RESEARCH PROJECT (Week 3)
DATE: November 27, 2012LAB SUMMARY: Using the rehydrated primers qPCR was run on the cDNA samples and analyzed for gene expression.
MATERIALS AND METHODS:
Primer Preparation:
- Make a stock (100 µm) solution of your primer by adding the appropriate amount of nuclease free water. The appropriate µL of water to be added is calculated by determining the nm measurement of your primer and moving that number one decimal place to the right (i.e. if 28.5 nm then add 285 µL of water).
- Make a working stock (10 µm) solution of your primer by adding 10 µL of your stock solution made in step 2 to 90 µL of nuclease free water.
- Prepare a qPCR mastermix using the table below.
- Add 23 µL of the qPCR mastermix to each qPCR tube.
- After cDNA samples have thawed add 2 µL of each sample to their respective wells (first 10 wells in each row) and 2 µL of PCR water to the "blanks" (last 2 wells in in each row).
- Cap the tubes and turn in to the TA to be run.
| Component |
Volume |
Final Conc. |
24 reactions |
| Sensimix |
12.5 µL |
1x |
300 µL |
| SYBR 25X |
1 µL |
2 µM |
24 µL |
| upstream primer, 10 µM |
1.25 µL |
2.5 µM |
30 µL |
| downstream primer, 10 µM |
1.25 µL |
2.5 µM |
30 µL |
| 0.1% DEPC water |
7 µL |
N/A |
168 µL |
RESULTS:Pacific Oyster hsc70 Control
| SampleNames: |
Cghsc70_1 |
Cghsc70_2 |
Cghsc70_3 |
Cghsc70_4 |
Cghsc70_5 |
Cghsc70_6 |
Cghsc70_7 |
Cghsc70_8 |
Cghsc70_9 |
Cghsc70_10 |
| Logistic_a: |
4.61247 |
3.89765 |
4.91938 |
4.69223 |
4.63266 |
4.39469 |
4.70995 |
4.4783 |
4.7086 |
4.44449 |
| Logistic_b: |
-17.1205 |
-16.8088 |
-16.6026 |
-17.2806 |
-14.2711 |
-16.8308 |
-17.8597 |
-19.7184 |
-18.0116 |
-19.4613 |
| Logistic_X0: |
28.6055 |
28.4579 |
27.9985 |
29.7496 |
25.8098 |
28.2543 |
30.2341 |
34.2575 |
30.1222 |
32.5787 |
| Logistic_Y0: |
0.845731 |
0.898102 |
1.01456 |
1.08991 |
1.36533 |
0.688272 |
0.567055 |
0.666371 |
0.756843 |
0.586394 |
| Logistic_Pvalue: |
7.47E-62 |
0 |
0 |
2.24E-64 |
6.39E-73 |
2.22E-16 |
1.11E-16 |
5.25E-53 |
7.19E-65 |
1.11E-16 |
| Noise(SPE): |
0.00295133 |
0.00184796 |
0.00269212 |
0.00226599 |
0.00169992 |
0.00328426 |
0.00325099 |
0.00216253 |
0.00212885 |
0.00286486 |
| EndofExpPhase(SDM): |
1.68803 |
1.60783 |
1.90858 |
1.948 |
2.1853 |
1.48867 |
1.43266 |
1.50085 |
1.62327 |
1.41313 |
| CP(SPE): |
18.6171 |
18.0491 |
17.8108 |
19.1247 |
14.8276 |
18.4223 |
20.1151 |
23.259 |
19.6425 |
22.3356 |
| CP(SDM): |
26.2079 |
26.0249 |
25.5726 |
27.2809 |
23.1748 |
25.8421 |
27.8126 |
31.7901 |
27.7316 |
30.199 |
| DynamicThreshold: |
1.26836 |
1.25389 |
1.46292 |
1.52009 |
1.77616 |
1.09011 |
1.00148 |
1.08469 |
1.19112 |
1.0012 |
| LowerCycleNumber: |
10 |
11 |
10 |
11 |
11 |
10 |
11 |
12 |
12 |
11 |
| UpperCycleNumber: |
17 |
18 |
17 |
18 |
18 |
17 |
18 |
19 |
19 |
18 |
| PointsForRegression: |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
| Number_OfRegressionWindows: |
138 |
135 |
133 |
138 |
134 |
141 |
140 |
132 |
137 |
136 |
| WeightedAverage_OfPvalue: |
0.00346283 |
0.00425426 |
0.00428261 |
0.00368622 |
0.00469677 |
0.00353112 |
0.00406129 |
0.00403847 |
0.00414033 |
0.00364858 |
| Stderr_OfWeightedPvalue: |
0.0010126 |
0.00114388 |
0.0011581 |
0.00112304 |
0.00109556 |
0.00110195 |
0.00124302 |
0.000985687 |
0.00110298 |
0.000995932 |
| WeightedAverage_OfEfficiency: |
0.932377 |
0.982364 |
0.93022 |
0.918504 |
0.98896 |
0.855057 |
0.917963 |
0.897806 |
0.951493 |
0.935903 |
| Stderr_OfWeightedEfficiency: |
0.00802643 |
0.0105101 |
0.00955769 |
0.00794954 |
0.0131236 |
0.00713596 |
0.00834875 |
0.00867499 |
0.00871964 |
0.00850997 |
| CT: |
25.0184 |
24.8214 |
24.3767 |
26.0523 |
21.9222 |
24.651 |
26.6007 |
30.5281 |
26.5308 |
28.9865 |
| TotalSampleNumber: |
10 |
|||||||||
| ReplicateSampleNames: |
Cghsc70_1 |
Cghsc70_2 |
Cghsc70_3 |
Cghsc70_4 |
Cghsc70_5 |
Cghsc70_6 |
Cghsc70_7 |
Cghsc70_8 |
Cghsc70_9 |
Cghsc70_10 |
| AverageEfficiency_OfReplicateSamples |
0.932377 |
0.982364 |
0.93022 |
0.918504 |
0.98896 |
0.855057 |
0.917963 |
0.897806 |
0.951493 |
0.935903 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
25.0184 |
24.8214 |
24.3767 |
26.0523 |
21.9222 |
24.651 |
26.6007 |
30.5281 |
26.5308 |
28.9865 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
Cghsc70 |
|||||||||
| AverageEfficiency_OfGenes: |
0.931065 |
|||||||||
| Stdev_OfGenesEfficiency: |
0.0123106 |
|||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
|||||||||
| MeanCV%_OfGenesCTs |
0 |
|||||||||
| SampleNames: |
Cghsc70_1 |
Cghsc70_2 |
Cghsc70_3 |
Cghsc70_4 |
Cghsc70_5 |
Cghsc70_6 |
Cghsc70_7 |
Cghsc70_8 |
Cghsc70_9 |
Cghsc70_10 |
| R(0) |
7.07651E-08 |
8.05603E-08 |
1.07948E-07 |
3.58372E-08 |
5.42879E-07 |
9.01199E-08 |
2.49806E-08 |
1.88436E-09 |
2.61565E-08 |
5.19695E-09 |
| normalizing gene R(0) |
3.88723E-05 |
6.83103E-05 |
5.7771E-05 |
3.2135E-05 |
6.79746E-05 |
1.13463E-05 |
1.85774E-05 |
6.18925E-05 |
3.43697E-05 |
8.28294E-06 |
| Cghsc70_1 |
Cghsc70_2 |
Cghsc70_3 |
Cghsc70_4 |
Cghsc70_5 |
Cghsc70_6 |
Cghsc70_7 |
Cghsc70_8 |
Cghsc70_9 |
Cghsc70_10 |
|
| normalized gene expression |
0.001820448 |
0.001179329 |
0.001868553 |
0.001115207 |
0.007986508 |
0.007942678 |
0.001344675 |
3.04457E-05 |
0.000761034 |
0.000627428 |
Pacific Oyster hsc70 Treatment (35°C)
| SampleNames: |
Cghsc70_33 |
Cghsc70_36 |
Cghsc70_39 |
Cghsc70_40 |
| Logistic_a: |
5.04855 |
3.12305 |
3.59984 |
3.91084 |
| Logistic_b: |
-18.7826 |
-24.0072 |
-17.0717 |
-19.898 |
| Logistic_X0: |
36.2225 |
38.0356 |
35.5184 |
36.906 |
| Logistic_Y0: |
1.50637 |
0.97691 |
1.0002 |
0.76448 |
| Logistic_Pvalue: |
0 |
1.11E-16 |
7.49E-42 |
2.22E-16 |
| Noise(SPE): |
0.00234063 |
0.00191052 |
0.0033882 |
0.00156564 |
| EndofExpPhase(SDM): |
2.44093 |
1.57263 |
1.65729 |
1.49409 |
| CP(SPE): |
24.071 |
27.9479 |
23.616 |
24.909 |
| CP(SDM): |
33.4742 |
35.8135 |
32.5321 |
34.2735 |
| DynamicThreshold: |
1.97482 |
1.27572 |
1.33044 |
1.13007 |
| LowerCycleNumber: |
10 |
7 |
11 |
9 |
| UpperCycleNumber: |
17 |
14 |
18 |
16 |
| PointsForRegression: |
8 |
8 |
8 |
8 |
| Number_OfRegressionWindows: |
131 |
130 |
119 |
131 |
| WeightedAverage_OfPvalue: |
0.00417791 |
0.00535271 |
0.00545111 |
0.00441052 |
| Stderr_OfWeightedPvalue: |
0.00103978 |
0.000864743 |
0.00115545 |
0.00108649 |
| WeightedAverage_OfEfficiency: |
0.924461 |
1.0197 |
0.793878 |
0.927356 |
| Stderr_OfWeightedEfficiency: |
0.00953762 |
0.0113664 |
0.00989551 |
0.00973769 |
| CT: |
32.0818 |
34.6383 |
31.0549 |
32.9239 |
| TotalSampleNumber: |
4 |
|||
| ReplicateSampleNames: |
Cghsc70_33 |
Cghsc70_36 |
Cghsc70_39 |
Cghsc70_40 |
| AverageEfficiency_OfReplicateSamples |
0.924461 |
1.0197 |
0.793878 |
0.927356 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
32.0818 |
34.6383 |
31.0549 |
32.9239 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
| GeneNames |
Cghsc70 |
|||
| AverageEfficiency_OfGenes: |
0.916348 |
|||
| Stdev_OfGenesEfficiency: |
0.0464282 |
|||
| MeanCV%_OfGenesEfficiencies |
0 |
|||
| MeanCV%_OfGenesCTs |
0 |
|||
| SampleNames: |
Cghsc70_33 |
Cghsc70_36 |
Cghsc70_39 |
Cghsc70_40 |
| R(0) |
8.6639E-10 |
1.64274E-10 |
1.68961E-09 |
5.01005E-10 |
| normalizing gene R(0) |
0.000228251 |
0.000339918 |
0.000243804 |
0.000325683 |
| normalized gene expression |
3.79578E-06 |
4.83274E-07 |
6.9302E-06 |
1.53832E-06 |
| ttest |
0.128665488 |
|
| control |
treatment |
|
| mean |
0.002467631 |
3.1869E-06 |
| standard deviation |
0.002947699 |
2.85251E-06 |
Olympia Oyster hsc70 Control
| SampleNames: |
Ochsc70_51 |
Ochsc70_52 |
Ochsc70_53 |
Ochsc70_54 |
Ochsc70_55 |
Ochsc70_56 |
Ochsc70_57 |
Ochsc70_58 |
Ochsc70_59 |
Ochsc70_60 |
| Logistic_a: |
4.47018 |
5.47537 |
6.72475 |
6.01244 |
6.09034 |
5.02154 |
5.67038 |
6.04049 |
4.79645 |
5.05103 |
| Logistic_b: |
-14.2301 |
-14.4401 |
-16.3352 |
-14.386 |
-14.414 |
-17.339 |
-16.4839 |
-17.055 |
-16.2933 |
-15.9593 |
| Logistic_X0: |
23.2856 |
23.5669 |
26.0445 |
23.4119 |
23.4001 |
27.43 |
26.3303 |
27.223 |
26.025 |
25.6369 |
| Logistic_Y0: |
1.17494 |
0.826837 |
1.03254 |
1.12255 |
0.785799 |
0.647771 |
0.272009 |
0.86001 |
0.714237 |
0.825052 |
| Logistic_Pvalue: |
2.22E-16 |
0 |
0 |
0 |
0 |
2.29E-40 |
4.40E-40 |
1.11E-16 |
5.98E-41 |
0 |
| Noise(SPE): |
0.00616007 |
0.00806866 |
0.011608 |
0.00857911 |
0.00895273 |
0.00836941 |
0.00904747 |
0.00842127 |
0.007137 |
0.00689192 |
| EndofExpPhase(SDM): |
1.96572 |
1.79812 |
2.25145 |
2.18836 |
1.86581 |
1.56656 |
1.30132 |
1.96243 |
1.58327 |
1.73708 |
| CP(SPE): |
14.6587 |
15.0055 |
17.6451 |
14.8483 |
14.8846 |
18.9689 |
17.8161 |
18.5153 |
17.4542 |
16.9582 |
| CP(SDM): |
20.9008 |
21.1915 |
23.748 |
21.0426 |
21.0369 |
25.1621 |
24.0313 |
24.9317 |
23.7239 |
23.3187 |
| DynamicThreshold: |
1.57341 |
1.31651 |
1.6478 |
1.65974 |
1.33028 |
1.11135 |
0.791187 |
1.41543 |
1.15232 |
1.28451 |
| LowerCycleNumber: |
8 |
8 |
8 |
8 |
8 |
8 |
9 |
9 |
8 |
8 |
| UpperCycleNumber: |
14 |
14 |
14 |
14 |
14 |
14 |
15 |
15 |
15 |
14 |
| PointsForRegression: |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
7 |
8 |
7 |
| Number_OfRegressionWindows: |
50 |
52 |
46 |
51 |
52 |
47 |
47 |
46 |
136 |
47 |
| WeightedAverage_OfPvalue: |
0.00554649 |
0.00552903 |
0.00585796 |
0.005333 |
0.00552529 |
0.00575024 |
0.00534921 |
0.00591711 |
0.00444784 |
0.00603472 |
| Stderr_OfWeightedPvalue: |
0.00246151 |
0.0024692 |
0.00237175 |
0.00242758 |
0.00247117 |
0.00228035 |
0.00209191 |
0.00219693 |
0.00119662 |
0.00217191 |
| WeightedAverage_OfEfficiency: |
0.952131 |
0.979029 |
0.966477 |
0.96428 |
0.976991 |
1.01608 |
0.972844 |
1.00971 |
0.998567 |
1.09886 |
| Stderr_OfWeightedEfficiency: |
0.0174659 |
0.0161545 |
0.0181611 |
0.01638 |
0.0161491 |
0.0167916 |
0.0143999 |
0.0190055 |
0.0115093 |
0.0215524 |
| CT: |
19.7767 |
20.0682 |
22.6301 |
19.9228 |
19.9198 |
24.0423 |
22.9085 |
23.8024 |
22.6023 |
22.1934 |
| TotalSampleNumber: |
10 |
|||||||||
| ReplicateSampleNames: |
Ochsc70_51 |
Ochsc70_52 |
Ochsc70_53 |
Ochsc70_54 |
Ochsc70_55 |
Ochsc70_56 |
Ochsc70_57 |
Ochsc70_58 |
Ochsc70_59 |
Ochsc70_60 |
| AverageEfficiency_OfReplicateSamples |
0.952131 |
0.979029 |
0.966477 |
0.96428 |
0.976991 |
1.01608 |
0.972844 |
1.00971 |
0.998567 |
1.09886 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
19.7767 |
20.0682 |
22.6301 |
19.9228 |
19.9198 |
24.0423 |
22.9085 |
23.8024 |
22.6023 |
22.1934 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
Ochsc70 |
|||||||||
| AverageEfficiency_OfGenes: |
0.993497 |
|||||||||
| Stdev_OfGenesEfficiency: |
0.0133764 |
|||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
|||||||||
| MeanCV%_OfGenesCTs |
0 |
|||||||||
| SampleNames: |
Ochsc70_51 |
Ochsc70_52 |
Ochsc70_53 |
Ochsc70_54 |
Ochsc70_55 |
Ochsc70_56 |
Ochsc70_57 |
Ochsc70_58 |
Ochsc70_59 |
Ochsc70_60 |
| R(0) |
1.18739E-06 |
9.71079E-07 |
1.65832E-07 |
1.07354E-06 |
1.07577E-06 |
6.25968E-08 |
1.36854E-07 |
7.38635E-08 |
1.69044E-07 |
2.24136E-07 |
| normalizing gene R(0) |
6.31341E-07 |
9.07273E-07 |
2.44817E-06 |
1.02064E-06 |
1.07034E-06 |
1.00175E-07 |
2.92339E-06 |
8.24903E-08 |
1.32972E-07 |
1.1628E-07 |
| normalized gene expression |
1.880739001 |
1.070327569 |
0.067737381 |
1.051826436 |
1.005072346 |
0.624876386 |
0.046813316 |
0.89542054 |
1.271275451 |
1.927557952 |
Olympia Oyster hsc70 Treatment (35°C)
| SampleNames: |
Ocjhc70_61 |
Ocjhc70_62 |
Ocjhc70_70 |
Ocjhc70_64 |
Ocjhc70_65 |
Ocjhc70_66 |
Ocjhc70_67 |
Ocjhc70_68 |
Ocjhc70_69 |
| Logistic_a: |
5.88134 |
5.33069 |
5.60916 |
5.8344 |
6.4425 |
5.99186 |
5.45806 |
5.56211 |
6.70563 |
| Logistic_b: |
-14.2002 |
-15.463 |
-14.8067 |
-14.2135 |
-13.9633 |
-13.8791 |
-14.2332 |
-15.2543 |
-14.6173 |
| Logistic_X0: |
23.3146 |
25.2046 |
24.0503 |
21.4344 |
22.5857 |
22.5782 |
22.9947 |
24.5787 |
23.7039 |
| Logistic_Y0: |
0.836216 |
0.257906 |
0.622727 |
0.801476 |
0.462878 |
0.487889 |
0.594238 |
0.402439 |
0.36173 |
| Logistic_Pvalue: |
2.22E-16 |
0 |
1.11E-16 |
0 |
0 |
0 |
1.15E-41 |
0 |
5.17E-41 |
| Noise(SPE): |
0.00648648 |
0.00766007 |
0.00739693 |
0.0158572 |
0.0097268 |
0.00795948 |
0.00749064 |
0.00885507 |
0.0101388 |
| EndofExpPhase(SDM): |
1.87622 |
1.21527 |
1.62237 |
1.83336 |
1.59842 |
1.54276 |
1.55982 |
1.39899 |
1.55396 |
| CP(SPE): |
14.4342 |
16.5079 |
15.3697 |
14.1475 |
14.1855 |
14.0108 |
14.4729 |
16.113 |
15.2024 |
| CP(SDM): |
20.9214 |
22.8462 |
21.6915 |
19.2365 |
20.2245 |
20.2021 |
20.6403 |
22.2448 |
21.3463 |
| DynamicThreshold: |
1.35946 |
0.740416 |
1.12624 |
1.32535 |
1.03551 |
1.0193 |
1.08077 |
0.905143 |
0.962914 |
| LowerCycleNumber: |
7 |
9 |
8 |
7 |
8 |
8 |
8 |
8 |
8 |
| UpperCycleNumber: |
14 |
15 |
15 |
12 |
14 |
14 |
15 |
14 |
14 |
| PointsForRegression: |
8 |
7 |
8 |
6 |
7 |
7 |
8 |
7 |
7 |
| Number_OfRegressionWindows: |
140 |
47 |
144 |
15 |
53 |
51 |
139 |
51 |
52 |
| WeightedAverage_OfPvalue: |
0.00427839 |
0.00524349 |
0.00379797 |
0.00714377 |
0.00536622 |
0.0056624 |
0.00426598 |
0.00527387 |
0.00532757 |
| Stderr_OfWeightedPvalue: |
0.00118674 |
0.00211807 |
0.00121152 |
0.00530734 |
0.00242938 |
0.00232566 |
0.0012066 |
0.00239851 |
0.00233613 |
| WeightedAverage_OfEfficiency: |
1.00119 |
0.926847 |
0.931367 |
0.985003 |
0.980648 |
1.00539 |
0.961806 |
0.996725 |
1.00882 |
| Stderr_OfWeightedEfficiency: |
0.0119465 |
0.0136762 |
0.00879906 |
0.0293803 |
0.0156297 |
0.0179463 |
0.0118599 |
0.0164665 |
0.0162072 |
| CT: |
19.7916 |
21.7108 |
20.5673 |
18.2113 |
19.1182 |
19.0892 |
19.5304 |
21.1262 |
20.228 |
| TotalSampleNumber: |
9 |
||||||||
| ReplicateSampleNames: |
Ocjhc70_61 |
Ocjhc70_62 |
Ocjhc70_70 |
Ocjhc70_64 |
Ocjhc70_65 |
Ocjhc70_66 |
Ocjhc70_67 |
Ocjhc70_68 |
Ocjhc70_69 |
| AverageEfficiency_OfReplicateSamples |
1.00119 |
0.926847 |
0.931367 |
0.985003 |
0.980648 |
1.00539 |
0.961806 |
0.996725 |
1.00882 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
19.7916 |
21.7108 |
20.5673 |
18.2113 |
19.1182 |
19.0892 |
19.5304 |
21.1262 |
20.228 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
Ocjhc70 |
||||||||
| AverageEfficiency_OfGenes: |
0.977533 |
||||||||
| Stdev_OfGenesEfficiency: |
0.0103389 |
||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
||||||||
| MeanCV%_OfGenesCTs |
0 |
||||||||
| SampleNames: |
Ochsc70_61 |
Ochsc70_62 |
Ochsc70_70 |
Ochsc70_64 |
Ochsc70_65 |
Ochsc70_66 |
Ochsc70_67 |
Ochsc70_68 |
Ochsc70_69 |
| R(0) |
1.37796E-06 |
3.7232E-07 |
8.11959E-07 |
4.04764E-06 |
2.18096E-06 |
2.22451E-06 |
1.64659E-06 |
5.54665E-07 |
1.02331E-06 |
| normalizing gene R(0) |
2.211E-07 |
2.02288E-07 |
2.68509E-06 |
2.28605E-06 |
7.23604E-07 |
4.52774E-07 |
1.54268E-06 |
2.71211E-07 |
8.90452E-07 |
| normalized gene expression |
6.232317139 |
1.84054876 |
0.302395149 |
1.770580132 |
3.014022062 |
4.913078252 |
1.067355513 |
2.045144382 |
1.149207768 |
| ttest |
0.033017142 |
control |
treatment |
|
| mean |
0.984164638 |
2.481627684 |
||
| standard dev |
0.636320887 |
1.933090743 |
Pacific Oyster hsp70 Control
| SampleNames: |
Cghsp70_1 |
Cghsp70_2 |
Cghsp70_3 |
Cghsp70_4 |
Cghsp70_5 |
Cghsp70_6 |
Cghsp70_7 |
Cghsp70_8 |
Cghsp70_9 |
Cghsp70_10 |
| Logistic_a: |
4.97206 |
4.89399 |
5.20761 |
4.6974 |
4.67184 |
5.09689 |
5.72558 |
5.80239 |
4.95987 |
5.00722 |
| Logistic_b: |
-16.5162 |
-15.6244 |
-15.0202 |
-15.5042 |
-14.8572 |
-16.0523 |
-15.5298 |
-14.3032 |
-15.141 |
-17.0796 |
| Logistic_X0: |
24.1153 |
25.3466 |
23.799 |
24.4837 |
23.9389 |
25.5243 |
24.87 |
23.3921 |
23.636 |
26.255 |
| Logistic_Y0: |
0.890553 |
0.923238 |
0.912911 |
0.994168 |
1.34987 |
0.620585 |
0.663421 |
0.669956 |
0.723392 |
0.553442 |
| Logistic_Pvalue: |
1.11E-16 |
0 |
4.84E-47 |
0 |
1.75E-47 |
1.11E-16 |
3.49E-45 |
1.02E-38 |
0 |
2.13E-54 |
| Noise(SPE): |
0.0118649 |
0.00266556 |
0.00654644 |
0.00643893 |
0.00557208 |
0.0054524 |
0.00864699 |
0.00698656 |
0.00793259 |
0.00437449 |
| EndofExpPhase(SDM): |
1.79339 |
1.80374 |
1.84339 |
1.83818 |
2.18298 |
1.54178 |
1.69247 |
1.69741 |
1.61087 |
1.46749 |
| CP(SPE): |
16.7334 |
15.6689 |
15.2574 |
16.0049 |
15.2183 |
16.6693 |
16.3708 |
14.6218 |
15.4508 |
17.3841 |
| CP(SDM): |
22.0141 |
23.0014 |
21.5008 |
22.1993 |
21.5996 |
23.2308 |
22.5538 |
21.0098 |
21.3734 |
24.0487 |
| DynamicThreshold: |
1.3479 |
1.36482 |
1.38142 |
1.41939 |
1.76921 |
1.08391 |
1.18227 |
1.18718 |
1.1711 |
1.01265 |
| LowerCycleNumber: |
8 |
10 |
8 |
8 |
8 |
9 |
8 |
8 |
8 |
9 |
| UpperCycleNumber: |
13 |
17 |
15 |
14 |
15 |
15 |
15 |
14 |
14 |
16 |
| PointsForRegression: |
6 |
8 |
8 |
7 |
8 |
7 |
8 |
7 |
7 |
8 |
| Number_OfRegressionWindows: |
14 |
137 |
137 |
50 |
135 |
49 |
140 |
52 |
53 |
145 |
| WeightedAverage_OfPvalue: |
0.00740119 |
0.00407445 |
0.00437955 |
0.00587903 |
0.0048294 |
0.00501874 |
0.00394702 |
0.00545851 |
0.00506384 |
0.00338625 |
| Stderr_OfWeightedPvalue: |
0.00594899 |
0.00109585 |
0.001183 |
0.0025241 |
0.00121635 |
0.00207705 |
0.00122765 |
0.00242271 |
0.002408 |
0.00116733 |
| WeightedAverage_OfEfficiency: |
1.0404 |
1.03645 |
0.97272 |
1.06164 |
1.02305 |
1.01032 |
0.935762 |
1.00499 |
1.03547 |
0.986515 |
| Stderr_OfWeightedEfficiency: |
0.0362567 |
0.0110996 |
0.0117174 |
0.0217681 |
0.0154844 |
0.0151322 |
0.00958023 |
0.0168633 |
0.016945 |
0.00836637 |
| CT: |
20.9935 |
21.8619 |
20.4009 |
21.0983 |
20.4827 |
22.1134 |
21.4381 |
19.8838 |
20.2911 |
22.9567 |
| TotalSampleNumber: |
10 |
|||||||||
| ReplicateSampleNames: |
Cghsp70_1 |
Cghsp70_2 |
Cghsp70_3 |
Cghsp70_4 |
Cghsp70_5 |
Cghsp70_6 |
Cghsp70_7 |
Cghsp70_8 |
Cghsp70_9 |
Cghsp70_10 |
| AverageEfficiency_OfReplicateSamples |
1.0404 |
1.03645 |
0.97272 |
1.06164 |
1.02305 |
1.01032 |
0.935762 |
1.00499 |
1.03547 |
0.986515 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
20.9935 |
21.8619 |
20.4009 |
21.0983 |
20.4827 |
22.1134 |
21.4381 |
19.8838 |
20.2911 |
22.9567 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
Cghsp70 |
|||||||||
| AverageEfficiency_OfGenes: |
1.01073 |
|||||||||
| Stdev_OfGenesEfficiency: |
0.0118238 |
|||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
|||||||||
| MeanCV%_OfGenesCTs |
0 |
|||||||||
| SampleNames: |
Cghsp70_1 |
Cghsp70_2 |
Cghsp70_3 |
Cghsp70_4 |
Cghsp70_5 |
Cghsp70_6 |
Cghsp70_7 |
Cghsp70_8 |
Cghsp70_9 |
Cghsp70_10 |
| R(0) |
4.28098E-07 |
2.33405E-07 |
6.47605E-07 |
3.97879E-07 |
6.1164E-07 |
1.95802E-07 |
3.13814E-07 |
9.2934E-07 |
6.99228E-07 |
1.08642E-07 |
| Normalizing Gene R(0) |
3.88723E-05 |
6.83103E-05 |
5.7771E-05 |
3.2135E-05 |
6.79746E-05 |
1.13463E-05 |
1.85774E-05 |
6.18925E-05 |
3.43697E-05 |
8.28294E-06 |
| Normalized Gene Expression |
0.011012913 |
0.003416838 |
0.011209874 |
0.012381497 |
0.008998074 |
0.017256914 |
0.016892213 |
0.015015383 |
0.020344313 |
0.013116385 |
Pacific Oyster hsp70 Treatment (35°C)
| SampleNames: |
Cghsp70_31 |
Cghsp70_32 |
Cghsp70_33 |
Cghsp70_34 |
Cghsp70_35 |
Cghsp70_36 |
Cghsp70_37 |
Cghsp70_38 |
Cghsp70_39 |
Cghsp70_40 |
| Logistic_a: |
3.25396 |
3.33071 |
4.41512 |
3.40371 |
4.30224 |
4.09296 |
4.86702 |
4.18028 |
3.92236 |
4.21299 |
| Logistic_b: |
-13.1789 |
-13.0419 |
-11.8601 |
-11.4691 |
-11.3299 |
-12.1064 |
-11.3398 |
-12.3601 |
-11.0042 |
-11.8356 |
| Logistic_X0: |
22.7309 |
24.5431 |
21.2232 |
20.9209 |
20.3502 |
20.6878 |
19.6533 |
21.76 |
20 |
20.331 |
| Logistic_Y0: |
2.37299 |
2.7002 |
2.49618 |
2.42603 |
2.12522 |
1.69895 |
2.0156 |
2.04556 |
2.09401 |
2.0411 |
| Logistic_Pvalue: |
0 |
1.91E-57 |
7.76E-57 |
1.23E-49 |
0 |
0 |
7.56E-52 |
0 |
0 |
1.49E-44 |
| Noise(SPE): |
0.00687664 |
0.00216936 |
0.00307781 |
0.00327407 |
0.00310332 |
0.00325281 |
0.00379922 |
0.00456491 |
0.00256225 |
0.00442619 |
| EndofExpPhase(SDM): |
2.93989 |
3.2792 |
3.24761 |
3.00068 |
2.84938 |
2.39889 |
2.835 |
2.76381 |
2.74937 |
2.75778 |
| CP(SPE): |
14.2464 |
13.9844 |
11.4993 |
11.4176 |
10.7471 |
11.4736 |
10.4574 |
12.5336 |
10.2714 |
11.3903 |
| CP(SDM): |
20.1994 |
21.7783 |
18.5694 |
18.2064 |
17.6739 |
18.1589 |
17.0711 |
19.16 |
17.2834 |
17.783 |
| DynamicThreshold: |
2.65988 |
2.99078 |
2.87343 |
2.71499 |
2.48885 |
2.05055 |
2.4272 |
2.40697 |
2.42297 |
2.40165 |
| LowerCycleNumber: |
6 |
9 |
9 |
7 |
8 |
8 |
7 |
7 |
8 |
7 |
| UpperCycleNumber: |
12 |
16 |
16 |
14 |
15 |
15 |
14 |
13 |
15 |
14 |
| PointsForRegression: |
7 |
8 |
8 |
8 |
8 |
8 |
8 |
7 |
8 |
8 |
| Number_OfRegressionWindows: |
48 |
129 |
136 |
137 |
147 |
149 |
156 |
52 |
156 |
144 |
| WeightedAverage_OfPvalue: |
0.00603533 |
0.0046863 |
0.00446769 |
0.0048245 |
0.00453795 |
0.00454082 |
0.00416746 |
0.00603378 |
0.00353238 |
0.0044074 |
| Stderr_OfWeightedPvalue: |
0.00236167 |
0.00112695 |
0.00113027 |
0.00124818 |
0.00132079 |
0.00115123 |
0.00110889 |
0.00248009 |
0.00105694 |
0.00126353 |
| WeightedAverage_OfEfficiency: |
1.03438 |
0.963708 |
0.969604 |
1.17626 |
0.968586 |
1.05511 |
0.945077 |
1.0697 |
0.905241 |
1.04634 |
| Stderr_OfWeightedEfficiency: |
0.0237034 |
0.0149601 |
0.014606 |
0.0200301 |
0.0145614 |
0.0120338 |
0.00916613 |
0.0254454 |
0.0071967 |
0.0156529 |
| CT: |
19.0383 |
20.4997 |
17.3782 |
17.0039 |
16.491 |
17.0171 |
15.93 |
17.9811 |
16.0942 |
16.6432 |
| TotalSampleNumber: |
10 |
|||||||||
| ReplicateSampleNames: |
Cghsp70_31 |
Cghsp70_32 |
Cghsp70_33 |
Cghsp70_34 |
Cghsp70_35 |
Cghsp70_36 |
Cghsp70_37 |
Cghsp70_38 |
Cghsp70_39 |
Cghsp70_40 |
| AverageEfficiency_OfReplicateSamples |
1.03438 |
0.963708 |
0.969604 |
1.17626 |
0.968586 |
1.05511 |
0.945077 |
1.0697 |
0.905241 |
1.04634 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
19.0383 |
20.4997 |
17.3782 |
17.0039 |
16.491 |
17.0171 |
15.93 |
17.9811 |
16.0942 |
16.6432 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
Cghsp70 |
|||||||||
| AverageEfficiency_OfGenes: |
1.0134 |
|||||||||
| Stdev_OfGenesEfficiency: |
0.0248822 |
|||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
|||||||||
| MeanCV%_OfGenesCTs |
0 |
|||||||||
| SampleNames: |
Cghsp70_31 |
Cghsp70_32 |
Cghsp70_33 |
Cghsp70_34 |
Cghsp70_35 |
Cghsp70_36 |
Cghsp70_37 |
Cghsp70_38 |
Cghsp70_39 |
Cghsp70_40 |
| R(0) |
1.63564E-06 |
5.882E-07 |
5.22688E-06 |
6.79209E-06 |
9.72499E-06 |
6.72964E-06 |
1.4401E-05 |
3.4277E-06 |
1.28377E-05 |
8.74241E-06 |
| Normalizing Gene R(0) |
0.000125093 |
2.27813E-05 |
0.000228251 |
0.000152362 |
0.00031752 |
0.000339918 |
0.000814844 |
0.000211674 |
0.000243804 |
0.000325683 |
| Normalized Gene Expression |
0.013075364 |
0.02581937 |
0.022899743 |
0.044578697 |
0.030627969 |
0.019797805 |
0.017673373 |
0.016193344 |
0.052656054 |
0.026843342 |
| ttest |
0.004187992 |
|
| control |
treatment |
|
| mean |
0.01296444 |
0.027016506 |
| standard deviation |
0.004791894 |
0.012684946 |
Olympia Oyster hsp70 Control
| SampleNames: |
hsp70_51 |
hsp70_52 |
hsp70_53 |
hsp70_54 |
hsp70_55 |
hsp70_56 |
hsp70_57 |
hsp70_58 |
hsp70_59 |
hsp70_60 |
| Logistic_a: |
2.48633 |
2.16606 |
2.83963 |
2.89718 |
2.83213 |
2.87614 |
3.15497 |
3.45032 |
3.46036 |
3.60513 |
| Logistic_b: |
-13.5068 |
-14.0953 |
-15.861 |
-15.9113 |
-14.9462 |
-18.0063 |
-16.2677 |
-16.5187 |
-16.6697 |
-16.3926 |
| Logistic_X0: |
25.4418 |
25.1616 |
27.4781 |
25.8175 |
26.2349 |
29.3395 |
28.9302 |
29.6302 |
29.2507 |
29.3615 |
| Logistic_Y0: |
0.801174 |
0.311068 |
0.244875 |
0.300861 |
0.214694 |
0.231036 |
0.132164 |
0.621713 |
0.403069 |
0.358554 |
| Logistic_Pvalue: |
2.22E-16 |
2.22E-16 |
5.61E-59 |
8.99E-36 |
0 |
4.05E-49 |
1.11E-16 |
7.31E-56 |
5.42E-66 |
0 |
| Noise(SPE): |
0.000889137 |
0.00113655 |
0.00233916 |
0.00836763 |
0.00285443 |
0.00408892 |
0.00156175 |
0.00260068 |
0.00150124 |
0.00125462 |
| EndofExpPhase(SDM): |
1.23653 |
0.693549 |
0.757073 |
0.823719 |
0.72028 |
0.760253 |
0.703639 |
1.24824 |
1.03234 |
1.01238 |
| CP(SPE): |
14.1379 |
14.7248 |
17.5613 |
17.8812 |
16.5355 |
20.3875 |
18.1208 |
19.1739 |
18.3834 |
18.064 |
| CP(SDM): |
22.6837 |
22.5579 |
24.9768 |
23.4754 |
23.6879 |
27.0103 |
26.3678 |
27.0489 |
26.7274 |
26.7823 |
| DynamicThreshold: |
1.01929 |
0.502877 |
0.502144 |
0.566474 |
0.468914 |
0.497689 |
0.418682 |
0.936278 |
0.718453 |
0.686096 |
| LowerCycleNumber: |
11 |
11 |
10 |
8 |
9 |
9 |
11 |
9 |
11 |
12 |
| UpperCycleNumber: |
18 |
18 |
17 |
13 |
16 |
16 |
18 |
16 |
18 |
19 |
| PointsForRegression: |
8 |
8 |
8 |
6 |
8 |
8 |
8 |
8 |
8 |
8 |
| Number_OfRegressionWindows: |
140 |
136 |
135 |
13 |
137 |
137 |
144 |
136 |
134 |
135 |
| WeightedAverage_OfPvalue: |
0.00422721 |
0.00403543 |
0.00403243 |
0.00668428 |
0.003973 |
0.00381899 |
0.00326621 |
0.00425879 |
0.00427882 |
0.00435005 |
| Stderr_OfWeightedPvalue: |
0.00121181 |
0.00119631 |
0.00110285 |
0.00584196 |
0.00124816 |
0.00113969 |
0.00117376 |
0.00119612 |
0.00124072 |
0.00128279 |
| WeightedAverage_OfEfficiency: |
0.868478 |
0.887834 |
0.910775 |
0.989557 |
0.87051 |
0.897178 |
0.86295 |
0.973682 |
0.918756 |
0.936913 |
| Stderr_OfWeightedEfficiency: |
0.00819808 |
0.00933662 |
0.00865585 |
0.028174 |
0.00875727 |
0.00791071 |
0.00716471 |
0.00967134 |
0.00979932 |
0.0102732 |
| CT: |
21.3921 |
21.3258 |
23.7595 |
22.3522 |
22.4682 |
25.8487 |
25.1101 |
25.7798 |
25.4813 |
25.5127 |
| TotalSampleNumber: |
10 |
|||||||||
| ReplicateSampleNames: |
hsp70_51 |
hsp70_52 |
hsp70_53 |
hsp70_54 |
hsp70_55 |
hsp70_56 |
hsp70_57 |
hsp70_58 |
hsp70_59 |
hsp70_60 |
| AverageEfficiency_OfReplicateSamples |
0.868478 |
0.887834 |
0.910775 |
0.989557 |
0.87051 |
0.897178 |
0.86295 |
0.973682 |
0.918756 |
0.936913 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
21.3921 |
21.3258 |
23.7595 |
22.3522 |
22.4682 |
25.8487 |
25.1101 |
25.7798 |
25.4813 |
25.5127 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
hsp70 |
|||||||||
| AverageEfficiency_OfGenes: |
0.911663 |
|||||||||
| Stdev_OfGenesEfficiency: |
0.0138691 |
|||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
|||||||||
| MeanCV%_OfGenesCTs |
0 |
|||||||||
| 51 |
52 |
53 |
54 |
55 |
56 |
57 |
58 |
59 |
60 |
|
| hsp70_51 |
hsp70_52 |
hsp70_53 |
hsp70_54 |
hsp70_55 |
hsp70_56 |
hsp70_57 |
hsp70_58 |
hsp70_59 |
hsp70_60 |
|
| R(0) |
9.55038E-07 |
9.96961E-07 |
2.05972E-07 |
5.1267E-07 |
4.75547E-07 |
5.31965E-08 |
8.58486E-08 |
5.56253E-08 |
6.74953E-08 |
6.61359E-08 |
| Normalizing Gene R(0) |
6.31341E-07 |
9.07273E-07 |
2.44817E-06 |
1.02064E-06 |
1.07034E-06 |
1.00175E-07 |
2.92339E-06 |
8.24903E-08 |
1.32972E-07 |
1.1628E-07 |
| Normalized Gene Expression |
1.512713524 |
1.098854993 |
0.08413302 |
0.502299859 |
0.444297363 |
0.531036978 |
0.029366103 |
0.674324851 |
0.507591559 |
0.568765239 |
Olympia Oyster hsp70 Treament (35°C)
| SampleNames: |
hsp70_61 |
hsp70_62 |
hsp70_70 |
hsp70_64 |
hsp70_65 |
hsp70_66 |
hsp70_67 |
hsp70_68 |
hsp70_69 |
| Logistic_a: |
3.23304 |
4.00051 |
0.0714255 |
3.42596 |
4.03185 |
4.00084 |
3.80749 |
3.93727 |
3.51469 |
| Logistic_b: |
-14.8575 |
-16.1587 |
-9.99194 |
-13.4434 |
-10.9076 |
-11.0757 |
-10.8325 |
-12.9489 |
-14.4179 |
| Logistic_X0: |
26.5173 |
28.3791 |
26.6276 |
25.4028 |
20.0946 |
20.3347 |
19.9997 |
25.1722 |
25.0252 |
| Logistic_Y0: |
0.315234 |
0.154143 |
0.108101 |
0.40319 |
0.283478 |
0.312134 |
0.559202 |
0.239445 |
0.236968 |
| Logistic_Pvalue: |
0 |
1.11E-16 |
1.11E-16 |
0 |
1.05E-56 |
1.12E-64 |
2.22E-16 |
4.44E-16 |
0 |
| Noise(SPE): |
0.00104654 |
0.00243068 |
0.000323469 |
0.00269601 |
0.00161723 |
0.00117058 |
0.00172459 |
0.00659418 |
0.00253635 |
| EndofExpPhase(SDM): |
0.89177 |
0.877975 |
0.119724 |
1.0025 |
0.95559 |
0.981714 |
1.19277 |
0.922851 |
0.860266 |
| CP(SPE): |
15.4401 |
17.9459 |
15.5217 |
14.9282 |
9.81034 |
9.75425 |
9.82531 |
15.3671 |
15.153 |
| CP(SDM): |
23.9261 |
25.8472 |
22.6015 |
22.6347 |
17.3383 |
17.5924 |
17.2354 |
22.3143 |
22.4986 |
| DynamicThreshold: |
0.604025 |
0.517274 |
0.114074 |
0.704192 |
0.620343 |
0.647509 |
0.876848 |
0.584445 |
0.549885 |
| LowerCycleNumber: |
12 |
11 |
10 |
11 |
9 |
11 |
9 |
9 |
9 |
| UpperCycleNumber: |
19 |
18 |
17 |
18 |
16 |
18 |
16 |
16 |
16 |
| PointsForRegression: |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
| Number_OfRegressionWindows: |
136 |
134 |
96 |
143 |
155 |
148 |
142 |
143 |
147 |
| WeightedAverage_OfPvalue: |
0.00412468 |
0.00404688 |
0.00503646 |
0.00385586 |
0.00379954 |
0.00439125 |
0.00479605 |
0.0038742 |
0.00370792 |
| Stderr_OfWeightedPvalue: |
0.00115414 |
0.00114012 |
0.00097747 |
0.00129051 |
0.00103745 |
0.00127707 |
0.001232 |
0.00122117 |
0.00109876 |
| WeightedAverage_OfEfficiency: |
0.934941 |
0.897453 |
0.336825 |
0.863336 |
0.924057 |
0.873781 |
1.05484 |
0.945257 |
0.936896 |
| Stderr_OfWeightedEfficiency: |
0.00940033 |
0.00850105 |
0.00973384 |
0.00824632 |
0.00795991 |
0.0103837 |
0.0169554 |
0.00915668 |
0.00770491 |
| CT: |
22.6808 |
24.6075 |
20.9546 |
21.3445 |
16.1331 |
16.3857 |
16.0301 |
21.0058 |
21.2975 |
| TotalSampleNumber: |
9 |
||||||||
| ReplicateSampleNames: |
hsp70_61 |
hsp70_62 |
hsp70_70 |
hsp70_64 |
hsp70_65 |
hsp70_66 |
hsp70_67 |
hsp70_68 |
hsp70_69 |
| AverageEfficiency_OfReplicateSamples |
0.934941 |
0.897453 |
0.336825 |
0.863336 |
0.924057 |
0.873781 |
1.05484 |
0.945257 |
0.936896 |
| StdErr_OfReplicateSamplesEfficiency: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CoefficientVariation(CV%)_OfReplicateSamplesEfficiencies: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| AverageCT_OfReplicateSamples: |
22.6808 |
24.6075 |
20.9546 |
21.3445 |
16.1331 |
16.3857 |
16.0301 |
21.0058 |
21.2975 |
| StdErr_OfReplicateSamplesCT: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| CV%_OfReplicateSamplesCTs: |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| GeneNames |
hsp70 |
||||||||
| AverageEfficiency_OfGenes: |
0.863043 |
||||||||
| Stdev_OfGenesEfficiency: |
0.0683228 |
||||||||
| MeanCV%_OfGenesEfficiencies |
0 |
||||||||
| MeanCV%_OfGenesCTs |
0 |
||||||||
| 61 |
62 |
70 |
64 |
65 |
66 |
67 |
68 |
69 |
|
| hsp70_61 |
hsp70_62 |
hsp70_70 |
hsp70_64 |
hsp70_65 |
hsp70_66 |
hsp70_67 |
hsp70_68 |
hsp70_69 |
|
| R(0) |
7.4324E-07 |
2.24125E-07 |
2.17564E-06 |
1.70698E-06 |
4.36986E-05 |
3.7343E-05 |
4.65908E-05 |
2.10743E-06 |
1.75763E-06 |
| Normalizing Gene R(0) |
2.211E-07 |
2.02288E-07 |
2.68509E-06 |
2.28605E-06 |
7.23604E-07 |
4.52774E-07 |
1.54268E-06 |
2.71211E-07 |
8.90452E-07 |
| Normalized Gene Expression |
3.361561639 |
1.107952449 |
0.810267225 |
0.746692189 |
60.39021534 |
82.47606743 |
30.20121472 |
7.770439305 |
1.973866587 |
| ttest |
0.366858231 |
average |
standev |
| Control |
0.595338349 |
0.438052559 |
|
| Heat Stress |
20.98203076 |
30.57204658 |
CONCLUSIONS: I was unsatisfied with my data from the hsc70 experiment so I chose to run an additional qPCR with the hsp70 gene. The data for the Olympia hsp70 was taken from Kate's experiment. I only ran the qPCR on the Pacific oysters for the hsp70 portion.
REFLECTION: I was pretty discouraged and doubted myself when I got my experimental results back for the hsc70 gene, as it felt like I was not able to achieve ideal results. However, I found out that many other people in lab had similarly unspectacular results and that it is more important in how I interpret and justify the data/outcome rather than getting hung up on ideal/non-ideal results. I did run the hsp70 experiments just to have a back up and to make sure that my lab techniques were not at fault for my hsc70 results.
RESEARCH PROJECT (Week 2)
DATE: November 20, 2012LAB SUMMARY: We finished RNA extraction, ran some nanodrop tests, and prepared the RNA samples for reverse transcription.
MATERIALS AND METHODS:
RNA Extraction:
- Turn on heating block to 55°C.
- Incubate your tissue sample at room temperature for 5 minutes.
- After adding 200 µL of chloroform to the sample, vortex for 30 seconds until the solution becomes a milky emulsion.
- Incubate the tube at room temp for 5 minutes then spin it in a refrigerate microfuge for 15 minutes at max speed. Carefully transfer most of ONLY the aqueous phase (top, clear liquid) to a fresh tube and dispose of the rest in the chloroform waste jar.
- Add 500 µL of isopropanol and mix by inverting the tube several times until the solution appears uniform.
- After incubating at room temp for 10 minutes, spin in refrigerated microfuge at max speed for 8 mins (a small, white pellet should form).
- Remove the supernatant (liquid portion), add 1 mL of 75% EtOH to the remaining pellet, then vortex briefly.
- Spin in refrigerated microfuge at 7500g for 5 mins.
- Carefully remove supernatant and ONLY the supernatant, making sure the pellet stays intact.
- Briefly (~15 s) spin the tube in the centrifuge to pool residual EtOH and remove it using a small pipette tip (using a clean KimWipe to soak up any residual EtOH inside the tube is ok as long as the pellet is not touched)
- Let the tube sit (no more than 5 minutes) with the top open to allow any remaining EtOH to evaporate.
- Dissolve the dry pellet in 100 µL of 0.1% DEPC-H2O and incubate the solution in 55°C for 5 mins.
- Remove the tube from the heat and flick a few times to mix the sample then place on ice.
RNA Quantification:
- Pipette 2 µL of 0.1% DEPC-H2O onto the Nanodrop pedestal and lower the arm.
- Click "Blank", to zero the instrument.
- Pipette 2 µL of your RNA sample onto the Nanodrop pedestal and lower the arm.
- Click "Measure" and record your RNA concentration and ratios.
- Raise the arm and wipe off the instrument using a KimWipe.
- Store sample in cold storage.
Reverse Transcription:
- Label a row of connected 0.2 ml PCR tubes.
- Pour 17.75 µL of each RNA sample into corresponding 0.2 ml PCR tubes.
- Store in ice.
RESULTS: I chose to run nanodrop on samples 35 and 69.
| Sample # |
A260/280 |
A260/230 |
Concentration |
| 35 |
1.95 |
1.34 |
417.8 ng/µL |
| 69 |
1.75 |
2.72 |
160 ng/µL |
New primers were designed for hsc70 and hsp70 of Olympia oyster:
hsc70 F - CTGACAGCTACTTTCACTTTCGT
hsc70 R - ATCAATGCCGATCGCTGGAG
hsp70 F - GTTCCGATTTGTTCCGTGCC
hsp70 R - TTGTCGCCATTTTCCTCGCT
CONCLUSIONS: Thesamples I tested with the nanodrop yielded sufficient concentrations. However, the ratios were slightly off from what is considered ideal for a clean RNA (1.8-2.0 for A260/280 and 1.5-2.0 for A260/230). There may have been a bit too much ethanol left over in the sample.
REFLECTION: I will be testing the newly designed primers next week. The hsc70 primer will be my first option. However, in the event that it is not an optimal choice I plan on going forward with my project focusing on the differences in the two oyster species' hsp70 production.
RESEARCH PROJECT (Week 1)
DATE: November 13, 2012LAB SUMMARY: We isolated and extracted RNA from the tissue samples - half of the samples were completed while the other half were only isolated (mixed with TriReagent, homogenized, then cold-stored).
MATERIALS AND METHODS:
RNA Isolation:
- Separate approximately 50-100 mg of tissue using a razor/scissors and tweezers and transfer to a 1.5 ml snap cap tube labeled "RNA." Weigh the first couple samples to get an idea of how much tissue is needed and eyeball the rest (make sure to sterilize the razor/scissors and tweezers using EtOH, bleach, and water in that order after each sample).
- Add 500 µL of TriReagent and homogenize the sample using a disposable pestle and a vortex. This may take a few minutes so be patient. Keep the tissues not being used on ice.
- After the sample is completely homogenized, add another 500 µL of TriReagent and vortex it for 15 seconds.
- Store away half of the samples in cold storage and proceed with extraction for the remaining half.
RNA Extraction:
- Turn on heating block to 55°C.
- Incubate your tissue sample at room temperature for 5 minutes.
- After adding 200 µL of chloroform to the sample, vortex for 30 seconds until the solution becomes a milky emulsion.
- Incubate the tube at room temp for 5 minutes then spin it in a refrigerate microfuge for 15 minutes at max speed. Carefully transfer most of ONLY the aqueous phase (top, clear liquid) to a fresh tube and dispose of the rest in the chloroform waste jar.
- Add 500 µL of isopropanol and mix by inverting the tube several times until the solution appears uniform.
- After incubating at room temp for 10 minutes, spin in refrigerated microfuge at max speed for 8 mins (a small, white pellet should form).
- Remove the supernatant (liquid portion), add 1 mL of 75% EtOH to the remaining pellet, then vortex briefly.
- Spin in refrigerated microfuge at 7500g for 5 mins.
- Carefully remove supernatant and ONLY the supernatant, making sure the pellet stays intact.
- Briefly (~15 s) spin the tube in the centrifuge to pool residual EtOH and remove it using a small pipette tip (using a clean KimWipe to soak up any residual EtOH inside the tube is ok as long as the pellet is not touched)
- Let the tube sit (no more than 5 minutes) with the top open to allow any remaining EtOH to evaporate.
- Dissolve the dry pellet in 100 µL of 0.1% DEPC-H2O and incubate the solution in 55°C for 5 mins.
- Remove the tube from the heat and flick a few times to mix the sample then place on ice.
RESULTS: Samples extracted this week were 14 (Pac control),15 (Pac control),31-33 (Pac 35°C),69 (Oly 35°C), and 70 (Oly 35°C).
| Sample Number |
Tissue Weight |
| 14 |
.042 g |
| 15 |
.045 g |
| 69 |
.011 g |
| 70 |
.050 g |
CONCLUSIONS: The tissue for sample 69 was not even close to 50 mg. I would guess that this is the case for many olympia samples since they are much smaller than pacific oysters. There were no quantifiable results this week. We should have data next week after nanodrop is completed.
REFLECTION: The methods and the whole lab in general were pretty easy, as it was the second time around. However, it took longer than expected. Also, if I recall correctly, I had bigger pellets the first time I did these procedures in Lab 3. I hope I had enough but I guess we won't know until next week when the nanodrop results are analyzed.
LAB 7: PROTEIN SDS-PAGE, WESTERN BLOT, ANALYSIS OF CONVENTIONAL PCR
(VIA AGAROSE GEL) & qPCR DATA
DATE: November 6, 2012LAB SUMMARY: Agarose gel electrophoresis was used to determine if amplification of the cDNA was successful. The results of the SDS/PAGE and western blot gave us a visual indication of which proteins were present in our extracted samples.
MATERIALS AND METHODS:
Agarose Gel Eletrophoresis:
- Place gel in gel box and fill with enough 1x TAE buffer that it fully submerges the gel.
- Remove combs from wells and load 7 µL of a 100 bp ladder in lane 1.
- Load 20 µL of your PCR sample into the gel.
- Run get at 100 V for approximately 1 hour and visualize the gel on the UV transilluminator.
SDS-PAGE Protocol:
- Begin boiling water on a hot plate.
- Add 15 µL of your protein + 15 µL of 2X Reducing Sample Buffer to a 1.5 ml screw cap tube.
- Mix sample by flicking and briefly centrifuge to pool liquid to bottom.
- Submerge the tube in boiling water for 5 minutes then centrifuge for 1 minute.
- Rinse gel wells thoroughly and load your entire sample into a well, making sure to note which wells were filled.
- Put the lid on, plug in the electrodes, and run for 45 minutes at 150 V.
- Turn off power, unplug everything, and remove the cassette, carefully cracking it open to expose the gel by itself.
- Trim wells at top of the gel and cut out a diagonal section at the top left corner to help identify its correct orientation later.
Western Blot:
- Soak the filter paper, membrane and gel in Tris-Glycine Transfer Buffer for 15 minutes.
- Assemble the blotting sandwich in the semi-dry blotting apparatus in this order: anode, 2 x filter paper, membrane, gel, 2 x filter paper, cathode.
- After transferring the blot for 30 minutes at 20 V, remove the gel from the sandwich and rinse off with transfer buffer.
- Wash membrane 2 times, for 5 minutes each, with 20 ml of pure water and put in a plastic box with 10 ml of Blocking Solution to incubate overnight on a shaker.
- Decant liquid.
- Rinse the membrane with 20 mL of water for 5 minutes, then decant. Repeat.
- Incubate the membrane in 10 mL of Primary Antibody Solution. Decant the solution.
- Rinse the membrane with 20 mL of Antibody Wash for 5 minutes, then decant. Repeat 3 times.
- Incubate the membrane in 10 mL of Secondary Antibody Solution for 30 minutes. Decant.
- Wash the membrane for 5 minutes with 20 mL of Antibody wash, then decant. Repeat 3 times.
- Rinse the membrane with 20 mL of pure water for 2 minutes, then decant. Repeat twice.
- Incubate the membrane in 5 mL of Chromogenic Substrate until a purple band appears. This will occur between 1-60 minutes after adding the Chromogenic Substrate.
- Dry the membrane on a clean piece of filter paper to the open air.
RESULTS:
SDS-PAGE
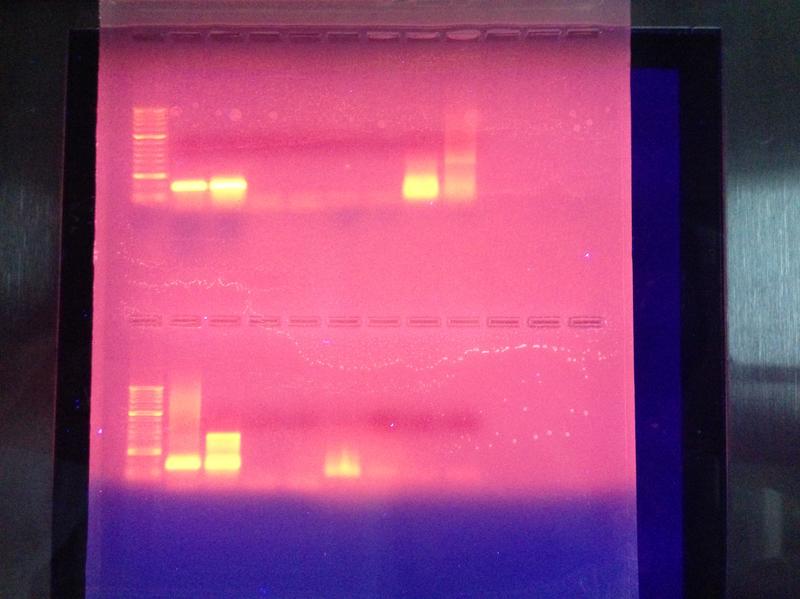
Agarose gel Electrophoresis
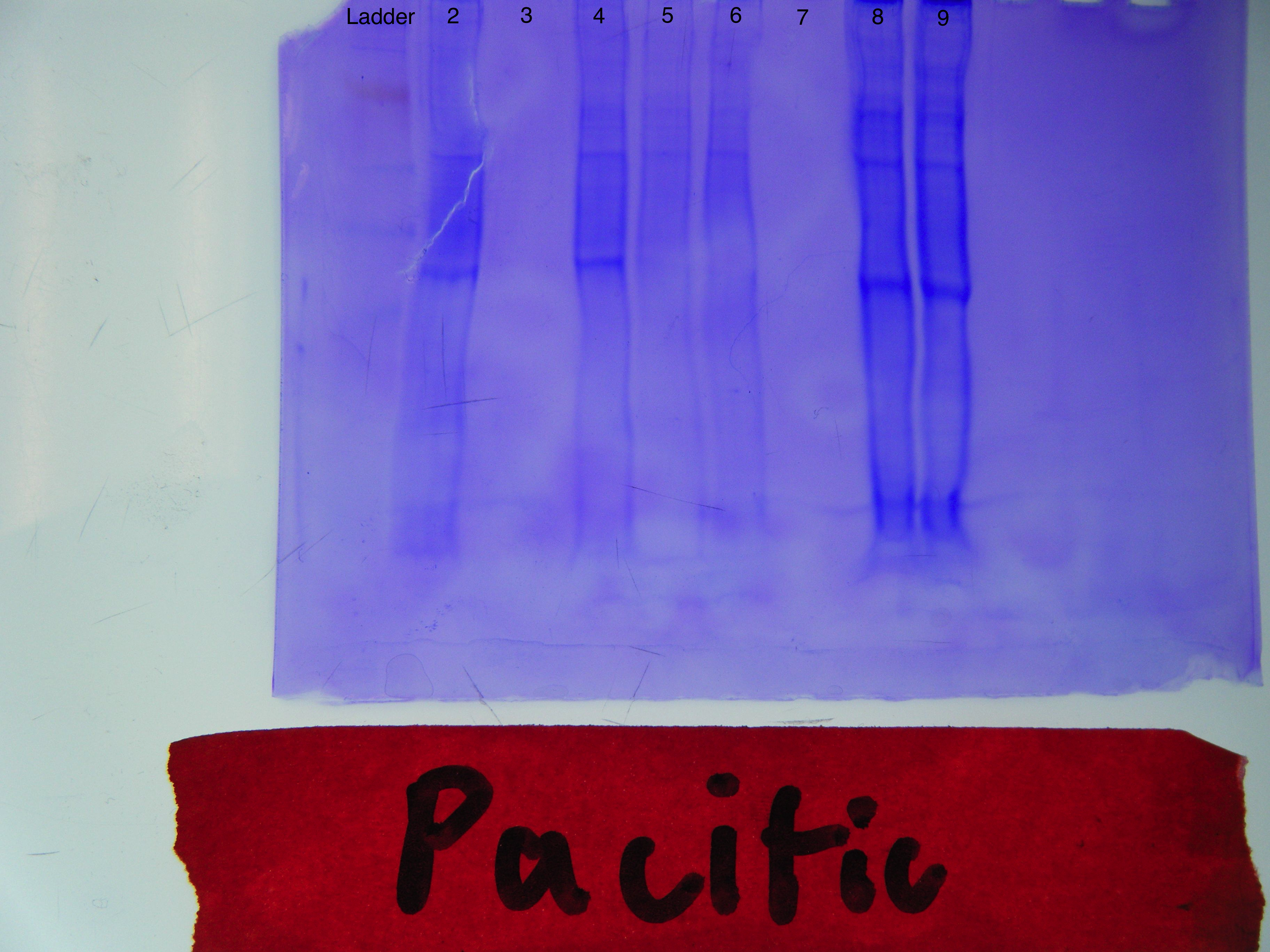
Western Blot

CONCLUSIONS: My samples from the SDS-PAGE are the first four on the bottom in this order: gDNA, cDNA, blank, blank. My blanks were clean as they showed no bands. My gDNA showed a solid line around 200 with faint traces all the way past 1000 while my cDNA showed solid lines from 200-500. Although my cDNA results were in line with what was to be expected, an increase in temperature at which PCR is run may yield better, more precise results for the gDNA next time. As for the agarose gel electrophoresis results, my samples were in lanes 8 and 9. It shows more distinct colorations than the other lanes, which reflects my previous results of relatively high protein concentrations.
REFLECTION: With the exception of the ladder, it was difficult for me to identify and distinguish the bands on the Western Blot results. It is even more difficult to do so with a captured image of it above. I honestly could not even label the lanes with confidence. It goes without saying, that I most likely will not use the Western Blot for my research project going forward and will instead rely on SDS-PAGE and agarose gel electrophoresis for my analysis.
LAB 6: CONVENTIONAL PCR, QPCR & PROTEIN ISOLATION
DATE: October 30, 2012LAB SUMMARY: In this week's lab we amplified DNA and cDNA using PCR and qPCR techniques and isolated protein from the gill tissue of Crassostrea gigas that had been exposed to heat shock at 35°C. The concentration of the protein isolated was determined by running a Bradford assay.
MATERIALS AND METHODS:
Conventional PCR:
- Make a reaction mastermix in a 1.5 ml microcentrifuge tube labeled "MM" and with your initials.
- Use the table below to calculate the total volume of each reagent required for the mastermix.
- Pipette 23 µL of your mastermix into each of your properly labeled 0.5 ml PCR tubes (clear w/attached lid).
- Add 2 µL of the appropriate template (1 cDNA, 1 DNA, and 2 blanks - water) to each tube and mix via pipetting.
- Turn in to TA for thermal cycling and cold storage.
| Reagent |
1 reaction |
5 reactions |
| Apex Red |
12.5 µL |
62.5 µL |
| 10 µM Fwd Primer |
1 µL |
5 µL |
| 10 µM Rev Primer |
1 µL |
5 µL |
| PCR water |
8.5 µL |
42.5 µL |
qPCR:
- Prepare another mastermix using the table below.
- Add mastermix to each of the qPCR (white with separate caps) tubes.
- Thaw cDNA samples and add 2 µL of cDNA template to each reaction and 2 µL of 0.1% DEPC water to the negative controls.
- Cap the tubes and turn in to the TA to be run and stored in ice.
| Component |
Volume |
Final Conc. |
5 reactions |
| Mastermix, 2X (Immomix) |
12.5 µL |
1x |
62.5 µL |
| Syto-13 dye (50 µM) |
1 µL |
2 µM |
5 µL |
| upstream primer, 10 µM |
1.25 µL |
2.5 µM |
6.25 µL |
| downstream primer, 10 µM |
1.25 µL |
2.5 µM |
6.25 µL |
| 0.1% DEPC water |
7 µL |
N/A |
35 µL |
Protein Extraction and Analysis:
- Thaw the tissues and separate a small piece (approximately 25 mg), making sure to record the exact weight.
- Homogenize the tissue with 500 µL of CellLytic MT solution in a properly labeled 1.5 ml tube, using a pestle.
- Close the tube and invert the tube several times, making sure it is completely homogenized.
- Spin the tube in a refrigerated microfuge for 10 minutes at max speed.
- Label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
- Transfer the supernatant (clear liquid portion) to the labeled tube and store on ice (if the supernatant is not present or lacking, then repeat step 4).
- Label a fresh 2 ml screw cap tube with the word "Protein", BA, your initials, and today's date.
- Dilute an aliquot of each of your protein samples by adding 15 µL of your protein sample, 15 µL of 0.1% DEPC water, and 1.5 ml Commassie into the 2 ml tube and inverting several times.
- Let the mixture incubate at room temperature for 10 minutes.
- Once the spectrophotometer has been zeroed with a "blank" mixture, transfer your mixture from step 8 into a cuvette, measure the absorbance at 595 nm, and record these values.
- Back-calculate your protein concentration using the following formula: y = 996.52x - 43.64 where x = absorbance and y = protein concentration in µg/ml.
RESULTS:
Absorbance: .974 and .968 for Sample 31
.956 and.941 for Sample 32
| Tissue Sample |
Tissue Weight |
Avg. Absorbance |
Protein Conc. |
| C.gigas gill (# 31) |
0.031 g |
.971 |
923.98 µg/ml |
| C.gigas gill (# 32) |
0.032 g |
.9485 |
901.56 µg/ml |
CONCLUSIONS: I made an error of adding 24 µL of my mastermix to each tube during my conventional PCR portion of the experiment. My absorbance readings were towards the higher end, which meant that my protein concentrations were also high.
REFLECTION: The purpose of this lab was to practice our PCR, qPCR, and protein isolation/quantification techniques, all of which will be used repeatedly throughout our research projects. I made the mistake of adding 24 µL of the mastermix instead of 23 µL during the conventional PCR process because I just looked at 48 µL for a 50 µL mixture and divided in half instead of seeing that the 2 µL of template added does not change whether the total volume is 50 µL or 25 µL. I found the qPCR section sort of confusing because basically everyone had a different number of reactions they were doing and all the numbers were completely different as well as the number of different qPCR tubes that were filled. It made it difficult to compare your methods with others'.
LAB 5: TISSUE DISSECTION, PRIMER RECONSTITUTION, END-POINT PCR
DATE: October 23, 2012LAB SUMMARY: This week's lab officially began our class experiment with the oysters being divvied up and exposed to their respective stressors. The entire class was divided in half with people working on Olympia oysters preparing the Olympia sample tissues and people working on Pacific oysters responsible for preparing the Pacific oyster tissues. The heat shocked oysters were exposed to their respective temperatures for one hour each.
MATERIALS AND METHODS:
Tissue Preparation:
- Label 1.5 ml PCR tubes with "#g1", "#g2", and "#m" where g1 and g2 signify gill tissue 1 and 2 and m signifies mantle tissue and the # is the sample number corresponding to the control or whatever stressor it is associated with from the following list: Control (Pacific) 1-15, Cyclodextrin (Pacific) 16-30, 35°C (Pacific) 31-40, 40°C (Pacific) 41-50, Control (Olympia) 51-60, 35°C (Olympia) 61-70, gradual 35°C (Olympia) 71-80, and P.S. 35°C (Olympia) 81-89.
- Measure and record the length and width of the oysters in millimeters.
- Pass down in a labeled and numbered (e.g. Pac 1, Pac 2, etc.) weigh boat to be shucked.
- Shuck the oysters using the same methods used in Lab 4.
- Using scissors and tweezers, section a small piece (approximately the size of a pencil eraser or slightly smaller) of mantle tissue and insert it into the correctly labeled ("#m" from step 1) tube and place on ice.
- Repeat step 4 with two separate pieces of gill tissue, making sure to put them into tubes labeled "#g1" and "#g2".
Primer Preparation:
- Collect your primer from the TA and make note of its molecular weight and concentration.
- Make a stock (100 µm) solution of your primer by adding the appropriate amount of nuclease free water. The appropriate µL of water to be added is calculated by determining the nm measurement of your primer and moving that number one decimal place to the right (i.e. if 28.5 nm then add 285 µL of water).
- Make a working stock (10 µm) solution of your primer by adding 10 µL of your stock solution made in step 2 to 90 µL of nuclease free water.
RESULTS:
Oyster Measurements
CONCLUSIONS: There really were no quantifiable results to analyze for this week's experiment. Next week's lab should provide us with some quantifiable data as we isolate DNA/RNA from these tissue samples.
REFLECTION: After mulling over it for weeks, we were finally able to get started on our class experiment project. The purpose of this lab was basically initial startup and preparation of the samples we will be testing over the next several weeks. After a few weeks of individually working on experiments, it felt different to work as a group and in teams on the first part of this lab.
LAB 4: REVERSE TRANSCRIPTION AND PRIMER DESIGN & PREP FOR EXPERIMENT
DATE: October 16, 2012LAB SUMMARY: The purpose of this experiment was to use reverse transcription to obtain cDNA from our RNA and get familiar with oyster shucking as we prepare for our upcoming class experiment. We also decided on the experiment each person is going to run as well as design primers to use in next week's lab.
MATERIALS AND METHODS:
Oyster Shucking:
- With oyster in one hand and the shucker in the other, carefully wedge the shucker between the shell opening near its hinge.
- Wiggle the shucker back and forth as you insert it further into the shell.
- Once the shucker is firmly into the shell, slice it open going horizontally along the opening.
- Discard the top shell and separate the oyster from its bottom shell by slicing off the adductor muscle.
Reverse Transcription Protocol:
- Let RNA thaw and then mix so that there is liquid only on the bottom of the tube.
- Label a 0.2 ml PCR tube with "cDNA" and your initials and combine 5 µL of your RNA + 1 µL of oligo dT + 4 µL of nuclease free water in the tube.
- Incubate the mixture for 5 minutes at 70°C then immediately transfer to ice.
- Add to the tube 5 of M-MLV 5X Reaction Buffer, 5 µL of dNTPs, 1 µL of M-MLV RT, and 4 µL of nuclease free water then let it incubate for 60 minutes at 42°C.
- Inactivate by heating at 70°C for 3 minutes, spin down the sample in a centrifuge, then store on ice.
Primer Design:
- After determining the gene you want to examine, research the gene online to find a good sequence of it.
- Using the NCBI website and its tools, design a primer of length 18-22 for the selected gene which should be set at 80-200 base pairs with a melting temperature of 60°C +/- 3 (you want to have high G/C content but want to avoid high G/C stretches).
- Copy and paste the forward and reverse sequences of the designed primer into the primer database.
RESULTS: The organism I chose to focus on is the Pacific Oyster (Crassostrea gigas). I am going to be quantifying any changes in the HSC70 gene in response to sudden temperature spikes - one hour exposure to 35°C and 40°C. Below is the primer I designed for this purpose:
Forward Primer - AGACACAGAAAGGCTGGTCG
Reverse Primer - ATTCTGGCAAGACGGGGATT
CONCLUSIONS: There really were no quantifiable results to analyze for this week's experiment.
REFLECTION: The purpose of this lab was to familiarize ourselves with the RNA to cDNA process as well as to finalize our preparations for the upcoming class experiment. Oyster shucking was not as difficult as I had anticipated.
LAB 3: RNA ISOLATION, PART 2
DATE: October 9, 2012LAB SUMMARY: The purpose of this experiment was to finish isolation of RNA, which was started in the previous lab. Also, during wait periods while the sample was in the centrifuge, we discussed the group experiments we wanted to do as a class.
MATERIALS AND METHODS:
RNA Extraction Protocol:
- Turn on heating block to 55°C.
- Incubate your tissue sample at room temperature for 5 minutes.
- After adding 200 µL of chloroform to the sample, vortex for 30 seconds until the solution becomes a milky emulsion.
- Incubate the tube at room temp for 5 minutes then spin it in a refrigerate microfuge for 15 minutes at max speed. Carefully transfer most of ONLY the aqueous phase (top, clear liquid) to a fresh tube and dispose of the rest in the chloroform waste jar.
- Add 500 µL of isopropanol and mix by inverting the tube several times until the solution appears uniform.
- After incubating at room temp for 10 minutes, spin in refrigerated microfuge at max speed for 8 mins. (a small, white pellet should form).
- Remove the supernatant (liquid portion), add 1 mL of 75% EtOH to the remaining pellet, then vortex briefly.
- Spin in refrigerated microfuge at 7500g for 5 mins. (the setting was only able to be changed by 200g at a time so it was actually set at 7600g for this step).
- Carefully remove supernatant and ONLY the supernatant, making sure the pellet stays intact.
- Briefly (~15 s) spin the tube in the centrifuge to pool residual EtOH and remove it using a small pipette tip (using a clean KimWipe to soak up any residual EtOH inside the tube is ok as long as the pellet is not touched)
- Let the tube sit (no more than 5 minutes) with the top open to allow any remaining EtOH to evaporate.
- Dissolve the dry pellet in 100 µL of 0.1% DEPC-H2O and incubate the solution in 55°C for 5 mins.
- Remove the tube from the heat and flick a few times to mix the sample then place on ice.
RNA Quantification:
- Pipette 2 µL of 0.1% DEPC-H2O onto the Nanodrop pedestal and lower the arm.
- Click "Blank", to zero the instrument.
- Pipette 2 µL of your RNA sample onto the Nanodrop pedestal and lower the arm.
- Click "Measure" and record your RNA concentration and ratios.
- Raise the arm and wipe off the instrument using a KimWipe.
- Give your labeled sample to the TA for cold storage.
RESULTS:
Tissue Sample - Gill of Pacific Oyster
Tissue Weight - 0.073 g
| Absorbance |
10.517 |
| A260/280 Ratio |
2.01 |
| A260/230 Ratio |
1.51 |
| RNA Concentration |
635.3 ng/µL |
CONCLUSIONS: The results obtained were again consistent with what is to be expected from these Nanodrop readings. For a clean RNA , the A260/280 range should have been 1.8-2.0 and the A260/230 in the 1.5-2.0 range. My A260/280 ratio was slightly above that range, possibly due to excess EtOH still being in the sample. However, my A260/230 ratio was 1.51 and fell in the ideal range for a clean RNA sample.
REFLECTION: The purpose of this experiment was to finish isolating RNA, which was started last week, so that we can continue to familiarize ourselves with these procedures. Also, it gave us a chance to compare/contrast DNA/RNA isolation techniques. The procedures performed in lab were used to measure RNA concentration and the ratios, which can be used to measure changes in gene expression of an organism. The entire experiment was pretty straightforward and having already done similar procedures last week made it run smoother and easier this week. Also, I appreciated the fact that ideal range for both ratios were provided for this experiment.
LAB 2: DNA ISOLATION; INITIATE RNA ISOLATION
DATE: October 2, 2012LAB SUMMARY: The purpose of this experiment was to isolate DNA and start the isolation of RNA from oyster tissue. A sample from the gill tissue of a Pacific Oyster was used to isolate DNA and RNA (only begun) from it. The tissues had to be kept cool in ice and the process relied on homogenizing them with various solutions (covered in detail below in the Materials and Methods section) and centrifuging afterwards.
MATERIALS AND METHODS:
RNA Isolation Protocol:
- Select a tissue sample to be used and separate 25-50 mg for DNA isolation and 50-100 mg for RNA isolation using a razor and tweezers.
- Weigh the tissue separated on a scale and record the exact weight (0.033 g for DNA and 0.073 g for RNA). Return any leftover tissue from the original sample to your TA.
- Label the 1.5 mL snap cap tubes containing the tissues with initials, date, and "RNA" and "DNA" so as to not confuse which samples are to be used for which process. Keep the tissues on ice.
- Add 500 µL of TriReagent to the snap cap tube containing the tissue and homogenize the sample using a disposable pestle and a vortex. This may take a few minutes so be patient.
- After the sample is completely homogenized, add another 500 µL of TriReagent and vortex it for 15 seconds.
- Give your labeled RNA sample to the TA for cold storage.
DNA Isolation (DNazol):
- Homogenize your tissue sample by mixing with 0.5 mL of DNazol and using a sterile pestle. Afterwards add another 0.5 mL of DNazol and mix well.
- Let the sample incubate at room temperature for approximately 5 minutes (it was closer to 8 or 9 minutes during my experiment).
- Spin your sample at 10,000 x g for 10 minutes using the microcentrifuge. Make sure to double check the settings for correct numbers and units prior to starting.
- Transfer your supernatant (the liquid portion) to a new, labeled tube.
- Add 0.5 mL of 100% ethanol to the sample and mix by inverting the tube 5-8 times.
- Let the sample sit in room temperature for 1 minute, at which time a cloudy precipitate should form.
- Centrifuge the tube at 5000 x g for 5 minutes.
- Remove the DNA and put it in a new tube using your pipette.
- Let the sample sit at room temperature for 1 minute and remove the lysate (liquid that is not DNA).
- Wash the DNA with 1 mL of 75% ethanol (invert the tube 6 times) and let it sit for 1 minute.
- Empty out the ethanol and repeat step 10 one more time.
- Empty out the ethanol and remove as much remaining ethanol as you can using a small pipette.
- Add 300 µL of 0.1% DEPC water to the DNA and pipette up and down until it homogenizes.
DNA Quantification:
- Pipette 2 µL of 0.1% DEPC water onto the Nanodrop pedestal and lower the arm.
- Select "dsDNA" from the pulldown menu (steps 1 and 2 were not done because they only need to be done once per class and I was not the first one to use the machine).
- Click "Blank" to zero the instrument.
- Pipette 2 µL of your DNA sample onto the Nanodrop pedestal and lower the arm.
- Click "Measure" and record the DNA concentration (337.9 ng/µL), A260/280 (1.83), A260/230 (0.60), and absorbance (11.219).
- Raise the arm and wipe off your sample with a KimWipe.
- Give your labeled DNA sample to the TA for cold storage.
RESULTS:
Tissue Sample - Gill of Pacific Oyster
| Isolation Process |
Tissue Weight (g) |
| RNA |
0.073 |
| DNA |
0.033 |
| Absorbance |
11.219 |
| A260/280 Ratio |
1.83 |
| A260/230 Ratio |
0.60 |
| DNA Concentration |
337.9 ng/µL |
CONCLUSIONS: The readings of my DNA sample that were obtained from the Nanodrop were generally consistent with what is to be expected. It is stated that a purified DNA should have A260/280 ratio of 1.7-1.9 to be considered good quality DNA. Mine was 1.83 which falls between those numbers. I did see a spike at the beginning of the graph prior to the 230 indicating a significant presence of either ethanol or other solvents. I believe that was due to the sample being part of the gill tissue (salts) rather than ethanol since I was careful not to leave behind any excess ethanol.
REFLECTION: The purpose of this lab was to practice and become familiar with the process of isolating DNA from a sample tissue. Also, by examining and remembering the different ratios obtained from different tissues it will be easier to determine in the future what part of an organism a tissue sample was collected without having to know all the details. The procedures performed in lab were used to measure the absorbance ratios with proteins absorbing at 280, DNA at 260, and ethanol or other solvents at 230 as well as DNA concentration measured in ng per uL. Isolating DNA can be useful in many ways. One scenario I can think of is combining this method with PCR to amplify a region in order to study a gene or a specific sequence of interest in an organism. This method is practical since this same technique can be used on most organisms, not just shellfish. There wasn't any procedure that was unclear but I wish an acceptable range of values for the A260/230 ratio would have been provided, as was for the A260/280.