Just for old times sake, I decided to post here. I'm building my very own website!
Yay!
6/30/2010 Wednesday
read the protocol for the new technique for measuring DNA methylation.
5/27/2010 Thursday
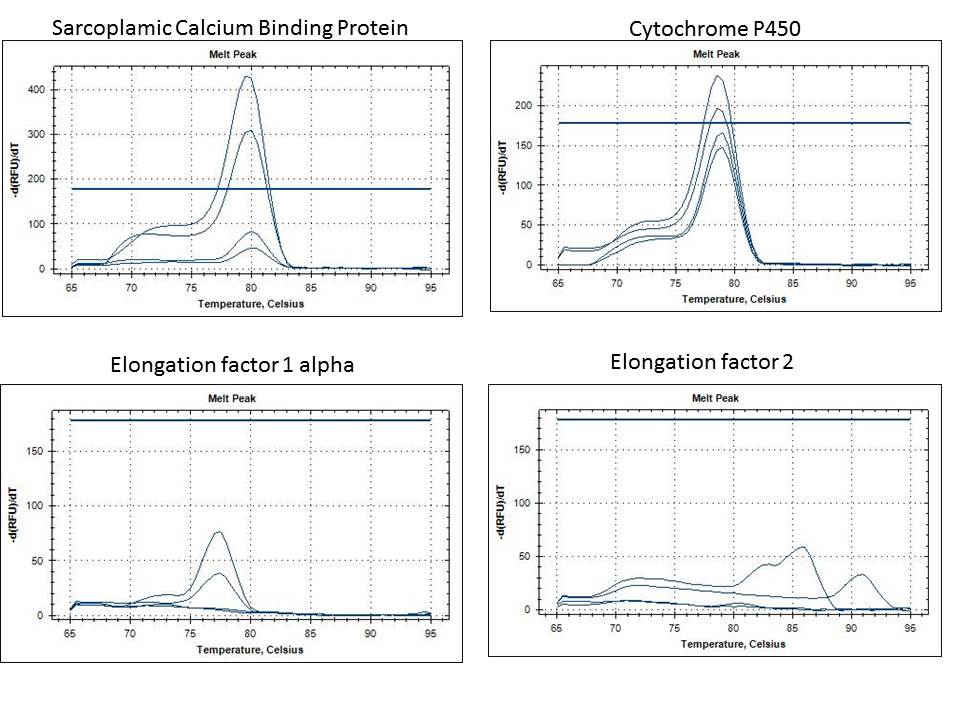
Results from qPCR are inconclusive
Well layout was like this:
Control: U H M H2O
Treated: U H M H2O
Melt curve
Nothing in the Control Undigested.
5/26/2010 Wednesday
Running a qPCR on the digested samples from 5/20/2010 to see if I can quantify any of the information from I have gathered from the gel
2x immomix 12.5 µL
Pf/Pr 1.0 µL
Syto-13 1.0 µL
H20 9.5 µL
Used the SYBR setting on the opticon.
Results:
5/21/2010 Friday
Running the digested DNA onto a gel
Not the results we expected or wanted to see, but the undigested looks to be where it should be.
Next step: Because the concentration is so high (and the amount put into each tube in the control case was so low) next time I may dilute the DNA 1 to 5 so I am pi-petting a greater volume into the tube. qPCR will quantify the results.
Another thing would be to use 2µg of DNA instead of just one.
5/20/2010 Thursday
Ran steps 2-5 using the setup under "next step" from yesterday. This time, after spec the samples I added Hepes (because the NaOH solution is alkaline (pH about 11.65)), this will ensure the DNA does not degrade in the high pH.
The concentrations of DNA were high:
So to make sure the NaOH was not absorbing at 260/280, i spec'd the 8mM NaOH
But the concentration here is close enough to 0 that we can assume this is not an issue.
I took 5µL from each of the isolated DNA and added 1µL of 5x loading dye and ran them onto a gel, results:
The gel looks good! The brightness of the band confirms that the concentration of DNA is quite high, and there is no random 900bp band on the gel!
Now to run restriction enzymes again:
Control: .930µg/mL DNA x _µL = 1µg about 1.075µL
Treated: .751µg/mL DNA x _µL = 1µg about 1.33µL
0.5µL MSPI uses buffer #4
1.0µL HPAII uses buffer #1
I made 50 µL solutions and put them to digest at 37° over night.
Next Step: Tomorrow I will heat stop the samples
HPAII at 65 degrees for 20 min
MSPI at 80 degrees for 20 min
5/19/2010 Wednesday
DNA isolation using another kit, this time not by Qiagen protocol as follows:
1. add 1000µL of DNAzol per 25-50mg of tissue homogenize until tissue is dissolved. Let sit for 5-10 minutes at room temp
2. Centrifuge 10,000g for 10 minutes pipette fluid into another tube
3. Wash with 100% EtOH, invert tube several times to ensure mixing. Either remove DNA with a pipette tip, or spin down the sample for 5min at 5000g and pipette EtOH out of tube
4. wash two times with 75% EtOH, letting ethanol run over over the pellet
5. Add pellet to a solution of NaOH to dissolve it.
made the solution of NaOH by first making a 4 Molar stock solution (80g of NaOH to 500 ml water). Then I took 200µL from that and added it to 998mL of nano pure water to make an 8mM solution. (c1v1=c2v2)
Results were not good, the concentration of DNA was very low.
Next step: add .5µL DNAzol to 25-50mg of tissue and add 20µL proteinase
k. let the enzymes work over night at room temperature. Run the rest of the steps the same way.
5/14/2010 Friday
Another DNA extraction done on WBgill tissue number 3, a control and a treated. I used the Robert's lab new extraction kit and worked at the undergraduate bench in room _ using the same pipettes that Sonia has been using, and comparing my protocol to hers.
5/13/2010 Thursday
qPCR on the samples digested 5/6/2010 using a primer that Mac found to show methylation.
Hoping to see a difference between the control and the treated.
Results from qPCR
It's odd that for U, H and M all treated samples had higher amplification compared to its control counterpart. This can be attributed to the fact that it is unknown how much gDNA was put into each test tube. It can only be an estimation. Another thing to note is that MSPI should be negative for both the treated and the control because it should cut the strand whether or not the sample is methylated. This leads me to believe I had incomplete digestion of the samples and that they should run longer to obtain decent results.
5/10/2010 Monday
Ran the two samples of isolated gDNA (using Horner-Devine Kit_) (that was not incubated at 37°C for two hours) onto a gel to determine whether this had an impact on the band seen in the undigested gDNA. (Sonia's gel showed no unusual band).
Result: a band is seen in the undigested genomic DNA for both the control and treated samples. The next step may be to extract DNA using the three kits as a comparison.
5/6/10 Thursday
Performed restriction digests on the samples and incubated them at 37°C for two hours (starting at 12:45 pm). The reactions will need to be heat stopped (HPAII at 65°C and MSPI at 80°C for 20 minutes). Some of the sample will be transferred to tubes where loading dye will be added and run onto a gel. The rest will be used in a qPCR reaction using a primer that Mac found to show methylation.
Results from gel:
Still have that odd band for the undigested samples. Not really sure why. (Horner-Devine Kit)
5/5/10 Wednesday
We have reason to believe that our DNeasy kit was to blame for the results seen last Wednesday (see Sonia's notebook for comparison of kits). Today I am extraction DNA from two samples (Control3 and Treated3) using the kit from the Horner-Devine lab.
The protocol from the DNeasy kit was used (see 4/21 for protocol). The samples were put at 56°C at 12:15 pm. Tomorrow these samples will be either undigested, digested with HPAI, MSPII and will be run onto a gel to determine if differences are seen between the two.
4/28/10 Wednesday
A .12% Agarose gel was made using
150 mL TAE, 1.8 g of Agarose and 12µL ethidium bromide
5µL DNA hyperladder was used
Layout is as follows:
condition #- U H M
Spaces were left between each condition.
Gel started at 1:00 pm
C5 will not be run onto the gel as it did not get removed from the water bath.
Results: (Robert's Kit #1)
Are not very conclusive. First of all, the undigested should be higher up, this mid-low range is not usual for genomic DNA, but was seen for both the control and the treated. This could mean that something was wrong with the starting DNA or if there was any DNase in when they were incubated at 37°C for 3 hours it could have started to deteriorate the DNA.
What we were hoping to find is no shift between the HPAII treated and MSPI treated samples. This would tell us that the 5-azacytidine caused demethylation to all samples.
It's difficult to tell from the picture. It appears that Treated3 and treated 5 show the same results for both HPAII and MSPI but treated4 looks as if there is a shift between the two, telling me that maybe that sample was still methylated.
I used the "ideal" concentration of azacytidine from the Daphnia paper, and because all of my oysters stayed a live, I am lead to believe that I could try a higher concentration on a different set and get different results (demethylation).
4/26/2010 Monday
Restriction digests on six of the ten samples- C3, C4, C5 and TX3, TX4, TX5.
Each sample was subjected to three different conditions- HPAII (if product forms= methylation), MSPI (product should not form= no methylation) or undigested (no enzyme added). See here
After water, buffer, enzyme and DNA had been added, they were put in a water bath at 37°C for 3 hours.
1:30 pm.
After 3 hours, the samples were removed from the water bath and the reaction was stopped, loading dye was added to each of these.
Next step: Run these onto a gel and look for methylation. We are hoping to see that 5-azacytidine causes demethylation in all the treated subjects.
4/22/2010 Thursday
Because I had three really low values of DNA and the absorbency was low I decided to do DNA extraction on those three again Control03, Treat01 and Treat02.
Same protocol as yesterday- 4/21/2010.
Results look much better:
Now, all values are within range and I can proceed with restriction enzymes.
4/21/2010 Wednesday
Extracted DNA from my 10 samples (5 control, 5 treated).
following the protocol in the DNeasy
1. In a 1.5 ml microcentrifuge tube 180 µL buffer ATL
2. 20 µL proteinase K mix (18.5 mg/mL)
3. Cut up sample and place into the tub with buffer
4. Incubate at 55 degrees C until fully dissolved, usually 1-3 hours. If needed, overnight incubation.
5. vortex. Add 200 µL buffer AL to sample. vortex
6. Add 200 µL of (96-100%) EtOH. vortex
7. pipet into a 2 ml DNeasy column with collecting tube
8. Centrifuge at 6000xg for 1 min. Discard collecting tube
9. Place column in new 2 ml tube and add 500 µL Buffer AW1. Centrifuge for 1 min at 6000xg
10. Place column in 2ml collection tube and add 500 µL buffer AW2. Centrifuge for 3 minutes at 20,000xg
11. Place column in a 1.5 or 2 mL micro centrifuge tube and add 100µL Buffer AE into DNeasy membrane. (keep the left over liquid and discard the DNeasy column) incubate at room temp and centrifuge for 1 min at 6000xg.
Control 03 had a significantly lower volume noticed during step 6 and 7. There is a chance the buffer did not get added.
Spec samples:
Control 03 had very little DNA as well as a low absorbance 260/280, not surprising.
What is surprising is the low values for treat01 and treat 02.
4/15/2010 Thursday
12:00 pm
All ten oysters are alive still. Temp: 6°
Today, gill tissue was extracted from each oyster. The samples were put at -80degrees to freeze until DNA isolation can be performed.
Next step: DNA isolation on each of the ten oysters. After isolation takes place the samples will be pooled.
4/14/2010 Wednesday
Checked oysters again, all remained closed upon stimulation.
11:30 am
Temp: 5°
4/13/2010 Tuesday
Checked oysters to determine if they are alive, did this by stimulating the shell. A defense mechanism of the oyster is to remain closed, when an oyster dies the muscle require to pull the sell closed no longer works so a dead oyster's shell will pop open.
All 10 oysters seem to be living.
Disposed of solid waste (gloves) appropriately in biohazard bag.
time checked: 2:30 pm
Temp: 5°
4/12/2010 Monday
Organized, prepared and labeled jars needed for experiment. Filtered 2 liters of .2 mm of seawater.
Setup picture to come.
1 Liter of .2mm filtered seawater was measured into each container.
In the fume hood, between 7.0 mg and 9.0 mg of 5-azacytidine was measured and placed into one of the 1 L seawater beakers (the range is due to fluctuation of the scale).
The beakers were placed in a 6° C room and air stones were added to each. The tops were covered with aluminum foil to eliminate air vapor. Five juvenille oysters gathered from Willapa bay were placed into each beaker.
They oysters will be checked on a daily basis to determine if they are alive. They will sit for 3 days in the solution and gill sample will be taken from each and pooled.
Start time 12:40 pm
4/9/2010 FRIDAY!
Did some data entering for Mac- the C.gigas Field data measurements for April 2010.
Label beakers for my experiment, which starts Monday.
HPAII at 65 degrees for 20 min using buffer 1
MSPI at 80 degrees for 20 min using buffer 4
4/8/2010 Thursday
Planned out my experiment. Decided to try just two treatments- a water control and one with a "high" concentration of 5-azacytidine.
5- azacytidine is labeled as harmful by the company Arcos who made it. Necessary safety precautions should be taken to avoid skin contact and inhalation. This means I will be mixing under the fume hood. Gloves and a face mask will be worn. The mixture will then be transported to the cold room where the oysters will be submerged and monitored throughout the day.
Each treatment will contain 1000mL of sterile seawater. The second condition will contain 7.4mg/L of 5-azacytidine, which was the nominal concentration shown in the experiment on Daphnia. Five oysters will be placed into each condition. They oysters will bathe for up to three days.
After that time, DNA will be isolated from each of the samples and like conditions will be pooled. The samples will be spec to see how much DNA is present. A portion of the pooled gDNA will be subjected to one of four conditions- undigested, or digestive enzymes MSPI and HPAII, H2O
Sample vol ng/µL with water volume and buffer volume to equal 1 µg
Undigested= sample+water
HpaII= sample + water+ 5µL buffer 1+ 1.0 µL enzyme
MSPI= sample+water+ 5µL buffer 4+ 0.5 µL enzyme
Samples work at 37 degrees for about 2 hours.
After this, traditional PCR
traditional PCR
95° for 10 min
95° 15 sec
55° 15 sec For forty cycles
72° 1 min
72° 10 min
4° forever
and finally samples will be run onto a gel and results will be recorded.
Note: Need to look up ideal temperatures for MSPI and HPAII
4/5/10 Monday
DNA isolation
1. In a 1.5 ml microcentrifuge tube 180 µL buffer ATL
2. 20 µL proteinase K mix (18.5 mg/mL)
3. Cut up sample and place into the tub with buffer
4. Incubate at 55 degrees C until fully dissolved, usually 1-3 hours. If needed, overnight incubation.
5. vortex. Add 200 µL buffer AL to sample. vortex and incubate for 10 min at 70 degrees C.
6. Add 200 µL of (96-100%) EtOH. vortex
7. pipet into a 2 ml DNeasy column with collecting tube
8. Centrifuge at 6000xg for 1 min. Discard collecting tube
9. Place column in new 2 ml tube and add 500 µL Buffer AW1. Centrifuge for 1 min at 6000xg
10. Place column in 2ml collection tube and add 500 µL buffer AW2. Centrifuge for 3 minutes at 20,000xg
11. Place column in a 1.5 or 2 mL micro centrifuge tube and add 100µL Buffer AE into DNeasy membrane. (keep the left over liquid and discard the DNeasy column) incubate at room temp and centrifuge for 1 min at 6000xg.
Spec to see how much DNA is present
Spec Results:
3/18/09 Thursday
Repeat procedure from Tuesday and Monday with six samples- NB07, NB08, NB09, NB10, NB11, NB12
results:
Also- made 50 ml of 75% EtOH using 37.5ml Ethyl alcohol and 12.5ml of 0.1%DEPC water.
3/16/2019 Tuesday
Repeat procedure from Monday with four more samples: NB03, NB04, NB05, NB06
Results:
3/15/2010 Monday
RNA isolation procedure:
1. 500 µL tri-reagant
2. cut up sample using clean razor blades place them into tube with tri-reagant
3. Grind the sample using a pestel until there is no longer a chunk of sample, KEEP ON ICE!
4. Add another 500 µL to the tube.
5. Mix on the vortex
6. Set in RT for 5 min
7. Centrifuge 12,000 g for 15 min at 4°C
8. Take aqueous layer off the top (without pipetting the middle layer.) into a new tube
9. Add 500 µL isopropanol mix
10. Let sit 5-10 min at RT
11. Centrifuge 12,000g for 8 min at 4°C
12. "Dump" off Isopropanol
13. If pellet is seen, add 1ml (1000µL) EtOH and vortex
If no pellet is seen add 1ml of EtOH DO NOT VORTEX
14. Spin 7500g for 5 min at 4°C
15. Pipette EtOH out of tubes, spin down and continue to pipette out
16. Add 100 µL DEPC water (less depending on size of pellet)
17. Put on heating block (55°) for 5 min (or longer if not dissolved)
18. Spec to see how much RNA present.
Spec results from the two NB samples
We are looking at how much RNA we have in each of these samples.
We are also looking at the 260/280 results which typically should be around 1.9 -2.1, both samples were low, especially NB02.
2/26/2010 Friday
Results from Wednesday's PCR:
2/24/2010 Wednesday
Ran samples from Monday onto a gel, layout as follows
AY-0003(542bp), DQ-6110(533bp) , AJ-9915(870bp), AB-5409(356bp), DQ-6108(323bp), AJ-4739(730bp), FJ-9527(395bp), DQ-6107(313bp)
Order is gNDA, H2O with blanks between EACH.
RESULTS:
Began conventional PCR on the remaining primers. Ran out of BB02 on Wednesday so I am rerunning primer DQ-6107.
All are treated with BB06 gDNA.
Primers are DQ-6107(313bp), FJ-7754 (617 bp), AJ-2213 (861bp), EU-2886(616bp), AY-0357(1054), AM-5317(454bp), AJ-9659 (325-19)?
Made 24 µL reactions using
12µL APEX (x3)
1 µL Primer (x3)
11 µL H2O (x3)
PCR settings were the same as Monday's:
95° for 10 min
95° 15 sec
55° 15 sec For forty cycles
72° 1 min
72° 10 min
4° forever
Next step: Will run these onto a gel on Friday.
2/22/2010 Monday
Began conventional PCR on other primer sets.
DQ-6110, AJ-9915, AB-5409, DQ- 6108, AJ-4739 and FJ-9527 as well as re-running AY-0003 to see if we get consistent results.
Made 50 µL reactions:
APEX 12 µL x2 = 25µL
Primer 1 µL x2 = 2 µL
H2O 10.5 µL x2 =21 µL
Used 2µL of BB02 gDNA.
Raising the annealing temperature back down to 55, hopefully this will account for the AY-0003 band shown on Mac's gel (2.8.2010) and mine from 2.17.2010 and 2.12.2010)
95° for 10 min
95° 15 sec
55° 15 sec For forty cycles
72° 1 min
72° 10 min
4° forever
2/17/2010 Wednesday
Using the left overs of the samples from Friday, run onto a gel. This time use hyperladder. For bands the correct size- cut them out using the same method as last time and store them.
Results are consistent with friday (2/12/2010).
As stated before AY-0003 came up with no band- inconsistent with Mac's (2/8/10). One explanation for this could be that her annealing temp was set to 55 and mine was at 60. We raised the temperature to increase the specificity but for that specific primer the temperature may have been too high to lay down.
Bands were cut out of five of the primer sets:
DQ-6098, AM-5551, AJ-3432, AJ-8882 and AJ-9316
2/12/2010 Friday
Running PCR samples from Friday onto a gel. if the bands are correct size- cut them out.
Gel is 1.2% agarose (150 mL TAE, 1.8 g Agarose) 12µL Ethidium Bromide.
5 µL Hyperladder
Lane ID as follows:
Gel- the first six lanes "broke" when I pulled the combs out.
problem- The hyperladder did not show up on the gel, there is a good chance I loaded wrong. There are definite bands but since it was impossible to determine their size- I was not able to cut bands out.
Note- bands between gDNA and H2O are Anna's samples so those can be ignored.
Analysis: A band is shown for primer DQ_6098, which is consistent with the gel from Feb 8, 2010 However the band is not as on Monday.
AY-0003 no definite bands here, inconsistent with Mac's where a faint band (2/8010)
AM-5551 and AJ-3432 my results appear to be consistent with Mac's (2/8/10)
AJ-8882 a faint band is shown as was AJ-9316
Nothing seen on AJ-5315
Because it is impossible to determine the sizes none of these anaylses are certain, just speculation.
Next step- repeat the traditional PCR using the same samples but use hyperladder next time.
2/10/2010 Wednesday
Conventional PCR on 7 primers using only undigested genomic DNA.
See ID and Expected band size here:
Made 50 µL reactions
12.5µL Apex x2= 25µL
1µL Primer x2= 2µL
10.5µL Water x2=21µL
We are upping the temp to avoid multiple bands. By increasing the temperature from 55 to 60 we can also increase the specificity at which the primers lay down:
95° for 10 min
95° 15 sec
60° 15 sec For forty cycles
72° 1 min
72° 10 min
4° forever
Next step: Run these out on a gel, skipping lanes. Use the UV camera to take a picture. Cut out bands that are the correct size
2/8/2010 Monday
Running a gel on primer A3 under U,H,M, H2O conditions with 1 and 2 µL samples
Primers A4 and A11 are meant to be negative controls, they contain no DNA
Primer DQ_6098 is also under the conditions U,H,M,H2O. The expected band size is 405 bp.
Gel:
150 mL TAE
1.2% agarose - 1.8g
12µL Ethidium Bromide
See lane ID here:
Results:
Correction in the picture AY-0003 is actually DQ_6098
Once again- no band for A3 undigested for either 1 or 2µL samples. Both A3s show bands on H and M (Although a little hard to see here. The waters were all clean.
AY-0003 showed bands for U, H, M and primer dimers in the water. The expected band size was 405bp and that's what we saw.
The negative controls (A4 and A11) which contained no DNA showed no bands but all A11 (MSPI and both H2O) showed primer dimers.
Conclusion: It's hard for me to tell what the problem is, when Mac and I ran samples side by side we got consistent results (a band for Primer A3 for each condition).
Next step:
Re-run primer A3 with same conditions. Also moving forward time- we're going to run some of the restriction digests Mac ran (2.8.2010).
2/5/2010 Friday
results from Wed: see Mac's notebook (Feb 4 2010)
Today- third time's a charm. conventional PCR on Primer A3 under conditions (Undigested, digested with HPAII and MSPI) using samples of 1 and 2µL. Waters were capped first so hopefully no contamination there.
| 12.5µL Apex |
12.5µL Apex |
|
| 10.5µL H2O |
9.5µL H2O |
|
| 24 µL rxn |
23 µL rxn |
|
| 1µL DNA |
2µL DNA |
|
| U |
U |
|
| H |
H |
|
| M |
M |
|
| H2O |
H2O |
95° for 10 min
95° 15 sec
55° 15 sec For forty cycles
72° 1 min
72° 10 min
4° forever
Next step: Run samples onto a gel (along with some primers from Mac's notebook)
Label picture of gel with the primers.
2/3/2010 Wednesday
Ran Mac's PCR out on a gel (see Mac's notebook 2/1/10)
Re-PCRing primer A3 under using samples (U,H,M) with 1 and 2 µL samples.
2/1/2010 Monday
Ran primers D7, D12, E9*, E11 and A3 under three conditions (U,H,M) out on a gel
Ran the corresponding waters as well.
See layout here:
Results:
Next step: Run primers B6, B12, C7, D1, D6, under three conditions (U,H,M) onto a gel with their corresponding waters.
1/29/2010 Friday
Ran gels from Wednesday.
See layout here
The results lead me to believe I either mixed up my samples (U, H, M) or was pipetting no sample into my wells.
Repeated exact procedure from 1/27/2010 Wednesday with primers B6, B12, C7, D1, D6, D7, D12, E9*, E11 and A3 (see in results above A3 should have shown a band in all three conditions U, H, M as we got previously so we are repeating it).
1/27/2010 Wednesday
Ran conventional PCR on 10 old primer sets and three conditions Undigested, HpaII and MspI on the same pooled samples of BB01 and BB04
Primers: A3, A4, A10, A11*, B1, A12, B2, B3, B4*, B5
Where the * indicates 2 µL of template rather than just 1.
See layout here:
The samples are ran on thermo controller
setting COLONYPC
95° for 10 min
95° 15 sec
55° 15 sec (40 cycles)
72° 1 min
72° 10 min
4° forever
Prepped gels for Friday to test again for methylation. Hoping for positive undigested and negative MspI!
Next step:__ Repeat for primers
B6, B12, C7, D1, D6, D7, D12, E9*, E11
For tonight... figure out the layout of my gel!
1/25/2010 Monday
Running the samples qPCRed by Mac (see her notebook) out onto a gel. The results were inconclusive so we're just testing to see whether or not we get a band. there are 4 primers A3,A4,A10,A11.
And pooled BB01 and BB04 samples for undigested, MSPI digested and HPAII digested.
First I add 5 µL of 5x loading buffer to each well. Then run them out on a gel putting a blank between each sample.
See lane ID here.
results: bands for primers A3 and A4 for all conditions (waters were clean).
A4 showed a band for undigested but not HspII or MSPI suggested no methylation (?).
A11 showed bands for undigested and HSPII but not MSPI (and it shouldn't have!) suggesting methylation (see circled bands below- above primer dimers).
Possible reasoning for A3 and A10 could be the amount of sample we added- may have been too diluted. We think the cutting site for primer A3 is not there.

1/22/2010 Friday
Summary- Restriction Digest on gDNA samples BB01 and BB04 with three conditions; undigested, enzymes HPAII and MSPI. Looking for methylation based on products formed.
First I diluted the samples 1:5 then I added them to buffer and water. The conditions were then added and kept on ice and put in a water bath at 37° (at 8:45 am). They will incubate to allow time for digestion to occur. We're hoping that the enzymes fully digest or cut at appropriate points.
Click here for Layout
1/20/2010 Wednesday
Running out samples from Friday on a gel...
Results:
Primer AM866427 came out funky the band size should be 315 instead it's all over the place
Primer AM860932 the expected band size was 309, It looks to be between 400-600
Primer EW778340 the expected band size was 248 and also looks about 400-600.
0932 and 8340 were cut out so the DNA can be separated.

01/15/2010 Friday
Traditional PCR on a sample of gDNA and 3 primer sets.
Primer layout here
AM866427
AM860932
EW778340
Next step- run these out on a gel and cut out bands.
01/13/2010 Wednesday
Made a stock solution of 3 primer sets (100 µM)
Made 10 µM dilution from that.
Next step: Restriction digests (HpaII, MspI and undigested) on samples, using old primer sets that worked.
Repeat using new primer sets.
See primer info here
12/11/09 Friday
Results: qPCR on gDNA using Primer A3 look better.

Still working on the table with c(t) and efficiency values.
Today: qPCR on primer set A11 only to see if different from original run.
See layout here
Next step: Analyze and run qPCR on primer set A10 (control)
Nanodrop

More info to come.
12/10/09 Thursday
Results from yesterday: interesting... possibly problems loading the wells...
Observations:
Waters are clean (good news!)
Saw some +/- for methylation, possibly saw some partial digestion.
Samples using primer A3 had a decent melting peak, we need to make sure the wells were loaded right
Samples in primers A10 and A11 both had low fluorescence and little amplification and were different in comparison to the positive control Mac Ran (See her qPCR)
Funky results for row F- MSPI showed amplification (should have been -), Undigested pooled also had some but that was supposed to be a control (nothing in the well).
--To have a better idea, re-ran qPCR on Primer A3 again Hoping for better results
Made a new stock of 10µM A3 primers, keeping PF and PR separate until they are mixed with the master mix.
c1v1=c2v2
100 µM (v1)= 10 µM (100 µL)
so 10µL of primer and 90 µL of water.
A3 Plate Layout
For tomorrow: Run samples for Primer A11 again, making a new stock.
Made sytol13 concentration
5mM = 5000 µM
5000µM (v1) = 50µM (300 µL)
12/9/09 Wednesday
Summary: qPCR on genomic tissue samples (either undigested, treated restrictive enzymes HPA II or MSP2) using primers A3, A11, A10. looking to see if primers amplify certain tissues and not others
Procedure:
See plate layout here
Ran the sample in the centrifuge 750 RPM, stopped when it reached 700.
Since each DNA is about 600-700 BP long we used a different setting on the qPCR.\
For Tomorrow: Analyze qPCR!
12/4/09 Friday
Summary: 25µL reaction Traditional PCR on primer sets A3,A11,A10 (see Mackenzie's qPCR ). Using undigested gDNA to test for methylation
Procedure:
Master mix:
Apex: 12.5 µL x 5 = 62.5
Primer: 1 µL (.5 Pf, .5 Pr) x 5= 5µL
Water: 10.5 µL x 5 = 52.5 µL
Total: 120 µL
Layout:
A3U, A3U, A3U, A3H20
A11U, A11U,A11U, A11H20
A10U, A10U, A10H20
1µL undigested gDNA into each (water is neg control)
ABC setting under Mac on the trad PCR.
Next step:
Analyze gel
12/3/09 Thursday
Made 1x TAE buffer using 300 mL of 50x stock solution and diluting it with 14.7 L of water.
Ran Agarose gel with samples from 12/2/09 and 9 other samples.
See lane ID here
Results:
Not great.
Amplification a band for water :(
Gel:

Few amplifications on bottom. Some positive results.
Some on top- annealing temp was low so primers were non specific. Not sure which on top were water.
12/2/09 Wednesday
Summary: Running trad.PCR on 12 samples using a primer that should amplify any Eukaryote cDNA. Based on our results (amplification or not) we can determine if the samples have cDNA or not. This should tell us information about the primers used to amplify as well.
Procedure:
See here for procedure and layout
Made Agarose gel -> saran wrap and refrigerate
Next step: tomorrow will load the gel and look for amplification.
learn skitch and put PCR pictures up.
note to me: write neater.
11/25/09 Wednesday
Based on the real PCR (see Mac's book 11/23/09) we see possible methylation for select samples. To test this further we are running traditional PCR on 8 total samples.
(ex: sample A3: A3= undigested, B3=H, C3=M, D3=H20)
25 µL samples
[10x] loading dye, we want 1/10 of that ---> [1x]
2.5 µL Loading dye
DNA marker: 5µL Hyperladder
Layout here
Sizes here
Next step: analyze gels, compare the sizes of amplification
Results:


11/20/09
Summary: genomic DNA exposing them to restriction enzymes. Restriction enzymes cut out specific portions of DNA backbone, without ruining nucleotides. The enzymes must be kept on ice because they are proteins and will denature at increased temperature
Looking to see if products are formed in digestion, HpaII and MSPI (no product should be formed here) this will tell us whether they are methylated or not.
Procedure:
9 samples total
sample vol ng/µL with water volume and buffer volume to equal 1 µg
Undigested= sample+water
HpaII= sample + water+ 5µL buffer 1+ 1.0 µL enzyme
MSPI= sample+water+ 5µL buffer 4+ 0.5 µL enzyme
Here for layout
Enzymes work at 37°C for about 2 hours
samples run real qPCR looking for amplification (that will tell us whether or not is is methylated)
11/19/09
Made Agarose gel for PCR sample from 11/18/09.
150 mL TAE
12% agarose per every mL of TAE = 1.8g
Heat for 2 min swirl, heat for 1 min
add 12µL Ethidium Bromide
gel sets in ~20 min
5 µL HyperLadder in well one, DNA marker
For tomorrow: Read background on genomic DNA and restriction digest
Enzymes we are using HpaII and MSP1
See Lane ID here
Results:
__Gel results__
Rhodopsin (top): Lanes 1,2,3,4,6 and 9 showed band size of 400 (about 40 ng/band)
Lane 6 also had band size of 600 (60 ng/band)
It is suggested that lane 6 had an isoform.
Lane 8 did not have any bands (not even primer dimers)
Opsin (bottom): No real bands. The lower bands are most likely due to primer dimers. Lanes 9, 10 and 11 should have shown primer dimers as well but they did not show up here.
Sample error or human error could explain these results.
Next step label PCR gel with lane ID
11/18/09
Summary: Ran traditional PCR on 9 sepia cDNA from various locations.
Procedure:
Master mix
Used PF/PR Rhodopsin and
PF/PR Sepia Opsin
2 samples of water (no cDNA added)
PCR set up here (date is wrong)
Next step: put it on a gel and analyze
11/18/09
Results from qPCR on 11/12/09
For HSP70: only well D showed any amplification.
Looking at the melting curve only well D showed a peak, could have been sharper
For GXP: Wells C and D showed amplification. The amplification of C was very weak and abnormal
Looking at melting curve, both had a peak, although C was not sharp
Fluorescence was extremely low for all results.
Pi-petting errors may be to blame
11/12/09
Summary: Running qPCR of a cg_cDNA gill. Looking at expression of HSP70 and GXP.
Procedure:
set up 24 well
Master Mix:
2x immunox 112.5 µL
PF 4.5 µL
PR 4.5 µL
50 µBcyto 9 µL
H20 85.5 µL
Total: 216 µL
1 µL into each well
1 µL cDNA into wells A-D for columns 1 and 2
1 µL H20 into wells E-H in columns 1 and 2
Ran qPCR
link to plate layout


























