger, 5 small colonies grew larger compared to
24hrs ago and turning green-yellow
Q2 none
Q3 none
Q4 none
NL3 TCBS
Q1 One large blob, appears wet and green, size has not increased in the last 24hrs
Q2 none
Q3 none
Q4 none
I made a new streak plate on TCBS. I used a metal loop and dipped in alcohol and flamed before and after every swab. I streaked out the large yellow colony from NL2 and the large blob colony from NL3. I will check the results after 24hrs.
I made a smear of the large yellow colony from NL2 and the large blob colony from NL3 onto separate slides. After the smears dried, I heat fixed them and did a Gram stain. Under the 100X oil immersion lens, I saw Gram negative short rods on both slides. Some looked like two rods linked together but I don’t see evidence that they are chained rods.
Reverse Transcriptase to get cDNA
I thawed the MCE2 RNA sample on ice. I transferred 18 µL to an RNA-free tube and heated it at 70o C for 5 minutes. Then iced the tube for less than 10 minutes. I added 7.25µL of the master mix (see below) and heated it at 42o C for one hour then 95o C for three minutes to get cDNA.
Master Mix:
5 µL 5x MMLV Buffer
1.25 µL 10mM dNTB
.5µL MMLV RTase
.5µL Oligo dT primero
Q-PCR
I prepared my cDNA for Q-PCR. I made two master mixes, actin and cjunk (c-jun N-terminal kinase):
Master Mix:
7.5 µL BSA
2.25 µL Forward primer
2.25 µL Reverse primer
62.5 µL 2x SyBR MM
40 µL sterile water (used the tube of nanopure water near the PCR tray)
By the time I went to fill out the chart, it was confusing because several people entered their information differently. There were a few of us waiting to fill the PCR tray so it was a rushed job. I pipetted 23 µ L of my master mix - cjunk and 20 µ L of my cDNA into the wells E5 and E6. I pipetted 23 µ L of my master mix - actin and 20 µ L of my cDNA into the wells E7 and E8. I ran out of master mix and did not have my negative controls. I didn’t pipette enough of something when I made my master mixes.
July 22, 2010
PCR was performed on the extracted DNA from the Littorina using universal primers. After PCR, the products went through gel electrophoresis.
Nudibranch Streak Plate results
No lesions #1 (NL1) Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 “ “, 3 colonies
Q3 “ “, 8 colonies
Q4 none
NL2 Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 Small white round colonies, >20 colonies
Q3 none
Q4 none
NL3 Marine Agar
Q1 Small white round colonies, >20 colonies
Q2 Small white round colonies, >20 colonies
Q3 none
Q4 none
NL1 TCBS
Q1 none
Q2 none
Q3 none
Q4 none
NL2 TCBS
Q1 2 yellow shiny round colonies that fermented sucrose, 4 green shiny round colonies
Q2 none
Q3 none
Q4 none
NL3 TCBS
Q1 One large blob, appears wet green
Q2 none
Q3 none
Q4 none
I didn’t get much growth on the plates. I was hoping to see little or no bacterial growth since I swabbed unbroken skin. I’m not sure if the bleaching and water rinsing of the loops caused problems with the results. I will look over the plates again tomorrow.
We followed the RNA Extraction procedure from the lab handout. The entire class went through each step at the same time because we were working with TriReagent. Steven dispensed the chloroform when we reached that step. The RNA is stored in -80o C.
July 21, 2010
Nudibranchs
We received a few unhealthy Armina sp. Nudibranchs from Jim Murray’s lab. The nudibranchs were collected from Tacoma and placed in sea tables. The animals develop irregular stripes and die soon after lesions appear.
Alanna, Marie, Sarah and I examined one of the unhealthy nudibranchs taken out of the diseased tank. The nudibranch was 5cm in length and 2.8cm in width. Marie and Alanna swabbed the leading edge of lesions while Sarah and I swabbed healthy skin. I swabbed a 1cm x 1cm patch of healthy skin near the posterior on the dorsal side and made 4 quadrant streak plates with TCBS and marine agar plates. After each swab I bleached the loop and rinsed it with tap water. I wrapped the sides of the plates with parafilm, inverted and let them incubate.
I looked at tissue presses of a lesion from the nudibranch. I saw ciliates which appeared egg shaped with cilia. We were shown a demonstration of Gram staining. A smear on a slide was heat fixed by moving the slide quickly over a flame a few times. Crystal violet was placed on the slide for about 1 minute and rinsed off with water. Then placed Gram’s Iodine for a 1 minute and rinsed off. A decolorizing solution of alcohol and/or acetone was placed on the slide for several seconds and rinsed off. Safranin was placed on the slide for 20 seconds and then rinsed off. The slide was blotted dry and viewed with the 100X objective lens with oil.
I took one of the Gram stained tissue presses and observed it under 100X oil immersion lens. I saw a few ciliates that appeared like pink and purple Easter eggs. There were also lots of Gram negative short rod bacteria. I showed my slide to Charley O’Kelly and he saw lots of bacteria, host cells that appear to be leukocytes and possible ciliates.
Snails
We extracted DNA from the dissected piece of snail foot. We should get DNA of the snail and the microbial community. We followed the instructions for the QIAmp DNA Stool kit but only used half of the enzyme tablet that eliminates inhibitors for PCR. I processed sample MCE2 which came from a healthy snail.
July 20, 2010
The class reviewed marine invertebrate histology slides. I looked at slides under the compound microscope.
Mytilus sp . (normal): I identified oocytes, digestive gland tubules, intestine with possible pieces of algae, muscle, connective tissue.
Abalone hemocytes:
Dissections and Processing for DNA and RNA
Method:
1. Crack shell and remove the animal from the shell fragments
2. Cut off the digestive gland and observe under the dissecting scope for parasites
3. Cut off the head
4. Remove operculum, slice a small piece of the foot and place in a tube for DNA analysis
5. Place the remaining body parts into a RNase free tube
6. Place the tube for DNA analysis in the refrigerator and the tube for RNA analysis in the dry ice box.
I dissected 4 Littorina sitkana and 2 Littorina scutulata . I did not see signs of infection by parasites. I had processed 4 L. sitkana and 1 L. scutulata for DNA and RNA analysis.
July 19, 2010
Alanna and I dissected the giant red sea cucumber, Parastichopus californicus. It measured about 30cm in length. It used tube feet on the ventral side to move. The
papillae on the dorsal side appear like soft large and small conical spikes. The mouth in located at the anterior end and ventrally. Feeding tentacles surround the mouth. We cut along the dorsal midline from the anus to the mouth. When fully opened and pinned, we can see the white ring canal with polian vesicles. The esophagus was connected to the posterior side of the ring canal. The stomach and the rest of the digestive system appeared to have separated from the esophagus. The intestines and respiratory trees had been pushed down towards the anus but had not eviscerated through the anus. The longitudinal muscles run anterior to posterior and were quite thick. The gonads were yellow and noodle like. The animal appeared to be healthy, no lesions, growths or internal parasites.
I cut a 1cm square of the skin and placed it in a vial of bleach to isolate the spicules. I let it sit for a few hours. I made a slide of the spicules and they look like elaborate tacks about 15um in length.
|
||
| Giant Red Sea Cucumber |
|
||
| Spicules |