Teamatode processed the 22-hour exposure treatment snails from the Vt challenge experiment this afternoon. We also extracted RNA from 5 control and 5 experimental snails from the 1-hour and the 22-hour treatments, using the same protocol.
Based on the results from the PCR and gel yesterday, James ran a PCR and gel on the samples from C6 and C11. Since there is little PCR master mix left from the differential expression kit, he used a "recipe" that Lisa Crosson provided that went as follows:
Amount per Reaction
4 ul 5x Buffer
1.2 ul MgCl
1 ul BSA
0.5 dNTP (10 mM)
0.5 ul F-Primers
0.5 ul R-Primers
0.5 ul Taq
10 ul H20
Since eight reactions were needed, the following recipe was used (making enough for eight and a half reactions):
34 ul 5x Buffer
10.2 ul MgCl
8.5 ul BSA
4.25 ul dNTP
4.25 ul F-Primer
4.25 ul R-Primer
4.25 ul Taq
85 ul H20
Since James could not locate a P100, and this master mix is intended for 20 ul reactions, he had to use a P10 twice to get volumes between 10 and 20 ul. This includes while loading the wells. The well containing C6-Control initially only received 9 ul of master mix, so it received master mix from two different tips. C11-Control also received master mix from two separate tips, one that it shared with all other samples and one it shared with all the controls.
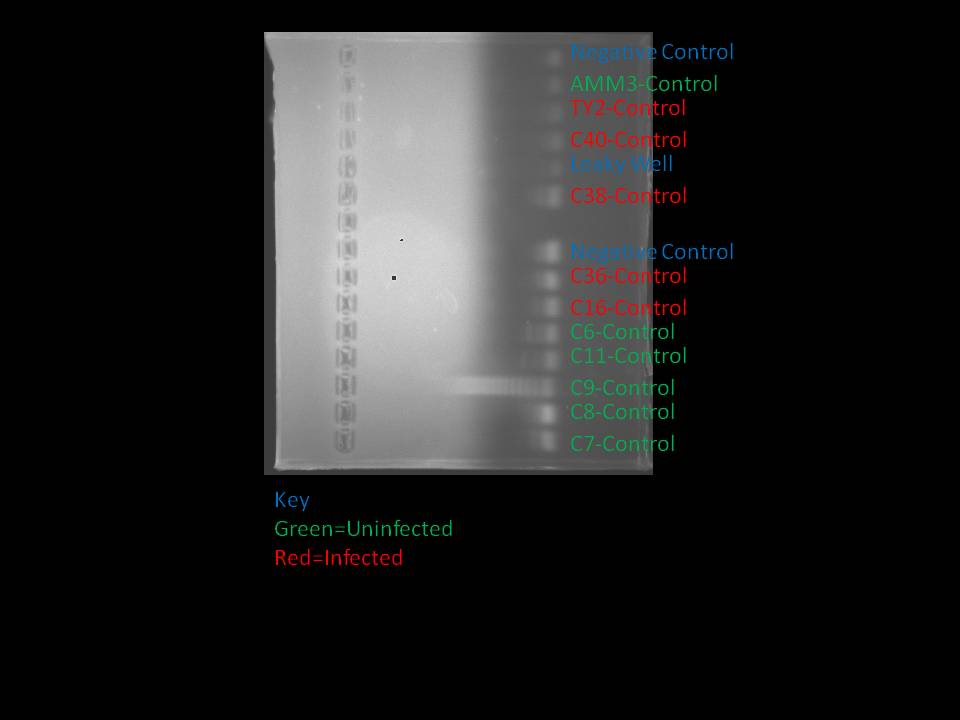
The PCR products were ran at 110 volts for a total of 23 minutes. The results were the following:

Note that C6 and C11 were both uninfected.
August 14, 2010
Today, Teamatode set up a Vibrio tubiashii challenge experiment w/ some of the L. sitkana from Jekyll cove. The goal of the challenge is to see whether/how Gprot and Cyclin (our 2 genes of interest from our trematode infected vs uninfected study).
- Vt suspension was made from one of Sarah's cultures (not sure what the starting volume was, need to find this out) by spinning down the bacteria, decanting the marine broth supernatant, and re-suspending in 20ml of sterile seawater (Steven did all of this for us - thanks Steven!)
- We plated serial dilutions of the culture to count colonies, but our plates didn't grow (probably b/c we had stored the culture in the fridge to slow its growth while we waited to locate plates), so we counted Elene's plates, which were made from the same original culture (but potentially re-suspended in a different amount of seawater) -
- 60 L. sitkana from Jekyll cove were divided between 6 250ml culture flasks that were filled with 282ml of seawater (filled to half-way up the neck of the bottle to ensure that the snails would remain submerged during the 1-hour exposure). The experimental snails (n=30 total) were exposed to a known dose of bacteria for 1 hour, and then examined at three time points post-exposure (immediately after 1 hour, at 10 hours, and 22 hours). We also maintained a control flask for each exposure treatment.
- To the 3 exposure flasks, we added 2ml of Vibrio tubiashi suspension, for a dose of --- per ml. After the 1-hour exposure, the flasks were emptied, rinsed, and refilled with seawater. 5 unexposed and 5 exposed snails were immediately measured, dissected (with a hammer) to determine sex and trematode infection status, and a 50-100mg sample of tissue was collected for gene expression analysis. The other 5 experimental and control snails from this treatment were moved to 300ml pyrex bowls filled with seawater, in order to monitor the longer-term effects of the exposure on mortality. Samples for the 10-hour and 22-hour treatments were processed in the same way.
At one point today, James found a second tray of restriction digested class samples in the Lab 5 refrigerator. The labels are virtually the same as the labels on the tubes found yesterday. It is assumed that these fridge samples are more reliable as they have been stored under better conditions, so from now on when restriction digested DNA from the class samples is needed we will use these samples.
James intended to run a PCR on the positive controls from both the class samples and the samples that Teamatode took from Cattle Point. However, there was a problem with the pippets; they do not seem to be calibrated properly according to Emma. As a result, the master mix did not contain the correct volume. However, she noted that she was still successful at running PCR with the pippet altered pippets so I continued with less reactions. All the Cattle Point positive controls were processed, but only two class samples. The intended master mix was the following:
206.5 ul PCR Master Mix (from the differential display kit)
13.2 ul F-primer
13.2 ul R-primer
146.85 ul PCR H20
The PCR product was put through a gel at 110 volts for a total of 25 minutes. The results were the following:
August 13, 2010
Since the methylation assay from yesterday was inconclusive, James and Carrie ran a new methylation assay using the restriction digested DNA from the class data (since these samples were known to generate bands in the initial assays). The PCR was ran as it was on August 12, 2010, except that the PCR master mix from the differential display was used instead of 2x Immomix. This PCR master mix was found to work effectively by Sarah Hu earlier in the day but an atypical solid at the bottle of the tube when Carrie and James were making the master mix. The master mix had the following proportion of reagants (NOTE: Enough master mix was made for seven reactions):
87.5 ul PCR Master Mix
5.6 ul F-Primer
5.6 ul R-Primer
62.3 ul PCR H2O
and unrefrigerated for an unknown period of time. The gel was ran at 110 volts in burst for a total period of around 28 minutes. Ethidium bromide was added directly into the gel, so no bath was necessary to visualize. After the gel was ran, it was difficult to see whether the dye was still on the gel. It is possible that the gel was ran for too long. The results were the following:
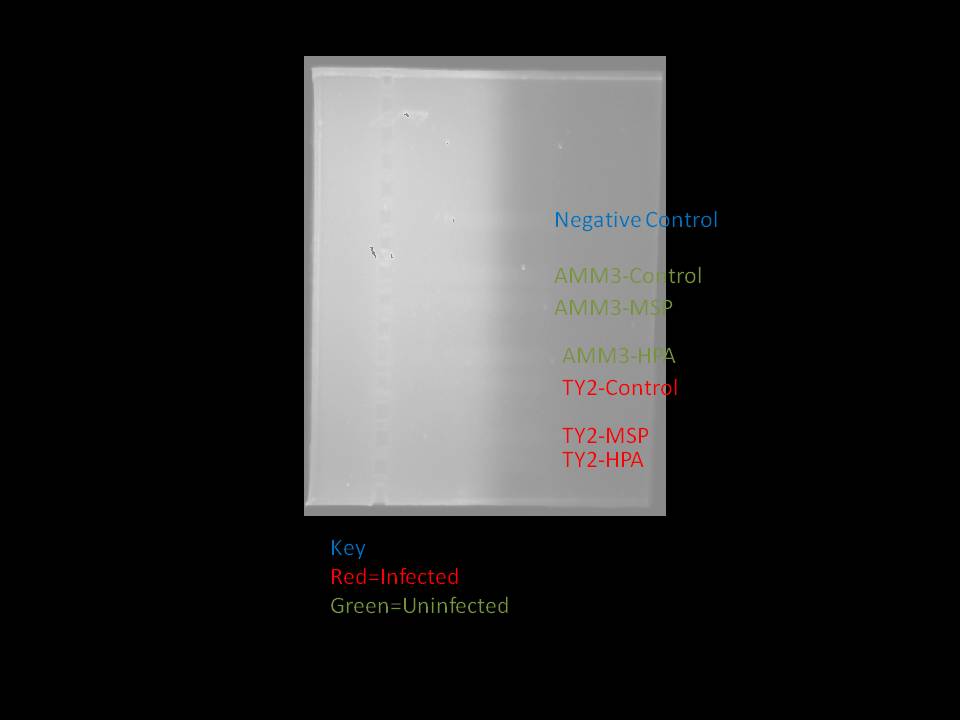
I was concerned that the lack of bands was a result of letting the gel run too long, so I ran another gel. This gel ran, in total, for 40 minutes. This is the result:
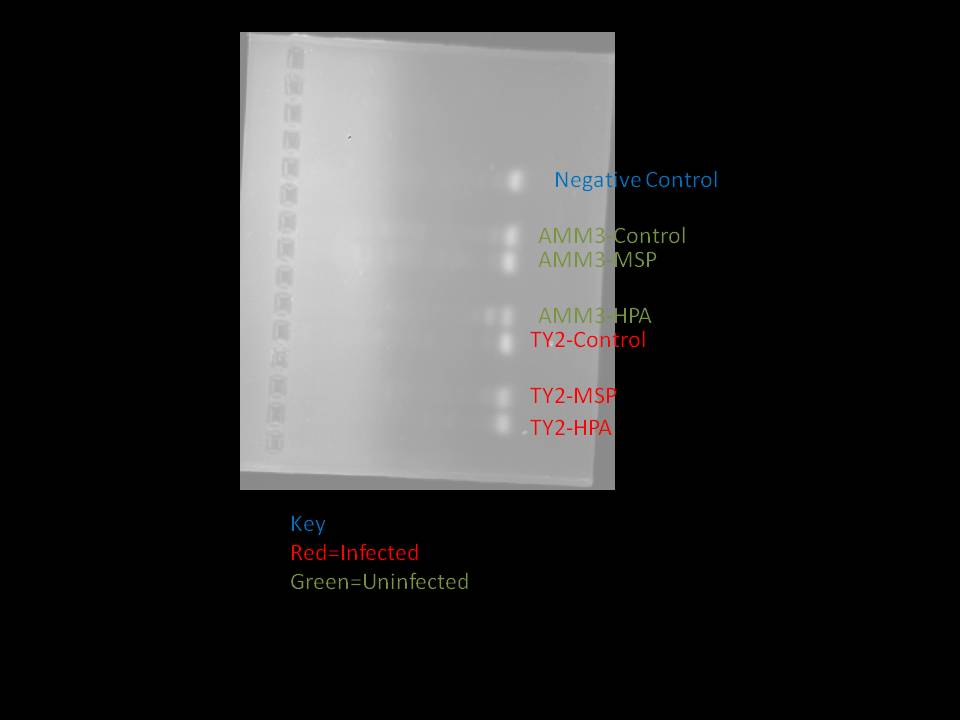
August 12, 2010
Morning: reading stuff!
When the reverse transcriptase gets here, we'll be setting up reactions for the 10 Cattle Point sitkana samples.
Reverse transcription Master Mix (for 25ul reactions):
5 ul 5X MMLV Buffer
1.25 ul 2.5uM dNTPs
0.5 ul MMLV Reverse Transcriptase
0.5 ul Oligo dTs <-- we used the last of the Oligo dT's - will need to use random primers in the future if we need to set up more of these reactions (substitute same volume of random primers that we have been using for oligo dt's)
7.25 total Master Mix per reaction
+ 5 ul RNA extract + 12.75 ul ddH2O
Promega protocol:
1. Incubate RNA+H2O (17.75ul) sample in 70C for 5 min, then in ice for 10 min
2. Add Master Mix (7.25ul)*
3. Mix gently by flicking tube(or vortex briefly and spin down)
4. Incubate for 1 hour at 42C
5. Incubate for 3 min at 95c
*Carrie added 7.25 master mix to C16,36,38,40,47 without Reverse Transcriptase - had to go back and add 0.5ul of enzyme to these tubes (other tubes received 6.75 of master mix w/out enzyme + 0.5ul of enzyme, so they should all have the correct ratios of master mix components)
James, Carrie and Tiffany processed the rest of the Reuben Tarte snails today.
Carrie and James set up qPCR reactions for the 10 Cattle Point L. sitkana that were DNased on 8/10, using the primers for Gprot, Cyclin, and the reference gene Actin.
qPCR master mix (per reaction)
12.5ul Sybr master mix
1.5ul BSA
8ul H2O
+2ul of cDNA
Since fresh G protein-mediated receptor primers came in today, James began a new methylation assay on the G protein-mediated receptor gene using the restriction digested DNA that Tiffany and Carrie made earlier. Only a subset of possible samples were put into the assay just in case something goes wrong in the process. The samples used in this assay were C9, C11, C16, and C36.
A few complications arose while generating the master mix for conventional PCR. First, due an arithmetic error, I generated a master mix that only contained enough reagents for 10 reactions. Then, while trying to add more reagents, I added too much water by accident. In the end, the master mix contained:
188.59 ul 2x Immomix
12.07 ul F-primer
12.07 ul R-primer
164.45 ul PCR Water
These values are at almost exactly the same proportions as the master mix used on August 10, 2010. The protocol was otherwise the same as on August 10, 2010. The wells were loaded into the thermocycler thus:
Strip 1
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
| C16-HPA |
C16-MSP |
C16-Control |
C9-HPA |
C9-MSP |
C9-Control |
C11-HPA |
C11-MSP |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
| C11-Control |
C36-HPA |
C36-MSP |
C36-Control |
Negative Control |
Negative Control |
-James mixed the master mix by repeatedly pumping liquid into the pippet and then ejecting the liquid again between Strip 1-Well 3 and Strip 1-Well 4. He did this because he could not remember if he had mixed it before beginning to load the master mix into wells.
-When adding master mixthe wells, James used one tip for Wells 1-6 of Strip 1 and a second tip for the rest.
-When adding DNA to the samples, James began to become concerned that the pippets were contaminating tubes. The centrifuge tubes that contained the DNA were so deep that the sides of pippet hit the sides of the tube. To contend with this issue, James began to bleach the pippet between wells. He began doing so after Strip 2-Well 1. Beginning after Strip 2-Well 2, he also rinsed the pippet with tap water after cleaning with bleach (before, we wiped the bleach off with a paper towel).
After the PCR was completed, James ran a gel with the PCR products. There was major evaporation in C16-MSP and less severe, but noticeable, evaporation in C9-HPA. The protocol was the same as on August 11, 2010, except that I used a different method for the loading dye. I put 2 ul blobs of dye on a piece of parafilm. There were as many blobs as there were wells to placed into the gel. Then I added 8 ul of product to these blobs such that every well on the PCR plate was placed into one blob. Then the mixture of product and dye were placed into a gel well. This procedure was done in the interest of preserving dye. An error was committed in C9-Control in which 10 ul of DNA was added to 2 ul of dye. Only 10 ul of the mixture was placed into a well, though. Also note something I have been forgetting to document: I ran the gel at 110 volts for 20 minutes. In the past, I might have run it for a little bit longer so that the dye was in the correct place. After the gel was ran, I used an ethidium bromide bath to visualizebands. The gel was soaked in the ethidium bromide bath for five minutes, and then placed into a water bath for 10 minutes. The results were as follows:

August 11, 2010
Carrie and Tiffany extracted DNA from the 10 Cattle Point snails (5 infected and 5 uninfected) that we have been using for the gene expression study. We used the complete tissue samples dissected from each snail (masses varied from .36g to .63g), and massed them by taking the mass of the entire tube, then subtracting the mass of the tube after the ethanol was pipetted out and the sample was allowed to dry in the tube for ~2min.
DNA extraction was carried out using the Quiagen kit, following the same protocol we used in class:
We used the Quiagen Stool Kit
- Assemble supplies - make sure reagents are diluted to correct concentration. Need 2-2ml microcent tubes, 2- 1.5ml tubes, 1 well-labeled microcent tube (for final conc)
- Cut a piece of tissue approx the size of a 1/2 pencil eraser and record tissue weight
- Cut sample into small pieces and put into a 2-ml centrifuge tube
- Add 1.4 mL of Buffer ASL as follows: add 700ul, mix with pestel (optional), vortex for 1 minute, then add another 700ul of ASL, then vortex continuously for 1 minute until the sample is thoroughly homogenized (try to homogenize as much as possible to get max DNA extracted)
- Heat sample for 5 min at 70
- Vortex for 15 seconds and then centrifuge at full speed for 1 min to pellet
- Pipette 1.2mL of the supernatant into a new 2ml cent tube. Discard the pellet
- Add 1/2 of an InhibitX tablet to the sample and vortex immediately and continuously for 1 min or until tablet is dissolved, then incubate at room temp for 1 min
- Centrifuge at full speed for 3 min to pellet inihibitors bound to InhibitEX
- Pipette all supernatant into new 1.5ml tube and discard pellet
- Centrifuge at full speed for 3 min
- Pipette 15ul Proteinase-K into a NEW 1.5ml tube
- Pipette 200ul of supernatant from step 11 into Pro-K tube
- Add 200ul of Buffer AL and vortex for 15 sec (Don't add Pro-k directly to the buffer!!)
- Incubate at 70c for 10 min
- Breifly centrifuge then add 200ul of Molec. Grade EtOH and mix by pulse vortexing for 15 sec, then briefly centrifuge
- Transfer full volume ~610ul to a new spin column (including white precipitate if this had formed), and centrifuge for 1min at 8000rpm
- Place spin column in a new collection tube
- Add 500ul of buffer AW1 without wetting the rim of the spin column, then centrifuge at 8000 for 1 min
- Place the spin column into a new collection sube
- Add 500ul of buffer AW2 wihout wetting rim, then centrifuge at full speed for 3 min (if there is still buffer in the top of the spin column, spin again)
- Place spin column into Final 1.5ml tube, add 100ul Buffer AE and incubate for 5 min at room temp
- Spin at 8000 for 1 min
- Voila DNA - store at -20, or in fridge if using in the near future (to avoid too much freeze-thawing)
The extracted DNA is currently being stored in the lab5 fridge b/c we are going to be using it this week for methylation tests.
We also carried out the HPAII and MSPI restriction digestion reactions on the DNA we extracted. We used 2ul of extracted DNA and 48ul of master mix for both enzymes, plus water. Carrie forgot to take the water/control reactions out of the 37c heat block (the HPA and MSP reactions were taken out of the other heat block after the correct 2hr increment), and James came to the rescue and took then out ~10pm. Since these samples had been in the heat block for 6 hours, Carrie re-pipetted some DNA controls (48ul H2O and 2ul DNA; they are in the freezer and labeled as "Cx (-) 2", and are in the same tube rack as the rest of the restriction digestion products) on 8/12 to use in the methylation tests, in case the other set had been degraded.
Restriction Digestion procedure:
HPAII master mix (per 50ul reaction):
5ul Buffer #1
1ul HPAII enzyme
42ul H2O
MSPI master mix (per 50ul reaction):
5ul Buffer #4
0.5ul MSP enzyme
42.5ul h20
Control = 48ul water and 2ul DNA
We added 48ul of master mix and 2ul of DNA to each cent tube, and incubated at 37 for 2 hours. (controls were also supposed to be incubated for 2 hours)
Once we have primers (G prot to replace the contaminated stock primer - do we know for sure that this is contaminated?), we'll run PCR for the restriction digestion products and see whether those intriguing banding patterns hold for our own snail samples!
Methylation investigation for class snail samples:
Since there were still bands on the negative controls on the band yesterday, we ran a new PCR and gel, but this time with actin primers in order to determine the bands were a result of contaminated stock primers. The master mix was exactly the same as on August 10, 2010 as there were again four negative controls. However, after the PCR I added 6.25 ul of loading dye each well. 6.25 ul was selected so that the 5x loading dye solution would be present at 1X. The calculations that I ran went thus:
concentration1*volume1=concentration2*volume2
5X*y=1X*(25ul+y)
where y=the volume of dye to be added
5X*y=25ulX+1X*y
y=6ul
The gel was ran as it was on August 10, 2010. The results were as follows:

James and Carrie dissected 8 more L. sitkana from Reuben Tarte to get some background info on parasite prevalence (in case we have time/materials to run an across-site comparison) - found 1 infection in Rt24, and took pics (posted on lab wiki by Steven). (see Littorine dissections spreadsheet).
August 10, 2010
Carrie ran a qPCR on 3 RNA samples that were extracted yesterday. 2 of the 3 showed amplification at CT 39 and 40, so she and Tiffany decided to do some rigorous DNAsing all of our samples. Reverse transcription was put on hold until more Reverse Transcriptase is received.
Each sample had 100ul, so to each tube we added:
10ul Turbo DNAse buffer
1ul DNAse
After we set up the 2nd 37C incubation, we noticed the heater had gone down to 33C. We put it back up to 37C for the next 30 minute incubation.
Of the samples, C47 got left behind, so it was processed one step behind the rest.
Due to the bands in the negative controls yesterday, James ran another gel with four negative controls with new PCR water and using a different dye. The components of the master mix were as follows:
Mastermix for PCR (per reaction):
2x Immomix 12.5 ul
F-primers 0.8 ul
R-primers 0.8 ul
PCR H20 10.9 ul
Since I wanted four reactions, the master mix was composed of the following amount of reagents:
2x Immomix 50 ul
F-primers 3.2 ul
R-primers 3.2 ul
PCR H20 43.6 u
Otherwise, the protocol was the same as on Aug. 9, 2010. The results were as follows:
[[image:file/view/081010_4NegControl2.jpg width="800" height="600"]]
August 9, 2010
In order to determine why there were bands in the negative controls on the August 7, 2010 gel, James ran another gel with four negative controls. PCR protocols were followed just as they were on August 6, 2010. The master mix was composed of the following reagents:
62.5 ul of Taq (=PCR master mix)
5 ul f-primer
5 ul r-primer
7.5 ul BSA
25 ul H20
While James was loading the wells to be placed in the thermocycler, he may have mistook the PCR master mix for his master mix. This sample was was loaded into the gel well that appears second from the top. Here are the results:
D'oh!
Meanwhile, Carrie and Tiffany extracted RNA from 5 infected and 5 uninfected individuals for Cattle Point. The following individuals were processed:
Infected:
CPL 16
CPL 36
CPL 38
CPL 40
CPL 47
Uninfected:
CPL6
CPL7
CPL8
CPL9
CPL11
Because all the infections occurred in females, only uninfected females were chosen as the controls. The protocol from the class website was followed (http://bio533.wikispaces.com/Lab_Littorina#Day3).
Starting tissue weights for each were as follows:
CPL 16 - 60mg
CPL 36 - 40mg
CPL 38 - 50mg
CPL 40 - 50mg
CPL 47 - 50mg
CPL6 - 50mg
CPL7 - 60mg
CPL8 - 50mg
CPL9 - 60mg
CPL11 - 50mg
When we got to step #17 where we had taken the supernatant from the tube and added isopropanol, we put the samples in the freezer (-20C) per Steven's suggestion for 2.75 hrs because we had to go to the larvae-disease discussion and dinner. When we were done, the samples were stored in the -80C.
8/7/2010
Plot of normalized Ct values for genes we've had any amplification for at an annealing temp of 60c. Plan is to try the other three (ferritin, cjunk, and lectin) at a lower annealing temp to see whether they might amplify w/ a little less primer specificity, but we're most likely be focusing on G prot (including trying to figure out whether it should be doing cool stuff involved w/ defense against trematodes!)
(having trouble inserting graph, will try after lunch!)
Due to the unusual band patterns in the ETS03 samples, James ran another PCR using the restriction digested ETS03 DNA. The procedures were the same as on August 6, 2010. The master mix was composed of the following reagants:
75 ul Taq
9 ul BSA
6 ul f-primer
6 ul r-primer
42 ul PCR H2O
Then he ran a gel using the PCR products. Here is an image of the result:

August 6, 2010
We looked at the qPCR results for the C-Jun K gene, and nothing amplified - hopefully, this means the gene just isn't being expressed at a high enough level to detect. Other possibilities are that the primers aren't quite right or we're not working w/ enough cDNA - these explanations are less likely b/c a)we got something (probably lots of genomic DNA) to amplify when we used this set of primers as a class, and b) we've gotten amplification for other primers from this batch for cDNA, which means there should be enough in there to amplify if the gene is being expressed at an appreciable level.
Tiffany and Carrie processed 8 more large snails from Cattle Point, in hopes of finding some infected ones. We were disappointed :( So far we have only found 1 snail in 31 dissected. James has estimated prevalence from our findings and the class data, and it looks like the high end of prevalence is probably ~12% for a 95%CI.
Drew generously offered to take Teamatode out to Jaekal (not sure if that's the right spelling) cove, which has been a high trematode prevalence site in the past. We collected ~65 snails haphazardly by hand from boulders and cobble in the high intertidal (we picked up snails from along a 200m stretch of shore) and brought them back to the lab on ice. We considered keeping them on ice and cracking them upon our return to the lab (which may be a better way to prevent picking up any artificial stress-induced gene expression changes in our data) but since we hadn't followed this procedure for the other sites, we instead decided to let them acclimate for ~24 hours before dissecting, which is closer to the way the other snails were treated. We will "process" these snails tomorrow, and will save tissue from 8 uninfected and (hopefully!) 8 infected individuals. The snails are currently chill'axing in the sea table (with a shorter pipe than we originally put in.........).
Today, Carrie set up qPCR for the final 2 genes we have primers for:
Ferritin heavy chain mRNA (Acc.# AY090096.1)
Metallothionein mRNA (Acc# AY034179.1)
We'll check out these results in the morning!
James ran a gel of the restriction digested DNA that he generated last night. This is an annotated image of the gel:

Click edit to see the second half of the gel.
Tiffany and Carrie processed 27 snails from Cattle Point for a total of 50 snails processed. We found 4 more infecteds, making the total 5/50 infected animals. All data is available at:
https://spreadsheets.google.com/ccc?key=0AtVQaQKSGjZxdG1EYlZoNktvMTh2clRFbkJ0dlg0b3c&hl=en&authkey=CJ_9pd0C
August 5, 2010
We checked out our qPCR for GProtein that we ran yesterday, and here are our results!!
https://spreadsheets.google.com/ccc?key=0AgycLcif4DQkdHh4OGg0ZTBELUtXU3pBMnlDMV9QT3c&hl=en&authkey=CLnBs64C
We also ran another PCR for Lectin. These results were difficult to interpret. There was amplification in some but not all samples, in 2 infected and 2 uninfected animals. It could be that it didn't work, or it could be that it did work and this gene is just not specific enough or is expression is just based on an individual. We decided to move on to another gene and come back to this later.
Carrie ran a qPCR for CJunK.
James and Tiffany did a restriction enzyme treatment to gDNA because the methylation samples were accidentally tossed out during the mass clean up of the Lab 5. We used the same restriction enzymes that were used on July 30, 2010.
MSPI
HPAII
Here is the gDNA we ran these restriction enzymes on:
*ETS03 = U. sit 6
*MM4 = I. sit 1
TY2 = I. sit 3
KJS8 = I. sit 2
AMM3
We also had a control for each sample, running 2ul gDNA with 48ul H2O.
*These samples were taken from the cDNA rack. There were 2 tubes labeled ETS03 and MM4, and we took the ones that were NOT labeled "cDNA". We think the gDNA tubes got misplaced in the cDNA rack.
As an addition complication, while preparing DNA for restriction digestion DNA was added to the a restriction enzyme tube of MM4 (James believes) twice.
In the evening, James ran a PCR on the restriction digested DNA using G protein-mediated receptor primers. The protocol followed was the same as on July 30, 2010. For verification purposes, the following master mix was used:
Number of Reactions: 15 treatments+2 controls+1 extra=18 reactions
Master Mix:
225 ul Taq
27 ul BSA
18 ul forward-primer
18 ul reverse-primer
126 ul H20
The following complications occurred in the process:
-The preparation of the master mix took longer than expected as the proper amount of reagents was not easy to obtain. Although the exact amount of time was not recorded, James began preparing the master mix at around 8:00 PM and placed the wells into the thermocycler at around 10:00 PM.
-Finding the proper amount of reagents was complicated. A small amount of Taq was donated to Tremateam from the anemone group. Water used in the negative controls was water that was left in the stock box in James's handwriting. He believes it contains PCR water that was left over from the first methylation assay. No other PCR water was available. The master mix contained PCR water that was labeled as PCR water but was generated by consolidating two separate containers. The Taq used was a consolidation of two separate, new aliquots of Taq plus the left overs from the anemone group.
The wells placed in the thermocycler were arranged thusly:
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
| Strip 1 |
TY2-Control |
TY2-MSP |
TY2-HPA |
KJS8-Control |
KJS8-MSP |
KJS8-HPA |
AMM3-Control |
AMM3-MSP |
| Strip 2 (A) |
AMM3-HPA |
MM4-Control |
MM4-MSP |
MM4-HPA |
ETS03-Control |
ETS03-MSP |
||
| Strip 2 (B) |
ETS-HPA |
|||||||
| Strip 3 |
- Control |
- Control |
Note: We should keep all DNA and cDNA in the refrigerator instead of freezing and thawing them all the time.
Aug 4, 2010
We went over our gel today with Steven, and we learned how to figure out the size of our PCR products. Using NCBI (http://www.ncbi.nlm.nih.gov/), you do a BLAST, then click on "nucleotide blast", then you enter the forward primer, followed by NNNNNNNNNNNN followed by the reverse primer.
Here is an example for what was typed in for GProt:
CTGTCTGCGCTATGGCCGGGNNNNNNNNNNNNNNNNNNNNNNNNNNCTCCTTTGACCGCCGCAGCA
You want to make sure the database is "others" and "nucleotide collection (nr/nt)" and you need to set the word count to some number less than the amount of bases in the forward primer (you want to be sure it is searching the database for a sequence that does not include any of the N's).
We BLASTed it, and then it came up with a single match:
Littorina littorea G protein-coupled receptor mRNA, complete cds, accession number EU557355.1
woo hoo!
The BLAST provides you with information that looks like the following:
Score = 38.1 bits (20), Expect = 4.2
Identities = 20/20 (100%), Gaps = 0/20 (0%)
Strand=Plus/Plus
Query 1 CTGTCTGCGCTATGGCCGGG 20
||||||||||||||||||||
Sbjct 946 CTGTCTGCGCTATGGCCGGG 965
Score = 38.1 bits (20), Expect = 4.2
Identities = 20/20 (100%), Gaps = 0/20 (0%)
Strand=Plus/Minus
Query 47 CTCCTTTGACCGCCGCAGCA 66
||||||||||||||||||||
Sbjct 1129 CTCCTTTGACCGCCGCAGCA 1110
From this, we take 1110-946 to get 164, which means our product should be 164bp. We used GProt in our methylation PCR, and looking at our gel, it looks like we have that size product for the TY2 HPA, MM5 HPA and MM5 control, so this is promising! :)We did the same thing for the LI ferritin PCR that was testing our RNA clean up, and the product in the DNA sample should be 171 bp. There is a faint band around that size in the positive control, although there is also other bands along the way, which is a little odd and suggests maybe some sort of contamination. However, these results also seem promising, as the other RNA samples are blank, so we are going to go ahead with the reverse transcription to get cDNA and run some qPCR's!
We will start with the GProt primers.
Reverse Transcription for cDNA!!!
We chose 4 infected and 4 uninfected Littorines.
I. sit 1
I. sit 3
I. scu 2
I. scu 3
U. sit 5
U. sit 7
U. scu 4
U. scu 6
2 negative controls
Master Mix:
5 ul 5X MMLV Buffer
1.25 ul 2.5uM dNTPs
0.5 ul MMLV Reverse Transcriptase
0.5 ul Oligo dTs
7.25 total Master Mix per reaction
+ 5 ul RNA extract
+ 12.5 ul ddH2O
25 ul product (cDNA)
We autoclaved 1.5 microcentrifuge tubes and PCR tubes for the class. :)
We followed the Promega protocol:
1. Incubate RNA+H2O sample in 70C for 5 min
2. Add Master Mix to sample as described above
3. Mix gently by flicking tube and incubate for 1 hour at 42C
4. qPCR!!!! Remember to run for gene of interest AND reference (Actin)!
qPCR Master Mix
- 2X SYBR 12.5ul
- BSA 1.5ul
- Forward primer .5ul
- Revers primer .5ul
- Sterile H2O 8ul
- Total mix to put in tube: 23ul
- 2ul of cDNA or Sterile H2O added to each
LI - Gprot_rec_946_965_F; Tm=63.3C, GC content=70%
LI_GProt_rec_1129_1110_R; Tm=63.1C, GC content = 65%
Using the qPCR machine:
- Turn it on
- click on MxPro in middle of screen on laptop
- click "cancel" for first pop up
- check box for SYBR Green with Dissociation Curve
- make sure it says "Lamp Ready" in bottom right corner
- set up thermal profile (40 cycles):
- 7 min 95C
- 10 sec 95C
- 30 sec 60C
- 30 sec 72C
- leave segment 3 of the profile alone - this is what gives us the dissociation curves (melting points of our products)
- 1 min 95C
- 30 sec 55C
- 30 sec 95C
- select where all samples are
- set "well type" to "unknown"
- check box for FAM - this is the filter reader for SYBR dye
- select wells with negative controls
- set "well type" to "NTC"
- click "Run" at top right
- click "Start Run" at bottom right
- Save to Teamatode folder on desktop, named: dategene (eg. 8410GPRO is the GProtein qPCR we ran on Aug 4, 2010)
- wait until the reactions start before leaving the machine
8/3 Running gels of DNased RNA extract and restriction-digestion PCR product
We poured gels for re-running our restriction-digestion PCR products (with appropriate primer concentrations), and for detecting large amounts of genomic DNA in our DNased RNA extracts - we poured 2 more gels after Carrie put the first ones in the freezer instead of the fridge at lunchtime (sorry again teammates!).
Picture from res-dig PCR looking for methylation differencesn and for DNased class-extracted RNA:
[[image:file/view/8.3._gel.jpg width="800" height="500"]]
Interpretation:
(need to run qPCR on RNA extracts regardless b/c traditional PCR will only have 1/100th the detection level for genomic DNA that qPCR will have)
8/2 Game plan (from Steven and Lisa)
- Focus on within-site infected vs uninfected first to see if the genes we have primers for are even involved in immune response to trematodes in the first place - do trematodes elicit an upregulation of immune genes?
- take the RNA extracts 6 infected and 6 uninfected from class exercise - both species
- DNA-ase everything
- PCR (primer pair doesn't matter) to check for genomic DNA carryover - shouldn't see bands if all RNA <-- update! Need to use qPCR to determine whether there's genomic carry-over b/c regular PCR isn't sensitive enough!
- Reverse transcription on cleaned (DNAsed) products
- qPCR to look for differences between infected and uninfected
- <-- update from class presentation: to REALLY get an idea of differences in expression, should go right to Differential Display (this way we are not depending on just the arbitrary immune gene primers that are available to us currently)
Extracted genomic DNA from the class RNA samples that were generated in the first week. To do so, we used the protocol found at http://aquacul4.fish.washington.edu/Protocols:Information%20Sheets/Commercial%20Protocols:Manuals/Ambion%20-%20Turbo%20DNA%20Free.pdf. We used the rigorous methods.
General "Rigorous" procedure:
1) add .1 volumes 10xTURBO DNase buffer (for our assumed 50ul volumes, this meant 5ul of buffer) to RNA
2) add 1[[image:file/view/i%2Fc.gif width="10" height="21"]]/2 of total TURBO DNase enzyme to be used (use 2-3ul total for rigorous procedure --> we added 1.5ul at this step)
3) mix gently and incubate at 37c for 30 min
4) add the other half of the DNase enzyme (we added 1ul at this step)
5) incubate at 37c for another 30 min
6) add 0.2 volumes (we added 12ul even though this was a little much) of DNase inactivation agent
7) incubate at room temp for 2 min, mixing 2-3 times
8) centrifuge at 10,000 for 1.5 min
9) transfer supernatant to a fresh tube and store at -80
Notes:
We ran a traditional PCR to see if there is any genomic DNA left in our RNA samples. We also renamed our samples.
RNA clean up with Ambion Turbo DNAse kit.
RNA clean up with Ambion Turbo DNAse kit
| Infected |
new ID |
comments |
Uninfected |
new ID |
comments |
| ETS 08 MM 5 CK SCUT 1 SH 2 MM 4 KJS 8 TY 2 |
I. scut 1 I scut 2 I. scut 3 I. scut 4 I. sit 1 I. sit 2 I. sit 3 |
no pellet when RNA was extracted very little RNA solution to start with |
KMM 4 TY 6 MCE 3 AMM 1 ETS 03 MCE 2 |
U. scut 5 U. scut 6 U. scut 7 U. sit 5 U. sit 6 U. sit 7 |
|
Traditional PCR on cleaned up RNA
| A B C D E F G H |
10 I. scut 1 I. scut 2 I. scut 3 I. scut 4 U. scut 5 U. scut 6 U. scut 7 neg control |
11 I. sit 1 I. sit 2 I. sit 3 pos control (ETS 03 DNA) U. sit 5 U. sit 6 U. sit 7 neg control |
12 X X X X X X X pos control (MM4 DNA) |
||
8/1
We ran another gel of the 1st PCR product from the restriction digestion - same procedure, same results (b/c still using the PCR product that had run with waay too much primer) - Steven looked at the picture from this on 8/2 and recognized the source of the problem, so we set up a new PCR (same master mix, except with the appropriate concentration of primers!) of our restriction digestion products on 8/2.
We dissected 23 large L. sitkana from Cattle Point, and found only 1 infection. Spreadsheet here (can't open the spreadsheet for some reason, will insert this link later!).
Saved snail (CpL 16) for DNA/RNA and also trematode for DNA (in Ethanol).
July 31, 2010
Loaded the PCR of restriction digestion product into an agarose gel to assess whether both, either, or none of the restriction enzymes cut the DNA at a restriction site. To generate the agorose gel, we followed the following protocol for a small gel rig:
1) Combine 50ml 1x TBE (we used TAE this time) buffer with .75g of agarose (our class supply was unavailable, so we stole a little bit of someone's fancy agarose...) and microwave til boiling. (if needed, swirl contents and microwave until it boils again) - careful not to let it boil over cause this makes a mess!
2) add 6ul Ethidium bromide and swirl to mix
3) let cool down a bit, or just pour into gel rig (make sure combs are in!) as-is, and let-cool until hardened (~15 min)
We ran this gel back in lab 5 w/ 110v for 15 min (didn't want it to run off the gel while we were at dinner), then for another 5 min at 115v.
Unfortunately I (Carrie) had gone back to the stock primers (10x the working concentration) instead of the working solutions Lisa had so kindly pipetted out for us a few days earlier - so, we ended up with huge bright glowing bands of primer-dimer and the rest of the results were difficult to make out - sorry Teamatode!
We also collected L. sitkana from Cattle Point on this day.
July 30, 2010**
Methylation Pilot Study
Began a pilot study to determine whether Littorina sitkana infected with trematodes methylate certain genes that are not methylated in uninfected individuals
(or vice-versa, right?)
. We decided to begin our search using primers for a G protein-coupled receptor mRNA from Littorina littorea. These primers are the following:
Forward:
Reference Numer: Ll_Gprot_rec_946-965_F
Sequence: 5'-CTG TCT GCG CTA TGG CCG GG-3'
Reverse:
Reference Number: Ll_Gprot_rec_1129-1110_R
Sequence: 5'-CTC CTT TGA CCG CCG CAG CA-3'
We chose this sequence for two reasons. First, it has many CCGG sequences within it, which is the sequence where methylation occurs. Second, Gorbuschin et al. (2009) found that Littorina littorea that were infected with Himasthla elongata had less mRNA for this gene than their uninfected counterparts.
The third reason was that we have primers for it already in the lab! :)
The following protocol was followed:
1)
Restriction Digestion using the enzymes MSP1 and HPAII:
Created two master mixes in 1.5 mL centrifuge tubes, one for MSPI and another for HPAII. The master mixes were created in the following manner:
From Amanda's notebook on Steven's genefish wiki (http://genefish.wikispaces.com/Amanda%27s+Notebook):
Undigested= sample+water
HpaII= sample + water+ 5µL buffer 1+ 1.0 µL enzyme
MSPI= sample+water+ 5µL buffer 4+ 0.5 µL enzyme
50ul reactions
Per reaction MSPI:
5ul buffer #4
0.5ul MSPI enzyme
2ul DNA (this is ballpark, as we have no idea what our starting concentrations are b/c we didn't have access to a spec when we did DNA extractions)
42.5ul water
Per reaction HPAII:
5ul buffer #1
1ul HPAII enzyme
2ul DNA
42ul H2O
Control: 2ul DNA + 48ul H2O
Procedure: keep enzymes on ice! incubate Rx at 37c for ~2hr (per protocol in Amanda's notebook)
Next step: PCR of DNA product to see whether enzymes digested it or not/whether there is a difference in methylation between the 2 groups!
Regular PCR Master Mix
25 ul reactions
12.5 ul of Taq Master Mix
1.5 BSA
1 ul f-primer
1 ul-r primer
7 ul of H20
+ 2ul of DNA product
We will have twenty reactions (3x the 6 samples we are testing, plus 2 negative controls). Adding enough for two more reactions, the final master mix will contain:
275 ul Taq Master Mix
33 ul BSA
22 f-primer
22 r-primer
154 of H20
Each tube received 23 ul of its respective PCR master mix and 2 ul of DNA extract. Tubes were briefly vortexed to ensure that the products were well mixed and centrifuged to ensure that all products were at the bottom of the tube.
Started PCR right after adding DNA, with program "OLY60" in the lab thermal cycler. Below is a map of the plate; wells that did not contain any reagants are left blank:
| A |
B |
C |
D |
E |
F |
G |
H |
|
| 1 |
||||||||
| 2 |
||||||||
| 3 |
||||||||
| 4 |
MM4 Control |
MM4 MSPI |
MM4 HPAII |
|||||
| 5 |
ETS03 Control |
|||||||
| 6 |
AMM3 Control |
Negative Control |
||||||
| 7 |
MM5 Control |
|||||||
| 8 |
KJS8 Control |
|||||||
| 9 |
TY2 Control |
Negative Control |
||||||
| 10 |
||||||||
| 11 |
||||||||
| 12 |
Teamatode! Checkin' out differences in (possibly) constitutive/un-challenged immune gene expression in Littorina sitkana from high and low parasite pressure sites
Rationale: I'll insert some papers about this stuff here later but here are the premises (including some big assumptions that would need to be tested in this system for us to make a compelling case):
1. Immune activity can be costly (in invertebrates as well as vertebrates)
2. In the presence of a parasite w. high fitness impacts, up-regulation of protective immunity (via constitutively expressed immune genes) should be favored even if this up-regulation comes with some fitness cost/trade-off. In the absence of the parasite, lower levels of expression might be better.
3. Immune gene expression depends on a) response to challenge, and/or b) heritable controls of gene expression (maternal effects).
Part "b" is what we're going to try to test in this project - methylation would be a possible mechanism for this kind of inheritance.