Lab 9: qPCR for hsp70 gene expression
Summary
Because I didn't get positive results for hsp70 protein expression with the three western blots, I did a qPCR using another group's primers and the cDNA samples that Halley isolated from our C. gigas gill tissue samples.
Materials/Methods
QPCR
1. Acquire cDNA template (I used Halley's samples)
2. Prepare master mix: (multiply each of the following amounts by 35, for 2 cDNA templates (for each sample x15) plus, 2 neg. controls and 3 for pipetting error)
- 12.5ul of 2X sensimix
- 1.25 ul forward and reverse primer
- 8ul ultra pure water
4. Add 2ul of cDNA sample to each positive reaction
5. Add 2ul of water to each negative control
6. Cap wells securely
7. Spin down so liquid pools at bottom
8. Load the plate and verify the PCR conditions, then start the run (done by Emma)
PCR conditions:
1. 95 C for 10 minutes
2. 95 C for 15 seconds
3. 55C for 15 seconds
4. 72C for 30 s (+plate read)
5. Return to step 2, 39 more times
6. 95 C for 10 Seconds
7. Melt curve from 65 C to 95 C at 0.5C for 5 seconds (+plate read)
Results
I haven't had time to look over the results of the qPCR yet
Conclusions
Hopefully if this worked, I can show that there was a general stress response to hypoxia, possibly slightly higher in the control and hyperoxic acclimated oysters.
Reflections (for discussion section of final paper)
1. Detail at least 2 reasons why your results turned out the way they did. This should be easy to do if your results are "unexpected", but even expected results can have multiple explanations. Really think about this, the answer "because I messed up in lab" (or any variation thereof) is not acceptable.
The western blot may not have turned out because of problems with the antibody solution, or because the protein concentration of my samples was night high enough. There also may not have been any heat shock proteins present (depends on qPCR results). Finally, on the last western blot that we did, I didn't add any protease inhibitor cocktail to the CellLytic solution so that could have messed up the proteins.
2. What are two obstacles that you encountered during your lab work and experimental design? Did these obstacles affect your results? Why?
One thing we encountered was lack of a really accurate DO meter (it tended to take a long time to stabilize when measuring, and any movement of the probe seemed to change the reading). Also, we ran out of oxygen during the acclimation phase, so we had to just add a bubbler to the high oxygen tank which didn't really push the DO as high as we had hoped. Finally, the nitrogen tank was also sensitive, and at one point turned off until we got to it the next day. The lack of control over the treatment variables might have affected our results in the sense that the oysters did show a difference in HIF-1a expression but it wasn't significantly different.
3. Explain at least one aspect of your research and its results that have a greater impact outside of your own personal learning experience. What would you tell a non-scientist who challenged the importance of your research?
It is important to understand how oysters and bivalves are affected by hypoxia and other stress especially for the commercial shellfish industry in our state which is worth $100 million dollars a year.
4. What part of your research and analysis has completely stumped you? Is there anything you can do to find the answer or will it always remain a mystery?
The Western Blot, especially the 3rd round where we got binding of proteins at the wrong length. I guess we could keep running them changing one thing at a time until it turns out right (as long as I confirm that there is positive gene expression for hsp70 with the qPCR)
5. In about 3 sentences each, summarize 2 papers that you are going to cite in your own paper that give insight into the results that you found.
Andreas Anestis, Hans O. Pörtner, Basile Michaelidis: Anaerobic metabolic patterns related to stress responses in hypoxia exposed mussels Mytilus galloprovincialis
This paper summarizes the relationship between hypoxia and expression of heat shock proteins. The looked at mussels exposed to hypoxia and higher ambient temperatures (to simulate high tide conditions). It illustrates nicely how hypoxia could actually induce expression of heat shock proteins and also that the combination between hypoxia and temperature stress accelerates the metabolic stress responses.
Mikulski, C. M., Burnett, L. E., & Burnett, K. G. (2000). The effects of hypercapnic hypoxia on the survival of shrimp challenged with vibrio parahaemolyticus
This paper discusses the how exposure to hypoxia affects shrimp exposed to vibrio. It is a good paper to relate how initial stress like hypoxia might not kill the organism, but it might make it more susceptible to disease outbreaks. If we find that hsp70 gene expression is positive, it will be important to discuss how a non lethal stress can actually still be relevant.
11/22/11
Lab 8: Finishing final Western Blot
Summary
I didn't actually attend lab because I had to go to the dentist, but I did perform one final western blot to see if I could find any hsp70 expression in the oyster gill tissues from our hypoxia experiment.
Materials/Methods
Protein extraction (repeated in an attempt to get higher concentration from 3 samples)
For each sample (4H, 4L, 4C):
1. Obtained previously frozen C. gigas gill tissue ~ (0.05g)
2. Transferred tissue to 1.5mL snap cap tube labeled "11/21 AT Protein (sample #, treatment)"
3. Added 500 microliters of CellLytic MT solution to the tube containing tissue sample
4. Homogenized tissue and solution with a sterile pestle
5. All 3 samples were placed in a refrigerated microfuge and spun down for 10 minutes at max speed.
6. Transfered supernatant to new tube labeled 11/21 AT Protein Stock (sample#, treatment)
SDS/PAGE: (For 6 of my samples: High DO #2 and #4, Control #2 and #4, Low DO #2 and #4)
7. Added 20ul of protein stock to a 1.5mL SCREW CAP tube labeled AT Protein (sample #, treatment) 11/21
8. Added 20ul of 2X Reducing Sample Buffer to each 1.5mL screw cap tube containing protein stock
9. Returned the rest of the protein stock to the -20C freezer for storage
10. Samples in screw cap tubes were mixed and then boiled for 5 minutes
11. After boiling, we centrifuged the samples immediately
12. Once the gel was set up in the rig, we loaded our samples - mine went into wells 7 through 12
13. Ran the gel at 150V for 20 minutes
Western Blot Immunodetection
1. Once the gel was finished running, we cracked the cassette and trimmed the wells at the top of the gel
2. We also notched the upper right corner in order to maintain correct orientation
3. We soaked the filter paper, membrane and gel in transfer buffer for 15 minutes and then assembled the blotting sandwich in the following order:
1. Annode +
2. filter paper
3. membrane
4. gel facing down
5. filter paper
6. cathode -
4. Connected the power supply and transferred the blot for 30 minutes at 20 Volts
Washing
1. Washed the membrane 2 times for 5 minutes each with 20 mL of pure water
2. Placed membrane in plastic box with 10mL of Blocking Solution and incubated overnight on a rotary shaker (1 Rev per Sec)
3. Remove liquid and rinsed membrane with 20 mL of water for 5 minutes
4. Incubate membrane in 10mL of Primary Antibody Solution (overnight) then decant (Primary Antibody Solution = 10mL blocking solution + 8uL primary antibody)
5. Rinse with 20mL of Antibody Wash for 5 minutes - REPEAT 3 TIMES
6. Incubate in 10mL of Secondary Antibody for 30 minutes, decant
7. Wash the membrane for 5 minutes with 20mL of Antibody wash - REPEAT 3 TIMES
8. Rinse with 20 mL of Pure water for 2 minutes, then decant - REPEAT 2 TIMES
9. Incubate membrane in 5mL of Chromogenic Substrate until there si a purple band (1-60 min)
10. Dry the membrane in open air on clean filter paper
Results
This western blot was different from the other two, but not in a good way. There appeared to be some binding but it was at the wrong size (near the bottom of the ladder). We are not really sure what might have caused this. The only thing we may have done wrong was not add any protease inhibitor cocktail to the CellLytic solution.
Here is a picture of the stained sds gel
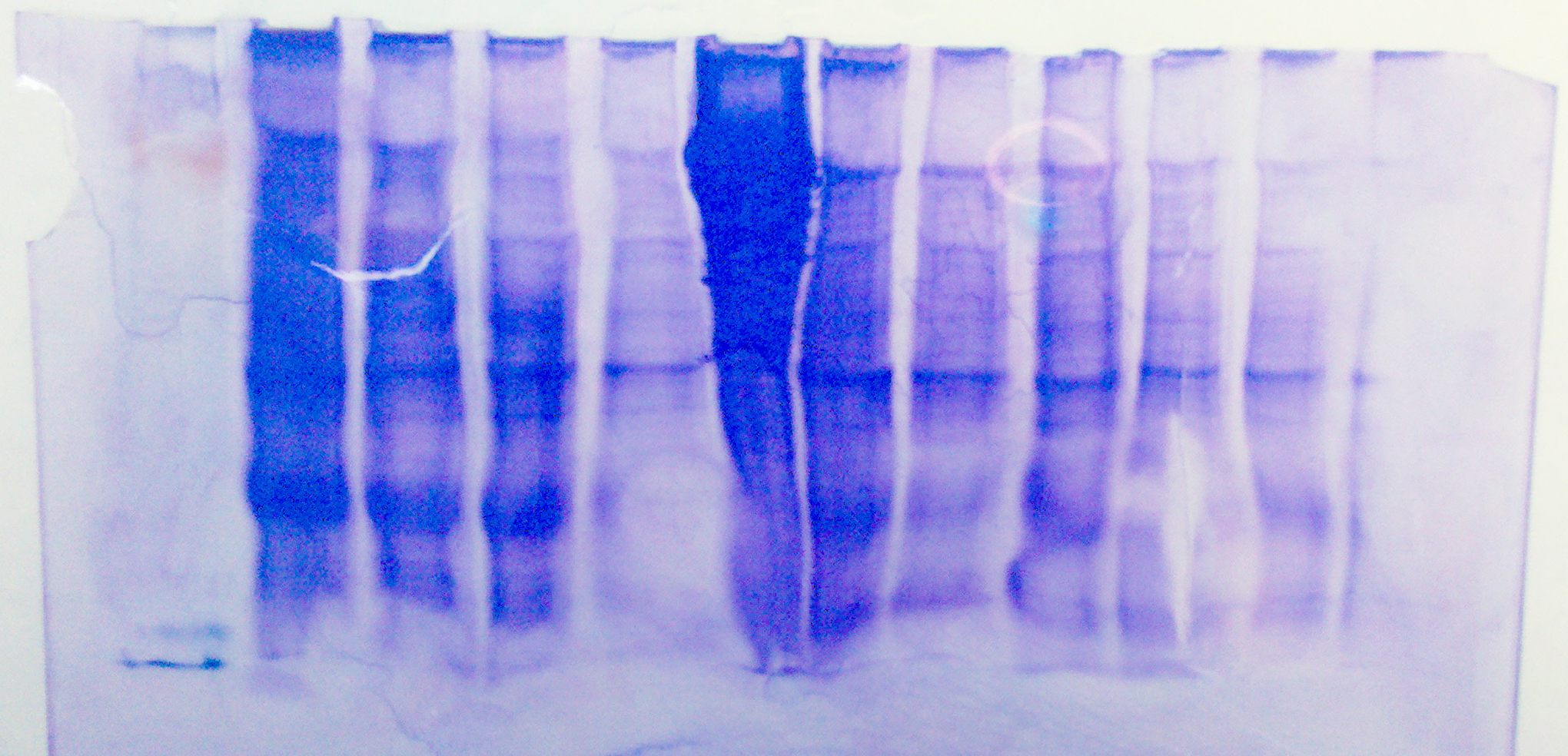
Here is a picture of the western blot: (ladder on left)

1 2 3 4 5 6 7 8 9 10 11 12
Conclusions:
There are definitely some proteins around the right length on our SDS gel, but again there was no binding even with higher protein concentration and higher antibody concentration. Not sure why there appears to be binding near the end of the gel.
Reflections
Could still be an antibody problem, but it is hard to say for certain, and it is really weird that there was binding at the wrong protein length. I will probably end up looking at hsp70 gene expression doing qPCR using the cDNA samples that Halley and Rizky extracted with primers specific for heat shock factor.
11/15/11
Lab 7: Q PCR, Epigenetics (Cytosine Methylation Dot Blot)
Summary
In this lab, I borrowed a DNA sample that Selena extracted from oyster tissue to do a Dot Blot. We also did Q PCR with cDNA samples extracted from oyster gill tissue (I used some of Halley's samples and primers)
Materials/Methods
Dot Blot
1. Made an initial dilution of DNA sample (168.8ng/ul) using C1V1=C2V2 formula to dilute to 50ng/uL with a total volume of 40uL. According to this calculation: V1=50*40/168.8 so V1 = 11.848
2. This means I used 11.848uL of DNA sample and 28uL of H2O
3. With the 40uL of initial dillution, we made the following dilutions:
- 0.8 ng/ul = 124ul H2O, 60ul of 20X SSC, 16ul of DNA (50ng/ul)
- 0.4 ng/ul= 132ul H2O, 60ul of 20X SSC, 8ul of DNA (50ng/ul)
- 0.2 ng/ul= 136ul H2O, 60ul of 20X SSC, 4ul of DNA (50ng/ul)
- 0.1 ng/ul= 138ul H2O, 60ul of 20X SSC, 2ul of DNA (50ng/ul)
- 0.05 ng/ul= 139ul H2O, 60ul of 20X SSC, 1ul of DNA (50ng/ul)
Preparing for the Dot Blot
1. Cut the nylon membrane to fit a 72 well manifold
2. For 10 minutes we soaked the membrane in 6X SSC buffer
3. Cut filter paper to size of membrane and wet with 6X SSC buffer
4. Put the membrane on top of filter paper in the manifold assembly
5. Denature DNA sample in SCREW CAP tubes for 10 minutes in boiling water
6. Transfer sample to ice
7. Apply 500ul of 6X SSC buffer to all the wells in the manifold and switch on vacuum to allow SSC to filter through
8. Spin your DNA sample down in centrifuge for 5 minutes
9. Put all volume of each DNA sample into separate wells without touching the membrane
10. Record where you put your samples
11. Let samples filter through the membrane, and if your sample is not filtering, pipette up and down
12. After filtration is complete, dismantle manifold and transfer membrane dot side up to filter paper (see step 13)
13. soak on denaturation buffer soaked filter paper for 10 minutes and then on neutralization buffer soaked filter paper for 5 minutes
14. Place membrane on dry filter paper to dry
15. Wrap dot blot in plastic and place DNA side down on the UV transluminator for 2 minutes at 120kj
Washing: (chromogenic immunodetection)
1. Prepare blocking solution and antibody solution
2. Place membrane in 10ml blocking solution for 30 minute incubation and place on rotary shaker
3. Decant blocking solution and rinse membrane with 20ml of water for 5 min, repeat
4. Incubate membrane with 10ml of primary antibody for 1 hour
5. Decant antibody solution and wash membrane with 20ml of 1x TBS-T for 5 minutes (repeat 3 times)
6. Incubate with secondary antibody solution for 30 minutes and decant
7. Wash membrane for 5 minutes with 20ml of TBS-T, repeat 3 times
8. Rinse with 20ml of water for 2 minutes, repeat twice
9. Incubate with 5 ml of Chromogenic Substrate until color develops, (1-60 minutes)
10. Rinse membrane with 20 ml of water for 2 minutes then decant, repeat 2 times
11. Dry membrane on filter paper
QPCR
1. Acquire cDNA template (I used Halley's samples)
2. Prepare master mix: (multiply each of the following amounts by 5, for 2 cDNA templates, 2 neg. controls and one for pipetting error)
- 12.5ul of 2X immomix
- 1ul of syto 13 dye
- 1.25 ul forward and reverse primer
- 7ul ultra pure water
4. Add 2ul of cDNA sample to each positive reaction
5. Add 2ul of water to each negative control
6. Cap wells securely
7. Spin down so liquid pools at bottom
8. Load the plate and verify the PCR conditions, then start the run (done by Emma)
PCR conditions:
1. 95 C for 10 minutes
2. 95 C for 15 seconds
3. 55C for 15 seconds
4. 72C for 30 s (+plate read)
5. Return to step 2, 39 more times
6. 95 C for 10 Seconds
7. Melt curve from 65 C to 95 C at 0.5C for 5 seconds (+plate read)
Results:
Results:
DNA methylation dot blot: positive results
 |
| Dot blot |
we will discussed q pcr results in next lab on 11/22/2011
Conclusions:
There does appear to be some methylation in the DNA extractions from oyster tissues based on the fact that there was binding of the antibody to methylated cytosines
Reflections:
The point of this lab was to test for DNA methylation in oyster , human and fly - we did find evidence of in the human and oyster, but not in the fly which is what we expected. I wouldn't really change anything about this lab, I thought it went pretty smoothly, besides everything taking a long time.
11/8/11
Lab 6: Protein extraction and expression from hypoxia experimental tissue samples
Summary
In this lab I extracted protein from 15 of our oyster gill tissue samples and looked for hsp70 expression using SDS/PAGE and Western blot procedures.
Materials/Methods
Protein extraction:
For each sample (1-15):
1. Obtained previously frozen C. gigas gill tissue ~ (0.03g)
2. Transferred tissue to 1.5mL snap cap tube labeled "11/8 AT Protein (sample #, treatment)"
3. Added 500 microliters of CellLytic MT solution to the tube containing tissue sample
4. Homogenized tissue and solution with a sterile pestle
5. All 15 samples were placed in a refrigerated microfuge and spun down for 10 minutes at max speed.
6. Transfered supernatant to new tube labeled 11/8 AT Protein Stock (sample#, treatment)
SDS/PAGE: (For 4 of my samples: High DO #1, Control #1, Low DO #1, Low DO #5)
7. Added 15ul of protein stock to a 1.5mL SCREW CAP tube labeled AT Protein (sample #, treatment) 11/8
8. Added 15ul of 2X Reducing Sample Buffer to each 1.5mL screw cap tube containing protein stock
9. Returned the rest of the protein stock to the -20C freezer for storage
10. Samples in screw cap tubes were mixed and then boiled for 5 minutes
11. After boiling, we centrifuged the samples immediately
12. Once the gel was set up in the rig, we loaded our samples - mine went into wells 3 through 6
13. Ran the gel at 150V for 20 minutes
Western Blot Immunodetection
1. Once the gel was finished running, we cracked the cassette and trimmed the wells at the top of the gel
2. We also notched the upper right corner in order to maintain correct orientation
3. We soaked the filter paper, membrane and gel in transfer buffer for 15 minutes and then assembled the blotting sandwich in the following order:
1. Annode +
2. filter paper
3. membrane
4. gel facing down
5. filter paper
6. cathode -
4. Connected the power supply and transferred the blot for 30 minutes at 20 Volts
Washing
1. Washed the membrane 2 times for 5 minutes each with 20 mL of pure water
2. Placed membrane in plastic box with 10mL of Blocking Solution and incubated overnight on a rotary shaker (1 Rev per Sec)
3. Remove liquid and rinsed membrane with 20 mL of water for 5 minutes
4. Incubate membrane in 10mL of Primary Antibody Solution (overnight) then decant (Primary Antibody Solution = 10mL blocking solution + 4uL primary antibody)
5. Rinse with 20mL of Antibody Wash for 5 minutes - REPEAT 3 TIMES
6. Incubate in 10mL of Secondary Antibody for 30 minutes, decant
7. Wash the membrane for 5 minutes with 20mL of Antibody wash - REPEAT 3 TIMES
8. Rinse with 20 mL of Pure water for 2 minutes, then decant - REPEAT 2 TIMES
9. Incubate membrane in 5mL of Chromogenic Substrate until there si a purple band (1-60 min)
10. Dry the membrane in open air on clean filter paper
Results
Here is a picture of the stained SDS/PAGE gel
1 2 3 4 5 6 7 8 9 10 11 12
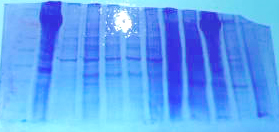
My samples in wells 3-6 showed some protein products. The sample in well 2 was our positive control (urchin gonad) that expressed hsp70 with the previous antibody solution that was used last week.
I developed the western blot from this gel and found the following results:
Positive control (urchin) in lane 2 showed hsp70 expression
Lanes 3-9 were blank
Lanes 10-11 (Igor's urchin gonad samples) showed hsp70 bands
Lane 12 was blank
Conclusions:
For my samples in wells 3-6 I concluded that there was either no expression, or no antibody binding. For the next samples that I run, we decided that we will run another SDS/PAGE gel with more protein stock in the wells (maybe 45ul instead of 30) and we will also add a higher concentration of primary antibody during the western blot procedure.
Reflections:
Although we didn't get hsp70 binding, we did get some protein products so it is worth running another western blot and adding a higher concentration of primary antibody to see if there is indeed hsp70. The other oyster samples from the OA experiment group didn't show binding either, so it seems more likely that the antibody just didn't bind to the oyster protein.
11/1/11
Lab 5: Agarose gel electrophoresis, protein SDS/PAGE and Western blot
Summary
In this lab we analyzed the cDNA templates we made in lab 4 using gel electrophoresis. We also extracted protein from the oyster tissue samples collected in our hypoxia experiment. Finally, we tested these protein extractions for the presence of hsp70 using SDS/PAGE and Western blot analysis.
Materials/Methods
Gel Electrophoresis with cDNA templates
1. Using our previously prepared cDNA samples that we created using PCR in lab 4, we loaded 25uL of each sample into wells 14-17 of an agarose gel as follows
14: Negative Control 1 HIF
15: Digestive Gland 1 HIF
16: Negative Control 2 HIF
17: Gill 2 HIF
2. We also loaded 7uL of a 100 bp ladder into the far left lane
3. We ran the gel at 100V for about 1hr
4. We looked at the results using the UV transilluminator
Protein Extraction and SDS/PAGE:
1. Obtained C. gigas gill tissue (0.023g)
2. Transferred tissue to 1.5mL snap cap tube labeled "11/1 AT Protein"
3. Added 500 microliters of CellLytic MT solution to the tube containing tissue sample
4. Homogenized tissue and solution with a sterile pestle
5. Sample was placed in a refrigerated microfuge and spun down for 10 minutes at max speed.
6. Transfered supernatant to new tube labeled 11/1 C.gigas AT Protein High DO
7. Added 15ul of the protein stock to a 1.5mL SCREW CAP tube labeled AT Protein High DO 11/1
8. Added 15ul of 2X Reducing Sample Buffer to the 1.5mL screw cap tube containing protein stock
9. Returned the rest of the protein stock to the -20C freezer for storage
10. Sample in screw cap tube was mixed and then boiled for 5 minutes
11. After boiling, we centrifuged the sample immediately
12. Once the gel was set up in the rig, we loaded our samples - mine went into well 6, labeled on the board "PT"
13. Ran the gel at 150V for only about 20 minutes and it was done!
Western Blot Immunodetection
1. Once the gel was finished running, we cracked the cassette and trimmed the wells at the top of the gel
2. We also notched the upper right corner in order to maintain correct orientation
3. We soaked the filter paper, membrane and gel in transfer buffer for 15 minutes and then assembled the blotting sandwich in the following order:
1. Annode +
2. filter paper
3. membrane
4. gel facing down
5. filter paper
6. cathode -
4. Connected the power supply and transferred the blot for 30 minutes at 20 Volts
Washing (Done by Emma)
1. Washed the membrane 2 times for 5 minutes each with 20 mL of pure water
2. Placed membrane in plastic box with 10mL of Blocking Solution and incubated overnight on a rotary shaker (1 Rev per Sec)
3. Remove liquid and rinsed membrane with 20 mL of water for 5 minutes
4. Incubate membrane in 10mL of Primary Antibody Sol, then decant
5. Rinse with 20mL of Antibody Wash for 5 minutes - REPEAT 3 TIMES
6. Incubate in 10mL of Secondary Antibody for 30 minutes, decant
7. Wash the membrane for 5 minutes with 20mL of Antibody wash - REPEAT 3 TIMES
8. Rinse with 20 mL of Pure water for 2 minutes, then decant - REPEAT 2 TIMES
9. Incubate membrane in 5mL of Chromogenic Substrate until there si a purple band (1-60 min)
10. Dry the membrane in open air on clean filter paper
Results
We had contamination in our first negative control (well 14) and our second negative control showed primer dimer. We did get product in our cDNA templates, but unfortunately there were multiple bands. On the western blot, it doesn't appear that we had any protein product bind to the antibody
Conclusions
Multiple bands on the agarose gel indicates either contamination, or multiple products - so its possible our primers weren't right. On the western blot, either we didn't have heat shock protein expressed in our samples, or the antibody we used wasn't correct.
Reflections:
I'm glad we had a practice run with the PCR and Western Blot procedures to show how difficult these processes can be so when it comes time to run the real samples from our hypoxia experiment we will be super careful and hopefully get better results!
10/25/2011
Lab 4: Tissue Dissection, Primer Reconstitution, and PCR
Summary
There were 2 parts to this lab:
1: Using the primers we ordered, we made a master mix and attempted to do a PCR on our cDNA samples from lab 3
2: We finished our oyster hypoxia experiment by sampling gill and mantle tissue from each oyster
Materials/Methods
PCR Reactions:
1. To make 50ul PCR reactions, we prepared a master mix large enough for 2 samples, 2 negative controls, and 1 extra to account for pipetting error
2. First we rehydrated our forward and reverse primers by adding enough TE buffer to make them 100uM stock solution:
- Forward primer HIF-1 - added 363ul buffer
- Reverse primer HIF-1 - added 280ul buffer
4. The master mix calculations were:
- 25ul GoTaq buffer *5 = 125ul GoTaq buffer
- 1ul of 10uM Forward primer *5 = 5ul Forward primer
- 1ul of 10uM Reverse primer *5 = 5ul Reverse primer
- 21ul Nuclease free H2O = 105ul Nuclease free H2O
- Total Master Mix volume = 235ul
6. We added 48ul of the master mix to each of the 4 PCR tubes labeled cDNA 1, cDNA 2, neg control 1, neg control 2
7. To each of the cDNA tubes we added 2ul of cDNA template. One was from our cDNA sample, and we got the other from a different lab group
8. To each of the negative control tubes we added 2ul of Nuclease free H2O.
9. All 4 of the PCR tubes were spun down.
10. We loaded the PCR tubes into the thermocycler to be put through the following steps:
- Denaturation (95C for 5 min)
- Denaturation (95C for 30 sec)
- Annealing (55C for 30 sec)* 40 cycles
- Extension (72C for 90 sec)
- Final extension (72C for 3 min)
- Hold at 4C
Oyster Dissection:
1. For each oyster we:
- Shucked the oyster
- Dissected gill tissue (3 pieces) and mantle tissue (3 pieces) and placed tissues in tubes labeled 1-5 with the correct treatment and tissue name
Results
By putting our cDNA through PCR we will hopefully be able to see our specific amplified DNA fragments that we designed primers for. Also now that we dissected our oyster tissues we can repeat the things we did in labs 1-4 on experimental samples to draw conclusions about how the oysters reacted to hypoxia.
Conclusions
We aren't sure if our primers will work properly because they were designed for DNA fragments that were too large for qPCR. Our oyster dissection went well except for a mixup with the tube labeling, but I think we have that all figured out.
Reflections
The purpose of this lab was to carry out the final step of analyzing gene expression by amplifying certain fragments (using primers) and making lots of copies (PCR). We also needed to finish our hypoxia experiment and take tissue samples for our group research project. I think everything in the procedure was pretty clear, I was just glad to be done with everything on time!
10/18/2011
Lab 3: Reverse transcription and primer design
Summary
In this lab we used reverse transcriptase to convert our extracted RNA (from lab 2) into cDNA. We also set up our group experiment for exposing oysters to hypoxia.
Materials/Methods
1. First we mixed up the stock sample of RNA
2. We combine the following components into a 0.5ml PCR tube labeled "HN/AT cDNA"
- 5 ul of RNA
- 1 ul of oligo dT
- 4 ul of nuclease free H2O
4. After incubation, we stored the sample on ice for 2 minutes
5. Then we mixed in the following components:
- 5ul of M-MLV 5X Reaction Buffer
- 5ul of dNTPs
- 1ul of M-MlV RT
- 4ul of nuclease free H2O
7. Inucbated in the thermocycler at 42C for 60 minutes and then heat inactivate at 70C for 3 minutes
8. Spun down the sample
9. Stored on ice in the -20C Freezer
Results
We didn't really get any numerical results for this lab. Hopefully we made some cDNA!
Conclusions:
Now we should be able to use this sample for PCR.
Reflections:
The purpose of this lab was to create cDNA from RNA using reverse transcription enzyme. Once you have cDNA you can amplify certain genes with specially designed primers and PCR. We spent most of our time in this lab setting up for our oyster hypoxia experiment so that we can sample our oysters next week and start the RNA extraction process that we learned in labs 1 and 2. Nothing was really unclear about the procedure, although since we were setting up our oyster experiment we didn't really learn much about how to design primers.
10/11/2011
Lab 2: RNA Extraction and Quantification
Summary
We continued the RNA extraction process with our sample of (C. gigas) digestive gland tissue. Once the RNA is completely separated from the DNA and other cellular components, we can quantify the amount of RNA present using the spectrophotometer.
Materials/Methods
1. Incubated RNA/Tri-Reagent sample from previous lab (see step 7 from 10/4/11) at room temperature for 5 minutes
2. Under the fume hood, added 200 microliters of chloroform with a pipette.
3. Vortexted sample at high speed until the solution appears milky
4. Incubated the sample another 5 minutes at room temperature
5. In a refrigerated microfuge set at max speed we spun down samples for 15 minutes
6. After removing tube from the microfuge I carefully separated the clear pipetted the aqueous phase away from the interphase and transferred the aqueous phase into a new microfuge tube labeled "AT/HN RNA 10/11"
7. Disposed of liquid waste (interphase and organic phase) from the old tube in the liquid waste jar under the hood. Disposed of solid waste (tube) into the garbage bag tray under the hood.
8. Added 500 microliters of isopropanol to the new tube that contains our RNA, closed the tube and mixed.
9. Incubated RNA/isopropanol solution at room temperature for 10 minutes
10. Spun down sample in a microfuge for 8 minutes until a small white pellet of RNA formed.
11. Pipetted off the supernatant until only the pellet remained in the tube
12. Added 1000 microliters of 75% EtOH (ethanol) and attempted to dislodge pellet from bottom of tube.
13. Spun in microfuge at 7500 rpm for 5 minutes
14. Pipetted off remaining EtOH until only the pellet remained.
15. Let air dry at room temp for up to 5 minutes
16. Added 100 microliters of 0.1%DEPC-H2O so the pellet would dissolve into solution
17. If pellet is not dissolved, incubate sample at 55 degrees for up to 5 minutes.
18. Place sample on ice until it can be quantified using the nanodrop spectrophotometer.
19. Blanked the spectrophotometer by adding 2 microliters of of 0.1%DEPC-H2O onto the nanodrop pedestal and lowering the arm, then pressing "blank" button on the computer screen
20. Wiped the blanking solution from the nanodrop pedestal and then added 2 microliters of our RNA solution to the pedestal.
21. Lowered the arm and clicked "measure" to determine 260/280 and 260/230 ratios as well as RNA concentration.
22. Placed clearly labeled "AT/HN RNA 10/11" RNA sample back on ice and gave sample to Emma for storage at -80 until next week.
Results
RNA Concentration: 193.6 ng/uL
260/280 ratio: 1.81
260/230 ratio: 1.02
Conclusions:
We did extract some RNA from our oyster digestive gland tissue sample although not very much. Our ratios indicate that our RNA solution was not pure because they were lower than described as normal in the nanodrop user guide. Normal pure RNA ratios for 260/280 would be ~2.0 and 260/230 would be 1.8 to 2.2. The fact that our ratios were lower indicates the presence of some contaminants.
Reflections:
The purpose of this lab was to complete our RNA extraction that we started last week and also to quantify the concentration and purity of our extracted RNA. Measuring the amount of RNA (and the purity of that RNA) could reveal details about gene expression in the tissue from which the RNA was extracted. Nothing was unclear in the procedure except that we didn't necessarily need to spin our samples in a refrigerated microfuge beyond the after the first round of spinning. I wish there was more information on what the shape of the curve means when the concentration was measured by the nanodrop.
10/4/2011
Lab 1: RNA Extraction and Protein Analysis (Part 1)
Summary
In this lab we practiced extracting RNA from the digestive gland tissue of a Pacific Oyster (C. gigas), by homogenizing the tissue with a substance called TriReagent. We also isolated protein from a sample of oyster gill tissue, and determined the protein concentration using the Bradford Assay.
Materials/Methods
RNA Extraction:
1. Obtained C. gigas digestive glad tissue (0.016g)
2. Transferred tissue to RNA free 1.5mL snap cap tube labeled "10/4 HN AT"
3. Added 500 microliters of TriReagent to 1.5mL tube containing tissue.
4. Homogenized tissue and reagent using a sterile pestle.
5. Added another 500 microliters of TriReagent
6. Vortexed sample for 15 seconds
7. Stored tissue at -80 C until next week.
Protein Extraction/Quantification:
1. Obtained C. gigas gill tissue (0.013g)
2. Transferred tissue to 1.5mL snap cap tube labeled "10/4 HN AT Protein"
3. Added 500 microliters of CellLytic MT solution to the tube containing tissue sample
4. Homogenized tissue and solution with a sterile pestle
5. Sample was placed in a refrigerated microfuge and spun down for 10 minutes at max speed.
6. Transfered supernatant to new tube labeled 10/4 HN AT Protein
7. Added 15 microliters of protein supernatant to new 2mL tube containing 15 microliters of DI water and labeled "Protein 10/4 HN AT"
8. Added 30 microliters of DI water to new 2mL tube and labeled "10/4 HN AT Blank"
9. Added 1.5 mL of Bradford Reagent to each of the 2mL tubes
10. Incubated samples at room temperature for 10 minutes
11. Added 1mL of well mixed protein sample to a cuvette with a P1000 micropipette
12. Added 1mL of well mixed blank sample to a different cuvette
13. Using a spectrophotometer set to read at 595nm, we placed our blank sample in the meter and hit "zero"
14. Then we recorded the protein sample absorbance value two times (the second recording was done after the sample was mixed once more with the pipette)
15. Our absorbance values were: 0.122 and 0.120
16. We back calculated to find the concentration using the equation: Concentration=(1013.9 * av. absorbance value) *2 [because of sample dilution]
Results
Protein Extraction/Quantification:
Average absorbance value = (0.122+0.120)/2 = 0.121
Protein Concentration = 245.36ug/mL
Conclusions
We will not know the results of the RNA extraction until we can separate the insoluble components from the RNA next week.
The protein extraction and quantification went as expected and our absorbance results fit the standard curve provided to us.
Reflections
The purpose of this lab was to practice protein and RNA extraction, in order to become familiar with these techniques so we can use them later in the quarter on our independent research project. The RNA extraction can be used to measure the amount of RNA present in the sample. For the protein extraction we used the Bradford Assay to measure the concentration of protein in our sample. The Bradford Assay is necessary because protein samples have different structures and absorb light a different wavelengths; the reagent in the Bradford Assay interacts with the proteins and will turn different colors of blue depending on the concentration of the protein present. These assays might be used when trying to determine the expression levels of certain genes or proteins in a specific tissue - maybe something that was exposed to an environmental disturbance such as change in temperature or pH. It might have been nice to have a picture or illustration of how TriReagent isolates RNA from DNA because there are a number of steps involved.