Analyzing Gel Electrophoresis
ELECTROPHORESIS PROCEDURE
- Place gel in gel box and fill with 1x TAE buffer (to fully cover wells)
- Remove combs from wells
- Load 5uL 100bp ladder in far left lane
- Load 25uL of your PCR sample into the gel
- Run gel at ~ 110V for ~ 20 min
- Visualize the gel on the UV transilluminator
Making an Agarose Gel
Supplies and Equipment:- Micropipettes (1-1000 μl)
- Sterile filter pipette tips (1-1000 μl)
- Tip waste jar
- 1L flask
- agarose
- 1X TAE
- Ethidium bromide
- Microwave
- Gel rigs
- Kimwipes
- Lab coat
- Safety glasses
- gloves
AGAROSE GEL POURING PROCEDURE
- Weigh 2g of agarose and mix with 150mL 1x TAE in a 1L flask
- Microwave solution for ~ 3 minutes. Keep an eye on the solution so that it does not boil over. You want the solution to be clear - no precipitate and no bubbles.
- Cool solution (you should be able to touch the flask for a few seconds), then add 12uL ethidium bromide(EtBr). WARNING: EtBr is a carcinogen be sure to wear gloves and appropriately dispose tip waste.
- Mix thoroughly by swirling, then pour into gel tray.
- Add gel combs. Using a clean pipet tip, pop any bubbles that could get in the way of your PCR product.
11/21/13
Sara was in lab today and attempted to perform qPCR. We were informed that our primers did not work.
11/19/13
Tissue samples were processed following the RNA extraction protocol provided (see below). I was responsible for processing all gill samples for each treatment for a total of 8 reactions. cDNA was also made today for gill samples and eye samples. We followed the cDNA protocol.
Nanodrop restuls:
11/17/13
Samples were taken from 2 specimens from each treatment on Nov.16th. Tissues were extracted from eyes, mantle, and gill tissue. There were 18 total tissue extractions performed. These will be processed during the next lab period. Tissues were stored in -80 degree Celsius freezer.
Note* Preliminary Nanodrop results were set to nucleic acid instead of RNA.
11/13/13
Experiment was set up. There were 9 scallops placed in both dark treatments, 10 scallops in the light control treatment, and 11 in the light/pH treatment. To simulate dark, the scallops were enclosed in a tarp. To simulate light, a light was placed above the containers and is being left on. Air stoned were placed in each treatment.
4 identical containers were used and filled with equal amounts of seawater. The pH of the treatment seawater was measured twice and read 7.41 and 7.46, respectively. The pH of the control water was also measured twice and read 7.81 and 7.87 respectively. The differences in pH readings likely occurred because the container differed in each reading, the former pH being in plastic and the later being in glass.
11/14/13
RNA extraction ensued (see protocol below). RNA was also quantified using a Nanodrop.
Table 1.) RNA quantification performed with Nanodrop. Tissue samples were obtained from C.Rubida under two treatments light (Li) and no light (NL). Tissues were samples from gills (G), mantle (M), eye (E), and adductor muscle (A).
| Li-G |
Li-M |
Li-E |
Li-A |
NL-G |
NL-M |
NL-E |
NL-A |
|
| lambda |
230 |
230 |
230 |
230 |
230 |
230 |
230 |
230 |
| Abs |
1.142 |
1.260 |
0.683 |
4.453 |
0.912 |
0.977 |
0.541 |
0.947 |
| 260/280 |
1.94 |
1.73 |
1.59 |
1.94 |
1.64 |
1.66 |
1.68 |
1.76 |
| 260/230 |
2.22 |
1.89 |
1.11 |
0.55 |
2.13 |
1.75 |
1.71 |
0.95 |
| ng/uL |
125.9 |
95.3 |
30.3 |
97.4 |
77.6 |
68.5 |
37.0 |
36.0 |
11/5/13
 |
| Sample size is approx. 40 organisms. Chlamys rubida |
Primers have been received:
Forward 1: SF_C.rubida_Gq-opsinB_355F
5'- ATCTGGCGGTCAGTGACCTCATCTTCTC-3'
Reverse: SF_C.rubida_Gq-opsinB_1100R
5'GATGGCTGAGAGCATATACCATGG-3'
Forward 2: SF_C.rubida_Gq-opsinB_RhOF2
5'-CATGAGACGCCAGGTCCACC-3'
We reconstituted primers with de-ionized water by multiplying the molecular weight. The amount of DI water added was 384mL, 291mL, and 259mL respectively. We vortexed these mixtures. The primers were then diluted using a 1:10. 100mL of each was made and placed in separate labeled tubes.These were labeled 355F-work, 1100R-work, and RhOF2-work. We vortexed these mixtures briefly.
From the tissue samples we performed RNA isolation (see protocol below). Samples were stored at -80 degree celsius. Set up did not occur today.
11/1/13
Went in and extracted 6 tissue samples from each experimental condition: mantle/eyes, abductor muscle, and gills.
LI-E NL-E
LI-G NL-G
LI-M NL-M
LI-A NL-A
The tubes were labeled as above. LI is short-hand for light, similarly NL is short-hand for no light. We are concerned that the mantle and eye tissue samples with be highly correlated. We were unaware prior to extraction how little tissue sample could be isolated from the eye tissue alone being that it is nearly inseparable from mantle tissue without proper dissecting scopes. The tubes were placed in a freezer at -80 degrees celsius.
10/31/13
Lab 5: Prep for Experiment and primers
Summary:
- Mock-up Experiment to prepare for the initiation of experiments
- Design and order primers
Reflections
We were able to obtain 40 scallops. They have been identified as Chlamys rubida, pink scallops. These species are approximately 5 cm in diameter. We learned that this species is a deep water species, occupying habitat at roughly 300 meters depth. We also modified our procedure. We have decided to do a preliminary test on two scallops in differing habitat conditions to determine if this species produces opsin in a positive correlation with light. Therefore, one scallop was placed in seawater with an air stone in a tank 50x25cm that is being exposed to light for a continuous 3 days. The other scallop was placed in the same size tank, also with an air stone but this scallop is being exposed to no light. We wrapped this tank in tarp that was double layered and on a shelf that was beneath the light-scallop. Friday Nov 1., we will be going in to obtain tissue samples from both specimen's mantle, gills, and eyes. We will then store this tissue in a super-chilled freezer at -80 deg C for 5 days before we being to analyze gene expression of opsin.
The overall experimental design has also been modified during this lab. This was after discussion some of the logistics of our experiment with Prof. Stevens and Claire. The experiment is now as follows: We will have four identical tanks each filled with eight specimens, an air stone, and sea water. Of the four tanks, two will be exposed to light and two will be in darkness. Furthermore, one tank from each lighting condition will have the pH lowered to see the effects of pH on opsin gene expression.
10/27
Lab 4: Protein SDS/Page and Western blot, analyze conventional PCR and qPCR
Summary:
- Run extracted total protein from previous labs on SDS-PAGE Gel/Western Blot
- Run conventional PCR samples on an agarose gel
- Download qPCR data and discuss analysis
Materials and Methods
PCR Protocol Electrophoresis Procedure
- Place gel in gel box and fill with 1x TAE buffer (to fully cover wells)
- Remove combs from wells
- Load 7uL 100bp ladder in far left lane
- Add 2uL of loading dye to PCR sample. Mix gently
- Load 20uL of your PCR sample into the gel: well 11 was used
- Run gel at ~ 100V for ~ 1hr
- Visualize the gel on the UV transilluminator
Protein Extraction and Analysis pt. 2:
Supplies and Reagents
- micropipettes (1-1000 μL)
- sterile filter pipette tips (1-1000 μL)
- sterile gel loading tips
- 1.5 mL screw cap tubes
- microcentrifuge tube rack
- lab coats
- safety glasses
- gloves
- lab pen
- timers
- heating block with water bath
- tube "floatie" (8 tube capacity)
- glass container for boiling water that can accommodate "floatie"
- protein gel box (SR provided)
- SDS/PAGE gels
- gel loading tips
- trays for staining gels
- power supply
- platform rocker/shaker
- plastic wrap
- 2X SDS reducing sample buffer
- protein ladder marker
- gel running buffer
- light box
- digital camera
SDS-PAGE Protocol
- Begin boiling water on hot plate.
- In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein stock and 15uL of 2X Reducing Sample Buffer. Return your protein stock to the box in the -20C freezer labeled protein samples.
- Mix sample by flicking.
- Boil sample for 5 mins.
- When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
- Slowly load your entire sample into the appropriate well using a gel loading tip: well 11 was used
- Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
- Turn power supply on and set voltage to 150V. Run for 45mins.
- Turn off power supply and disconnect gel box from power supply.
- Remove lid from gel box.
- Disengage the tension wedge.
- Remove gel from gel box.
- Carefully crack open cassette to expose gel.
- Trim wells at top of gel with razor blad
- Notch a designated corner of the gel to help you remember the correct orientation of the gel *Note: ladder was on the opposite side of the cut. Sample was on gel 1*
WesternBreeze Chromogenic Western Blot Immunodetection
Supplies and Reagents
- Nanopure water
- gel staining tray
- Blocking Solution
- rotary shaker
- Primary Antibody Solution
- Antibody Wash
- Secondary Antibody Solution
- Chromogenic Substrate
- timers
- lab coats
- safety goggles
- gloves
- SDS-PAGE gel
- Tris-Glycine transfer buffer
- filter paper
- nitrocellulose membrane
- semi-dry transfer station
WesternBreeze Chromogenic Western Blot Immunodetection
Protocol
- Soak the filter paper, membrane and gel in Tris-Glycine Transfer Buffer for 15 minutes.
- Assemble the blotting sandwich in the semi-dry blotting appartus:
- Anode (+++)
- filter paper
- membrane
- gel
- filter paper
- cathode (---)
- Transfer the blot for 30 minutes at 20V
- Remove the gel from the sandwich and rinse off adhering pieces of gel with transfer buffer.
- Wash membrane 2 times, for 5 minutes each, with 20 mL of pure water.
- Put the membrane in the plastic box and add 10 mL of Blocking Solution. Cover and incubate 30 min on a rotary shaker set at 1 revolution/second.
- * TA will do the rest of the steps. After class tomorrow you can come and see your results.*
- Decant liquid.
- Rinse the membrane with 20 mL of water for 5 minutes, then decant. Repeat.
- Incubate the membrane in 10 mL of Primary Antibody Solution. Decant the solution.
- Rinse the membrane with 20 mL of Antibody Wash for 5 minutes, then decant. Repeat 3 times.
- Incubate the membrane in 10 mL of Secondary Antibody Solution for 30 minutes. Decant.
- Wash the membrane for 5 minutes with 20 mL of Antibody wash, then decant. Repeat 3 times.
- Rinse the membrane with 20 mL of pure water for 2 minutes, then decant. Repeat twice.
- Incubate the membrane in 5 mL of Chromogenic Substrate until a purple band appears. This will occur between 1-60 minutes after adding the Chromogenic Substrate.
- Dry the membrane on a clean piece of filter paper to the open air.
Results
| Well 11 was used |
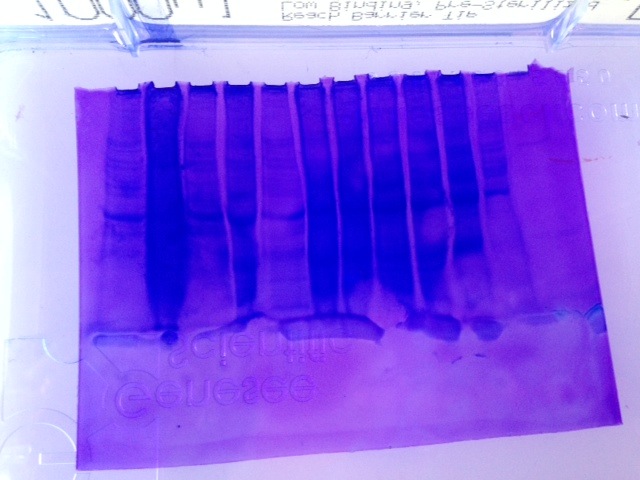
Fig 1.) Western Blot
Fig 2.) SDS-PAGE Gel * Note that the gel was reversed. Ladder is far right, well 12 is far left*
My sample was in well 11 which is second from the left.
Conclusions
The results from PCR (lanel 11) displayed a solid fluorescing band at 300 bp which indicated that amplification of cDNA was successful and thus what was expected. The results from the Western Blot were unclear. I need to see the sample again in person to clarify which was lane 11. The results from the SDS-PAGE gel were underwhelming and unclear (Fig. 2) Well 11-second from the left- shows a dark, semi-solid band indicative of a lot of protein and perhaps too much. Less sample should have been loaded initially and could have been prevented had protein quantification been done in lab 3.
Reflections
The purpose of this lab was to gain an understanding of how to analyze proteins, read PCR results from an agarose gel as well was perform Western Blot.
10/20/13
Lab 3: qPCR and protein extraction
Purpose
- Previously done: Reverse transcribe RNA to cDNA
- Perform qPCR on cDNA samples
- Run qPCR samples on agarose gel
Summary
The purpose of this lab was to become familiar with performing and analyzing quantitative PCR (qPCR). qPCR is a laboratory technique and is used to amplify and simultaneously quantify a targeted DNA molecule. It enables both detection and quantification of one or more specific sequences in a DNA sample. The procedure is similar to that of PCR except its key feature is that the amplified DNA is detected as the reaction progresses in real time.
!!Safety!!
- Wear clean gloves! Proteases are present on you skin and are detrimental to the integrity of your samples.
Materials and Methods
qPCR Supplies and Equipment
- PCR Plates (white); optically clear caps
- 1.5 ml microfuge tubes (RNAse free)
- Nuclease Free water
- filter tips
- Opticon thermal cycler
- kim wipes
- 2x Immomix Master Mix
- SYTO-13 Dye
- microfuge tube racks
- ice buckets
- timers
- cDNA samples
qPCR Protocol
Each template of cDNA will be run in duplicate. In addition to one negative controls (no template) will be run. Four total reactions will be taking place
1. Prepare master mix: Prepare enough master mix for your number of reactions +1 to ensure sufficient volume recovery.
For one 25μl reaction volume: Note this needs to be multiplied by four to ensure enough master mix.
| Component |
Volume |
Final Conc. |
| Master Mix (SsoFast EvaGreen supermix) |
12.5µL |
1x |
| upstream primer, 10μM |
0.5μl |
2.5μM |
| downstream primer, 10μM |
0.5μl |
2.5μM |
| Ultra Pure Water |
10.5uL |
NA |
2. Add 24 uL master mix to each well of white PCR plate
3. Thaw cDNA samples.
4. Add 1uL cDNA template to each reaction. Except negative control!
5. Add 1uL of ultra pure water to the negative control wells.
6. Cap the wells securely. Label.
7. Ensure the lids are clean and place strips on ice. (I like to wipe the lids with a clean kimwipe)
8. Load the plate, verify the PCR conditions and start the run (this will be done by your TA).
PCR Conditions
1. 95°C for 10 minutes2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 15 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s
Protein Extraction and Analysis pt.1supplies and equipment: Pacific Oyster-HSP 70 gene
- micropipettes (1-1000uL)
- sterile filter pipette tips (1-1000uL)
- sterile (RNase free) 1.5mL microcentrifuge tubes
- sterile 2 mL screw cap microcentrifuge tubes
- sterile disposable pestles
- spectrophotometer
- cuvettes for spectrophotometer
- microcentrifuge (refrigerated) or in fridge
- ice buckets
- gloves
- Kim wipes
- lab pens
- safety glasses
- CelLytic MT Cell Lysis Reagent (with Protease Inhibitor Cocktail added)
- Coomassie Protein Assay Reagent
- DI water
Protein Extraction Protocol
- Label the snap cap tube containing your tissue sample with your initials and the date using a lab marker.
- Add 500 ul of CellLytic MT solution to the 1.5mL snap cap tube containing your cut piece of frozen tissue.
- Homogenize the tissue with a sterile pestle.
- Close the tube. Invert the tube several times.
- Spin the tube in a refrigerated microfuge for 10 mins at max speed.
- While spinning, label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
- Carefully transfer supernatant to labeled tube and store tube on ice
Results
Need to clarify with Claire.
Conclusion
See above
Reflection
10/13/13
Lab 2: RNA isolation pt. 2
Purpose
- Complete RNA isolation procedure
- Prepare RNA for transcription to cDNA: Reverse transcription
Summary
The purpose of this lab is to obtain high-quality RNA which is the first and often the most critical step in performing many molecular techniques such as PCR and cDNA library construction. This is made possible through reverse transcriptase which transcribes RNA into single-stranded cDNA. The resulting cDNA is more stable than RNA and can undergo processes such as PCR amplification and has downstream application such as gene expression analysis.
!!Safety Considerations!!
- Wear safety glasses! Several of the reagents being worked with in lab are irritants.
- Wear clean gloves! For your own safety as well as the integrity of your RNA samples, you must wear gloves throughout this week's lab. Phenol and chloroform are nasty, caustic chemicals, so gloves are necessary when handling anything that comes in contact with either reagent. Additionally, RNases are constantly secreted from your skin and can easily enter, and subsequently degrade, your RNA sample.
Materials and Methods
All test tubes were labeled with initials (AF), and the date 10.8.13
Supplies and Reagents
- micropipettes (1-1000 μL)
- sterile filter pipette tips (1-1000 μL)
- 1.5 mL microcentrifuge tubes
- PCR Tubes
- RNA samples
- Thermocycler
- microcentrifuge + PCR tube rack
- lab coats
- safety glasses
- gloves
- lab pen
- timers
- ice buckets
- phenol/chloroform waste containers (liquid/solid)
- vortex
- hot water bath
- chloroform
- isopropanol
- 75% ethanol (EtOH)
- 0.1% DEPC treated water
- Oligo dT
- Master Mix: 14uL
- M-MLV 5X Reaction Buffer (5uL)
- dNTPs (5uL)
- M-MLV-Reverse Transcriptase (1uL)
- Nuclease free water (4uL)
RNA Extraction Protocol
- Add remaining 500uL TriReagent.
- Turn on heating block to 55° C - this was done prior to the start of lab.
- Incubate homogenized tissue from Lab 1 at room temperature for 5 minutes.
- In the fume hood, add 200uL of chloroform. Close lid quickly.
- Vortex vigorously for 30 seconds. Solution should become milky.
- Incubate tube for another 5 minutes at room temperature.
- Spine tube in refrigerated microfuge for 15 minutes at max speed. Remove gently
- Transfer the aqueous phase to a new microfuge tube. Do not transfer any of the interphase.
- Add 500uL isopropanol to the new tube containing RNA. Close tube.
- Mix by inverting several times until solution appears uniform.
- Incubate at room temperature for 10 minutes.
- Spin in refrigerated microfuge for 8 minutes at max speed. Make sure hinge side points out.
- Remove supernatant from pellet. Discard into separate microfuge tube.
- Add 1000uL of 75% EtOH to pellet. Vortex briefly.
- Spin in refrigerated microfuge for 5 minutes at 7500g.
- Remove and discard supernatant.
- Briefly spin tube and remove pooled EtOH with fine micropipette.
- Leave tube open and allow pellet to dry at room temperature for ~3 minutes.
- Re-suspend pellet in 100uL of 0.1% DEPC water. Pipet up and down until pellet is dissolved. Note: 1000uL of 0.1% DEPC water was added instead of 100uL.
- Incubate tube at 55° C for 5 minutes.
- Remove tube from heat, flick a few times to mix. Place sample on ice.
- Obtain a 0.5 mL tube. Label with initials and date
- Place 5uL of RNA sample, 1uL of oligo dT, 4uL of RNase-free water. Close lid
- Incubate at 70° C for five minute. Briefly centrifuge.
- Add 14uL of Master Mix. Store on ice.
Reflection
The purpose of this lab was to obtain high quality RNA and begin the steps necessary to transcribe RNA to cDNA which is used for PCR and gene analysis processes. I need to be clear about how many mL of 0.1% DEPC water to use so as to not over-dilute the RNA sample when is comes to personal experiments. Additionally, none of the procedure was of concern or caused confusion as these techniques have been employed in other courses such as FISH 340.
10/6/13 (Seahawks vs. Colts):
Lab1: DNA isolation; initiate RNA isolation

Purpose
- Select tissue for DNA, RNA and protein extraction.
- Start RNA extractions (add Tri-Reagent to samples, homogenize, and then re-freeze for next week).
- Isolate and quantify DNA from tissue.
Summary
The purpose of this lab was to become familiar with molecular lab techniques, namely the extraction of DNA and RNA. We are interested in looking at DNA to see what genes are present in the population as well as see how these genes change coincidently with environmental gradients. The reason we are interested in looking at RNA is to look more specifically at the genes present in an individual within a population.The tissue sample for DNA extraction was provided and came from the gills of an Olympic Oyster while the tissue sample provided for RNA extraction came from the gills of a Pacific Oyster. The technique used for DNA extraction involved dissolving the tissue sample in DNazol to obtain a homogenous solution. The DNA was then precipitated and isolated out of solution using several washes of ethanol. The amount of DNA that was obtained was quantified with Nanodrop. The technique for isolating RNA is similar to that of DNA except TriReagent was used to separate RNA from other cellular components.The RNA sample was re-frozen. In the next lab we will isolate the RNA.
!!Safety Considerations!!
- Wear safety glasses! Several of the reagents being worked with in lab are irritants.
- Wear clean gloves! For safety as well as the integrity of RNA samples. Phenol and chloroform are nasty, caustic chemicals, so gloves are necessary when handling anything that comes in contact with either reagent. Additionally, RNases are constantly secreted from your skin and can easily enter, and subsequently degrade, your RNA sample.
Materials and Methods
All test tubes were labeled with initials (AF), the date, and either PAC 47-G for RNA or OLY 87-G for DNA.
RNA Supplies and Reagents
- micropipettes (1-1000uL)
- sterile filter pipette tips (1-1000uL)
- sterile (RNase free) 1.5mL microcentrifuge tubes
- sterile disposable pestles
- vortex
- ice buckets
- gloves
- lab pens
- safety glasses
- TriReagent
RNA Isolation Protocol
- Label the snap cap tube containing tissue sample with your initials (AF) and the date (10.3.13). Keep the sample stored on ice until you are ready for homogenization.
- Add 500uL of TriReagent to the 1.5mL snap cap tube containing your tissue. Store on ice.
- Carefully homogenize the tissue using a disposable pestle.
- (This step was not completed and was saved for the next lab per Dr. Stevens). After the sample is completely homogenized, add an additional 500uL of TriReagent to the tube and close the tube tightly.
- Vortex vigorously for 15 seconds-the sample was actually vortexed for 25 seconds.
- Label tissue sample to and at -80ºC for one week.
DNA Supplies and Reagents
- micropipettes (1-1000 µL)
- sterile filter pipette tips (1-1000 µL)
- 1.5 mL microfuge tubes
- microcentrifuge tube rack
- microcentrifuge (room temperature)
- vortexer
- DNazol
- 100% ethanol
- 75% ethanol
- 0.1% DEPC water
- kim wipes
- Nanodrop
DNazol Extraction Protocol (Adapted from MRC manual)
- Using a sterile pestle, homogenize your tissue sample in 0.5 mL of DNazol in a 1.5 mL sterile microfuge tube. After the tissue is homogenized, add 0.5 mL more of DNazol and mix well.
- Let your sample incubate for 5 minutes at room temperature.
- Spin your sample at 10,000 x g (room temp) for 10 minutes. Pellet should form.
- Extract excess liquid with pipette from pellet and discard.
- Add 0.5 mL of 100 % ethanol to your sample.
- Mix sample by inverting 5-8 times.
- Store sample at room temperature for 1 minute.
- DNA should form a cloudy precipitate. Extract excess liquid with pipette and discard
- Let sample sit at room temperature for 1 minute. Remove excess liquid-not DNA- and discard.
- Wash DNA with with 1 mL of 75% ethanol: Pipette the ethanol into your DNA tube, invert 6 times, and let sit for 1 minute. Remove the ethanol from the tube and repeat.
- Remove excess ethanol with pipette and discard.
- Add 300 µL of 0.1% DEPC water to your DNA and pipette up and down multiple times to dissolve.
- Bring DNA sample to Nanodrop to quantify.
DNA Quanitification
- Pipette 2µL of your DNA sample onto the Nanodrop pedestal and lower the arm
- Click "Measure". Record your DNA concentration (ng/µL), A260/280 ratio and A260/230 ratio. NOTE: The Nanodrop uses the Beer-Lambert Law to calculate DNA concentration for you.
- Raise the arm and wipe off you sample with a KimWipe
- Clearly label your stock DNA sample with the word "DNA", source organism/tissue, your initials (AF), today's date (10.3.13) and the concentration in ug/uL.
- Store sample at -20ºC.
Results
These results were obtained from the Nanodrop.
Lambda: 230
Abs. 0.913
A-260 10 mm path: 0.259
A-280 10 mm path: 0.143
260/280: 1.82
ng/µL: 13.0
Conclusion
The results from DNA quantification were sufficient. The A260/A280 ratio that was obtained from the Nanodrop for my sample was 1.82. This indicates that the DNA sample was of good quality. (A purified DNA sample should a A260/A280 of 1.7-1.9). Based on these results we will analyze the DNA and identify proteins of interest.
Reflections
The purpose of this lab was to become familiar with molecular lab techniques. We are interested in looking at DNA to see what genes are present in the population as well as see how these genes are altered with environmental gradients. The reason we are interested in looking at RNA is to look more specifically at the genes present in an individual within a population. The greater implications of this type of research is to understand how organisms and populations adapt to environmental stressors (i.e temperature fluctuations for intertidal species such as oysters). None of the procedure was of concern or caused confusion as these techniques have been employed in other courses such as FISH 340.
Heat shock protein 70 (HSP 70)- may help explain why Pacific oyster can tolerate high temperatures as HSP family is expanded and highly expressed when in high temperatureInhibitors of apoptosis proteins (IAPs)- anti-apoptosis system may be critical for oyster’s amazing endurance to air exposure and other stresses.
- Possible sources for research paper:
http://www.genomics.cn/en/news/show_news?nid=99223