Results and commentary
Lab Discussion Notes
7 January 2009
Lab 1
Started RNA isolation procedure and extracted protein from fish brain tissue.
RNA Extraction:
Original sample from herring Clupea sp., majority was lost due to inanimate brain jumping wildly from storage tube onto floor. 15 mg were retained and combined with 81mg juvenile rainbow trout (RBT), Oncorhynchus mykiss brain sample (total 96 mg brain tissue) for RNA sample #1. RNA sample #2 contained 60mg of RBT brain tissue. The following protocol was followed (except first step which was not necessary for this part of the procedure):
RNA ISOLATION PROTOCOL
1. Turn on heating block to 55C. Also turn on spectrophotometer.
2. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.3. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg andadd to tube containing TriReagent.4. Carefully homogenize the tissue using a disposable pestle.5. Add an additional 500uL of TriReagent to the tube and close the tube.6. Vortex vigorously for 15s.
----- Stop here for Lab 1 and freeze sample at -80
Samples were labelled with my initials and 1 or 2, referring to sample # indicated above. Gloves were worn at all times and unused portion of samples were stored on dry ice during processing.
Protein Extraction:
Brain sample from RBT.
The following protocol was followed:
PROTEIN EXTRACTION PROTOCOL1. Add 0.5mL of CelLytic MT solution to a 1.5mL snap cap tube.2. Add 25mg of your tissue to the tube.
Total RBT brain tissue used: 30 mg
3. Homogenize the tissue with a disposable pestle.4. Close the tube and invert the tube several times.5. Spin the tube in a refrigerated microfuge for 10mins. @ max speed.
Max speed = 11,000 rpm
6. While spinning, label a fresh tube with the word "Protein", source organism/tissue, yourinitials, and today's date.7. Carefully transfer supernatant to labeled tube and store tube on ice.8. To a fresh tube, add 1.5mL of Bradford reagent.
Bradford reagent = Coomassie Blue
9. To this same tube, add 30uL of your protein extract.
Blank made with 1.5 mL of Coomassie Blue + 30uL of CellLytic MT and incubated at same time as sample.
10. Invert the tube several times and then incubate at RT for 10mins.
11. Mix the tube several times and transfer 1mL to a plastic, disposable cuvette.
Followed for both blank and sample. Blank inserted to spec. first for calibration.
12. Measure the absorbance at 595nm and record the value.13. Remove the cuvette from the spectrophotometer. Using a P1000 set to 1mL, carefullypipette the solution in the cuvette up and down a couple of times to mix.14. Measure the absorbance at 595nm and record the value.15. Repeat steps 13 and 14.
Sample value recorded 3 times:
1.284 A
1.276 A
1.273 A
16. Average the three absorbance values you recorded.
Average of 3 recordings: 1.278 A
17. Plug your average absorbance that you just calculated into the following equation to determine the concentration of protein in your sample:
UPDATE FROM PROTOCOL: It appears that in lab all of our protein values were outside of the curve.
There on Wednesday (or before if you would like) we need to dilute an aliquot of your sample and run again.
Please disregard the previous formula. Use the above formula: y=1011.9x
18. Write the concentration on your tube and place tube in TA's ice bucket. Your sample willbe stored @ -80C.
Concentration not yet determined so was not written on tube.
13 January 2009
Primer designing:
I identified nAG1 as my gene of interest because it has recently been identified as an important part of salamander limb regeneration (Kumar et al. 2007). It is homologous to XAG-2, a gene involved in Xenopus cement gland development P55869 (Aberger et al. 1998). Steven gave the following instructions to locate homologs in Oncorhynchus mykiss:
SR: Lets use sequence comparison to see if we can find homologs in [rainbow trout]:
1) go to BLAST
2) choose tBLASTn
3) Enter P55869 where is says enter Accesion Number
4) Change database to "est_others"
5) Under organism type "Salmonidae(taxid:8015)"
6) click BLAST.
this will provide you with any abalone ESTs that might be similar.
This resulted in 58 hits, many of them repeats. This was the first hit:
LOCUS CA342864 683 bp mRNA linear EST 05-NOV-2002
DEFINITION 672821 NCCCWA 1RT Oncorhynchus mykiss cDNA clone 1RT60J12_D_E06 5',
mRNA sequence.
ACCESSION CA342864
VERSION CA342864.1 GI:24588035
KEYWORDS EST.
SOURCE Oncorhynchus mykiss (rainbow trout)
ORGANISM Oncorhynchus mykiss
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Actinopterygii; Neopterygii; Teleostei; Euteleostei;
Protacanthopterygii; Salmoniformes; Salmonidae; Salmoninae;
Oncorhynchus.
REFERENCE 1 (bases 1 to 683)
AUTHORS Rexroad,C.E. 3rd, Lee,Y., Keele,J.W., Karamycheva,S., Brown,G.,
Koop,B., Gahr,S.A., Palti,Y. and Quackenbush,J.
TITLE Sequence analysis of a rainbow trout cDNA library and creation of a
gene index
JOURNAL Cytogenet. Genome Res. 102 (1-4), 347-354 (2003)
PUBMED 14970727
COMMENT Contact: Rexroad CE
USDA, ARS, National Center for Cool and Cold Water Aquaculture
11876 Leetown Road, Kearneysville, WV 25430, USA
Tel: 304 724 8340 x2129
Fax: 304 725 0351
Email: Caird.RexroadIII@ars.usda.gov
Single pass sequencing. Bases called with phred v0.020425.c and
trimmed with the aid of the trim_alt option. Vector identified by
cross_match v0.990329.
Seq primer: AGCGGATAACAATTTCACACAGGA.
FEATURES Location/Qualifiers
source 1..683
/organism="Oncorhynchus mykiss"
/mol_type="mRNA"
/db_xref="taxon:8022"
/clone="1RT60J12_D_E06"
/tissue_type="pooled"
/lab_host="DH10B"
/clone_lib="NCCCWA 1RT"
/note="Vector: pCMV SPORT6; Site_1: NotI; Site_2: SalI;
Library made from pooled tissue from brain, gill, liver,
spleen, muscle, and kidney."
ORIGIN
1 ttggcaggct ccagacactc ctcatacgcc tgtgcaatag aacttgtgaa cgccatgtat
61 cggtggtctc tcttgccttg ctctttgtca cctgcatgga agtgtccata cagaagaaaa
121 caaagaaagg ccctcaaact ctctcaagag gatgggggga tgacattact tgggtccaga
181 cctatgagga agccctgatg acaatgacag aaagtaagaa gcccctgatg gtcattcatc
241 acatggagga ttgtcctcat agtcaagctc tgaagaaggc gtttgctgct gataaagccg
301 tacaggaact tgcccaagag gattttgtca tgctcaattt gatacatgag actacagact
361 ccaatctggc accagatggc cactacgttc caagaattct ctttgttgat ccatccatga
421 ctgtgcgtgc tgagcttgtt gggaagtaca gtaaccacat gttcacctac aagccgagcg
481 acatcccgta cttggccgag aacatgaaga aagccaagcg tctactgcac actgaactgt
541 aacctcacag ccttttcaat aatgtaccca taatctactg ggttcctccc tgttccagag
601 gggttcactg agtcttggac attcttatgt ttacacagaa tgtatacaat agatcaatca
661 ataaagatga atgacggtgt gca
FASTA format:
gi|24588035|gb|CA342864.1|CA342864 672821 NCCCWA 1RT Oncorhynchus mykiss cDNA clone 1RT60J12_D_E06 5', mRNA sequence
TTGGCAGGCTCCAGACACTCCTCATACGCCTGTGCAATAGAACTTGTGAACGCCATGTATCGGTGGTCTC
TCTTGCCTTGCTCTTTGTCACCTGCATGGAAGTGTCCATACAGAAGAAAACAAAGAAAGGCCCTCAAACT
CTCTCAAGAGGATGGGGGGATGACATTACTTGGGTCCAGACCTATGAGGAAGCCCTGATGACAATGACAG
AAAGTAAGAAGCCCCTGATGGTCATTCATCACATGGAGGATTGTCCTCATAGTCAAGCTCTGAAGAAGGC
GTTTGCTGCTGATAAAGCCGTACAGGAACTTGCCCAAGAGGATTTTGTCATGCTCAATTTGATACATGAG
ACTACAGACTCCAATCTGGCACCAGATGGCCACTACGTTCCAAGAATTCTCTTTGTTGATCCATCCATGA
CTGTGCGTGCTGAGCTTGTTGGGAAGTACAGTAACCACATGTTCACCTACAAGCCGAGCGACATCCCGTA
CTTGGCCGAGAACATGAAGAAAGCCAAGCGTCTACTGCACACTGAACTGTAACCTCACAGCCTTTTCAAT
AATGTACCCATAATCTACTGGGTTCCTCCCTGTTCCAGAGGGGTTCACTGAGTCTTGGACATTCTTATGT
TTACACAGAATGTATACAATAGATCAATCAATAAAGATGAATGACGGTGTGCA
I followed these instructions to find primers:
1) go NCBI's Primer BLAST
2) enter accession number
3) uncheck "Specficity check"
4) click "Get Primers"
5) Select whichever primer pair you would like (maybe shoot for ~200bp)
6) enter that information for a primer pair (or multiple pairs) into the primer order form sheet.
After great muddling through the wholly new-to-me language of genetic databases, I settled on the following primer because it has 205bp and <2 degree difference between melting temps:
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
TTGCCCAAGAGGATTTTGTC |
Plus |
20 |
310 |
329 |
60.05 |
45.00% |
| Reverse primer |
GCTTTCTTCATGTTCTCGGC |
Minus |
20 |
514 |
495 |
59.96 |
50.00% |
| Internal oligo |
Plus |
||||||
| Product length |
205 |
||||||
| Product Tm |
|||||||
| Product Tm - min(OLIGO Tm) |
|||||||
14 January 2009
Lab 2
Continued the RNA isolation protocol from Lab 1 where we left off at step 6:
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube. NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
Both samples became a milky pink emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
I have 2 samples, Sample 1 contains both herring and trout brain and sample 2 contains only trout brain (as described in Lab 1 above). I prelabeled the new tubes for each sample 1a and 2a. Using my astute scientific skills I put sample 1 into new tube labeled 2a and sample 2 into new tube labeled 1a. Therefore Sample 1a is trout only, sample 2a is trout plus herring.
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
Solution did not appear lumpy after inverting several times.
17. Incubate at RT for 10 mins.
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
Both tubes had a small white pellet stuck to the side near the bottom of the tube.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
Used a kimwipe to carefully extract the rest of the EtOH from the side of the tube. After kimwiping the white pellet in my combo sample (2a) was no longer visible and I fear I lost it- will have to wait until next week's lab to know for sure.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer.
Spec was not operational for this part of the procedure. Sample was placed on ice for quantification next week.
Protein dilution
Due to last week's concentrations exceeding the calculated curve, I diluted my sample using the following ratios: 1:1, 1:3, 1:5. The protein and diH20 volumes used for each dilution are:
1:1 20ul Protein:20ul diH20 ; 1:3 20ul Protein:60ul diH20 ; 1:5 20ul Protein:100 ul diH20
I also made one dilution blank with 30ul diH20 in 1.5mL Coomassie blue
I followed steps 8-17 in the protein extraction protocol from lab 1. I had one false start in which I missed step 9 and added the Bradford reagent directly to my dilutions instead of extracting 30ul from my dilutions to add to the reagent. I caught this prior to running the samples on the spec, re-did the dilutions and had the following spec results:
1:1 (1) 0.789 (2) 0.795 (3) 0.786 Average for 3 readings: 0.790 x 2 (dilution factor) = 1.58 x 1011.9 (curve calculation) = 1598.8 x .001=1.599ug/ul
1:3 (1) 0.553 (2) 0.541 (3) 0.529 Average for 3 readings: 0.541 x 4 (dilution factor) = 2.16
1:5 (1) 0.417 (2) 0.410 (3) 0.405 Average for 3 readings: 0.4107 x 6 = 2.464
Protein gel protocol
Protein gels were prepared during the waiting periods of the RNA isolation procedure using the following protocol:
1. Begin boiling water on hot plate.
2. Thaw you protein extract from last week. Mix well by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly as demonstrated.
Pre-formed gels were used. Prior to loading into the gel box we rinsed the gels to remove residual preservative from the gel packaging. 1 x Tris-HEPES SDS Buffer diluted from 10x Tris was poured over the gels after the pre-formed gels were inserted into the gel box, fronts facing the interior well of the box. A gel-loading pipette was used to flush each well with tris to further remove preservatives.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well using a gel loading tip.
Jenn recorded the well configuration for our box, John and I were in the front gel:
1.Ladder
2. Amy
3. Amy
4. Ladder
5. John
6. John
7. Blank
8. Blank
9. Blank
10. Blank
11. Blank
12. Blank
We placed a ladder on the edge of our gel and between our samples in an asymmetric configuration.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL (does not have to be measured - just need enough to cover the gel) of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
We notched the top left corner, above the ladder on the edge.
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Be careful not to dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible. This may need to incubate O/N. If so, cover container with plastic wrap and leave on shaker/rocker.
Gels were incubated overnight. Mac took pictures the next day.

Ladder - Amy - Amy - Ladder - John - John
Ladder info from http://products.invitrogen.com/ivgn/en/US/adirect/invitrogen?cmd=catProductDetail&entryPoint=adirect&productID=LC5925&messageType=catProductDetail
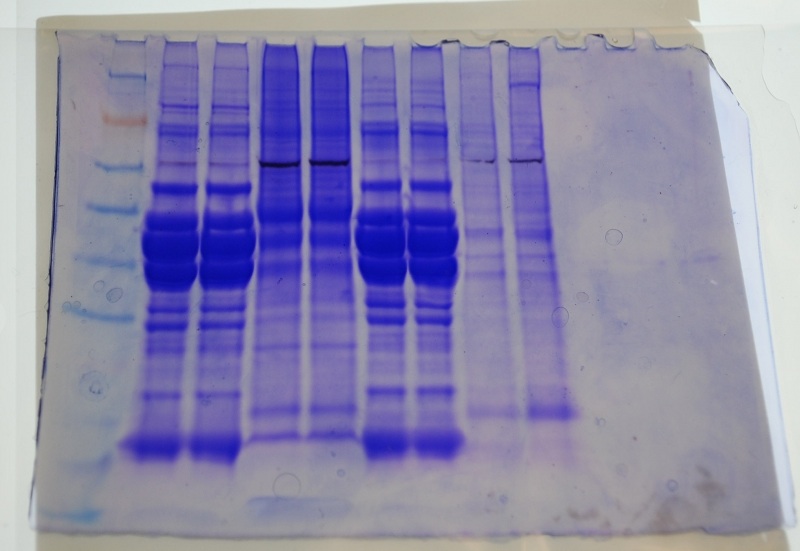
The three darkest bands in my protein sample appear to weigh out between Alcohol dehydrogenase and Glutamic dehydrogenase.
21 January 2009
Lab 3
We started this lab with RNA quantification using a NanoDrop. The NanoDrop procedure was simpler than the spec we were using for the protein quantification. The samples were still frozen when we started trying to read them, so I thawed mine in my hands for a few seconds, then placed 2ul of each sample on the NanoDrop for reading.
Results: Sample 1a: 0.752ug/ml (rainbow trout brain) ; Sample 2a (labeled ay1b on Nanodrop file): 0.703ug/ml
260/280: 1a: 1.99 ; 2a: 1.99
According to the NanoDrop T009-Technical Bulletin, a ratio of ~1.8 is "pure" for DNA and a ratio of ~2.0 is "pure" for RNA. In light of that, I think I did an ok job of isolating RNA since my ratio is 1.99 for both samples.
260/230 : 1a: 2.12 ; 2a: 1.85
This ratio is expected to be in the range of 2.0-2.2. My 1st sample (1a) fits nicely in this range, supporting further that this sample is "pure" nucleic acid. Conversely, the 2a sample falls below this range, indicating that there may be contaminants that absorb at 230 (such as EDTA, carbohydrates, and phenol).
We were given the following protocol for reverse transcription, which was modified as noted:
REVERSE TRANSCRIPTION PROTOCOL
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
Since my RNA quantity was below 2.5ug/ml, I did not have to dilute my samples for this step. In order to prevent the RNA from sitting in the thermal cycler for too
long, we started this procedure by making our Master Mix, then processed our samples as described below.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix
PER RXN- I have 2 samples and 1 blank, so made 3x the following amounts in one tube for my Master Mix. I should have made 4x to have a little extra because my blank Master Mix fell a tiny bit short.
4 ul 5x Buffer (AMV RT Buffer) x 3 = 12ul
8 ul dNTPs (10 mM total) x 3 = 24ul
1 ul AMV RTranscriptase x 3 = 3ul This was in freezer until ready for use and I added an additional 3ul of the Buffer due to misunderstood labelling, so I had to re-do the mix.
1 ul Oligo dT Primer x 3 = 3ul
1 ul RNase free water x 3 = 3ul
Total = 15 ul
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave on ice or store at –20C
PCR
Polymerase Chain Reaction involved amplifying a DNA (genomic or complementary) target using a polymerase, primers (short oligonucleotide), and dNTPs (A, C, T, Gs). In general the reaction is placed in a machine (thermocycler) where a series of temperature changes are performed [Denature (~94), Anneal (primer specific ~50-60), and Extention (~72)].
For this lab you will be using Promega's GoTaq Product. Please Read!
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| GoTaq®Green Master Mix, 2X |
25 |
1x |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| DNA template |
1–5μl |
<250ng |
Added 10ul of each primer stock to one new tube each with 90 ul of water to make 10uM solution.
I have 2 samples, 1 blank that has run through the reverse transcription procedure (for errors in RT), and one blank of plain diwater (for errors in PCR). Each of these was run in duplicate, so I made enough reaction stock for 9 samples to ensure enough for each of my samples.
GoTaq 25ul x 9 = 225ul
Up-primer 1 ul x 9 = 9ul
Down-primer 1 ul x 9 = 9ul
diH20 21ul x 9 = 189 ul
For each of 4 samples in duplicate, 48ul of this master mix was added to a new tube with 2 ul of one of the following: Sample 1a: cDNA trout brain; Sample 2a: cDNA trout + herring brain; RT Blank; and diH20 for PCR blank.
Load reactions into thermocycler. Set to cycle 40 times at 95 degrees for 1 minute, 50 degrees for 1 minute, 72 degrees for 2 minutes.
Make Agarose Gels
2 g agarose added to 150ml Buffer TAE. Microwave for ~4min, swirling every minute, until mix is just at boiling point but not yet boiling. Let cool then add 12 ml ethidium bromide (nasty stuff- allow to cool to avoid vaporization of ethidium bromide into face). Ethidium bromide binds to DNA and fluoresces it.
Place combs in gel box and pour agarose mix in before it solidifies. Try to avoid bubbles.
Next Day:
Run out PCR products on agarose gels and photograph.
22 January 2009
Poured buffer over gel. Loaded 15ul 100bp ladder and 25ul samples:
1 Ladder
2-9 John's samples
10 space
11 Ladder
12 Sample 1a 1
13 Sample 1a 2
14 Blank- breached wall while loading 13
15 Sample 2a 1
16 Sample 2a 2
17 RT Blank 1
18 RT Blank 2
19 PCR Blank 1
20 PCR Blank 2
Ran for 1 hour at 100 volts. Placed in UV box and bands were apparent. Photographed. On photo below my samples are in the lower right corner, starting with the ladder. John's are upper right, and Jenn's are on the left.
Both of my samples show distinct single bands and the controls show no distinct bands. Both Jenn & John's gels show 2 bands and I only have 1, however all of our bands are at approximately the same location indicating they have similar weights. This makes sense since we were instructed to identify primers with ~200 bp. Mine had 205bp and the location of my bands is at approximately 200 on the gel.
I cut one band from each of my samples and placed each in a tube in -20 degree freezer for later processing.
28 January 2009
Lab4 - Western Transfer - Immunoblots
- Run out protein samples as done previously
- Transfer proteins from gel to nitrocellulose membrane
- Probe membrane with antibody (HSP)
Run Protein Gel [as in Lab 2]
Mac started the protein gels prior to the start of lab. My protein was loaded in the back gel, which had the following configuration:
1. Ladder
2. John_a
3. John_a
4. Amy_a
5. Amy_b
6. Jenn_a
7. Jenn_b
8. Bob H17
9. Bob H23
Mac loaded samples in duplicate.
Transfer Proteins to membrane
1. Cool the transfer buffer to 4°C.
2. Soak the filter paper, membrane and gel in Transfer Buffer for 15 minutes.
3. Assemble the blotting sandwich in a semi-dry blotting apparatus as follows:
Air bubbles were smoothed out of the layers at the steps I indicate with bold type. Smoothing was done by rolling a tube across the layer from the center out to the edges to push air out of the layers.
• Anode (+++)
• Filter paper
• Nitrocellulose Membrane
Steven loaded one gel, John loaded the other. Steven marked the membranes with a single notch for the front gel and 2 notches for the back gel.
• Gel
• Filter paper
• Cathode (– – –)
4. Transfer the blot for 30 minutes at 20V.
5. Remove the gel from the sandwich and rinse with transfer buffer.
6. Use a cotton swab to remove any adhering gel from the membrane.
After transferring proteins to the membrane, we stained the gel with coomassie blue following the staining protocol from lab 2. Gels were soaked in acetic acid overnight.
Western Blotting Protocol
Western Breeze Manufacturer's Protocol
General Guidelines
• Avoid touching the working surface of the membrane, even with gloves.
• Work quickly when changing solutions as membranes dry quickly. If the membrane dries, re-wet the membrane with methanol and rinse with water before proceeding.
• Add solutions to the trays slowly, at the membrane edge, to avoid bubbles forming under the membrane. Decant from the same corner of the dish to ensure complete removal of previous solutions.
1. Prepare 20 mL of Blocking Solution
Ultra filtered Water 14 ml
Blocker/Diluent (Part A) 4 ml
Blocker/Diluent (Part B) 2 ml
Total Volume 20 ml
2. Place the membrane in 10 ml of the appropriate Blocking Solution in a covered, plastic dish provided in the kit. Incubate for 30 minutes on a rotary shaker set at 1 revolution/sec.
3. Decant the Blocking Solution.
4. Rinse the membrane with 20 ml of water for 5 minutes, then decant. Repeat once.
5. Prepare 10 mL of Primary Antibody Solution (1:3000 dilution)
Blocking Solution 10 ml
HSP 70 antibody 3.3 µl
Total Volume 10 ml
Primary antibody solution is specific for HSP70 (protein of interest).
6. Incubate the membrane with 10 ml of Primary Antibody Solution for OVERNIGHT
29 January 2009
NEXT DAY
I started the procedure working with Jenn at step 4 and followed through to step 5. At that point I went to class and Lisa and John were in the lab to continue working with Jenn.
Decant Primary Ab into 50ml blue cap tube, saved at 4C.
 |
| external image 20090131-8h4penekfh59y9cd6baiehb67c.jpg |
Results were posted by Mac:
Protein gel:
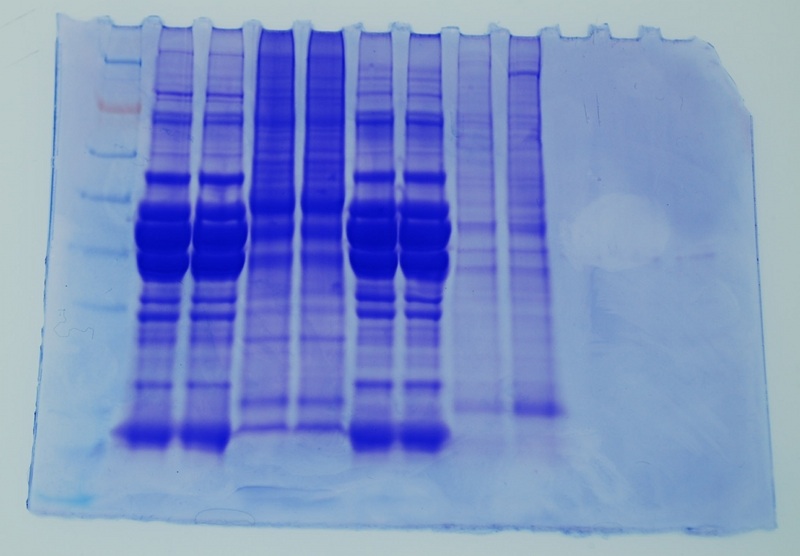
Ladder-John-John-Amy-Amy-Jenn-Jenn-Bob-Bob
It is interesting that John and Jenn's muscle tissue have a very similar banding pattern. Also my protein in this gel looks different than in the first gel- here the 3 bands seen in my first gel are present though more difficult to distinguish and there is a lot more color at the top of the lines than in the previous gel.
Membrane 45 minutes after development (note 2 notches in upper right corner):
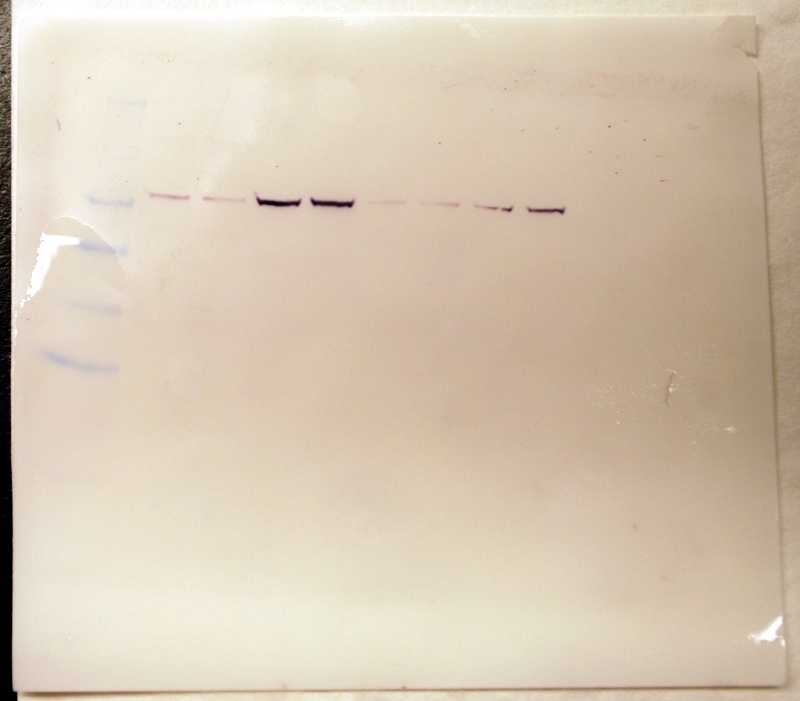
Membrane 1.5 hours after development- my samples show the darkest bands.
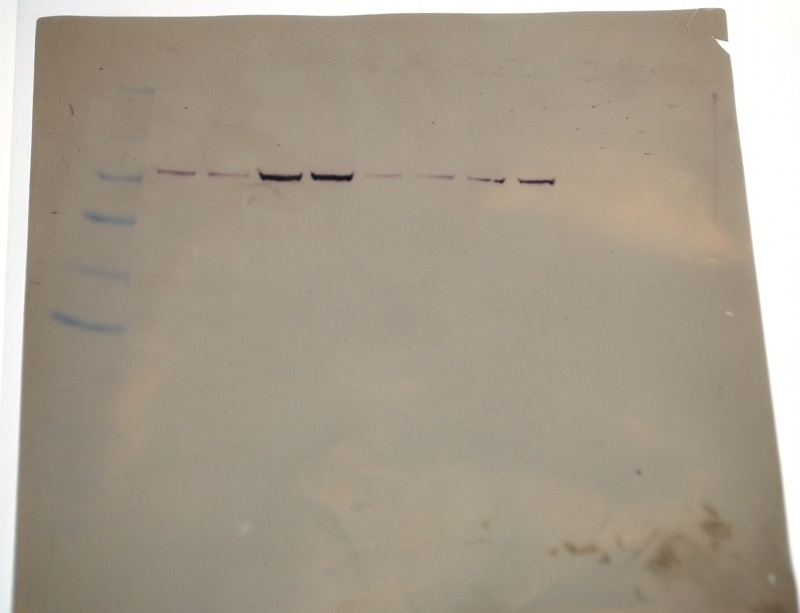
Even though my protein looks different than in the first run, there appears to be a strong signal for HSP70 as indicated by the dark band in the overlay above.
4 February 2009
Quantitative PCR lab using the following protocol:
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
I have 2 samples and 1 primer set, so I prepared enough master mix for 7 samples (with 1 extra to ensure enough for each sample).
1. Prepare master mix: Prepare enough reactions to run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template). Add 1 additional reaction to the total to ensure sufficient volume recovery.
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| Master Mix, 2X (Immomix) |
25µL |
1x |
| Syto-13 dye (50uM) |
2-5µL |
2 - 5µM |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| Ultra Pure Water |
to 48uL |
NA |
Final volumes I prepared:
Master Mix, 2X Immomix : 175ul
Syto-13 dye (2 uL): 14 ul
upstream primer, 10uM (2 uL): 14 ul
downstream primer, 10uM (2uL): 14 ul
Ultra Pure Water (17 uL): 119 ul
This was added to one tube for the Master.
2. Add mastermix to wells of a white PCR plate
3. Thaw cDNA samples.
4. Add 2uL cDNA template to each reaction.
5. Add 2uL of ultra pure water to the negative control wells. (I used 2 total negative control wells)
6. Cap the wells securely.
Well order:
(G) Control-1a-1a-2a-2a-Control (AY)
G is the row designation pre-stamped on the left side of my wells row. AY are my initials in sharpee on the right side of the wells.
7. If necessary, spin the strips to collect volume in the bottom of the wells. (Spun in a salad spinner with the rest of the class' samples).
8. and ensure the lids are clean (wipe with a Kim Wipe) before going into the Opticon. (Samples were placed in refrigerator for 45 min before Steven was available to load them in Opticon)
9. Load the plate, verify the PCR conditions and start the run. Run at 55 degrees C.
While spinning the samples we discussed the analysis of the QPCR peaks and melting curve and the problem of genomic contamination.
Primer-dimers would peak at the beginning (because they are small). If present the sample can be optimized by reducing the primer concentration.
Samples that fluoresce first have a higher concentration of double-stranded material (hopefully only cDNA).
Melting curve analysis: Melting Curve is produced by slowly raising the temp and taking frequent (e.g. ~1 sec.) readings. At a certain temp product(s) denature (double strand breaks)- time to denaturing determined by length and GC's because GC's are bonded stronger than AT's. The melting curve can be converted (by differentiation) so the graph shows a peak where the slope changes, corresponding to the point of denaturing.
In order to determine if genomic contamination is present, run RNA in a PCR. If a product results, then there is contamination because RNA is not stable enough to last the PCR process.
6 February 2009
QPCR results:
G1 36.04 negative control
G2 31.01 JRT brain
G3 25.66 JRT brain
G4 26.85 JRT+Herring brain
G5 20.51 JRT+Herring brain
G6 36.68 negative control
John, Jenn, and I had contaminated negative controls. One source of contamination may have been the water since we tended to share materials. My replicates are also very far off from each other: 5.35 difference in JRT reps, 6.34 difference in JRT+Herring reps, an approximately 20-fold difference in both sample reps. This could be an issue with my poor loading technique. It's possible that my cDNA was loaded at an angle and stuck to the side of the tube. If the sample was stuck too high in the tube then part of the sample was potentially caught in the cap because the caps were placed prior to spinning the material down.
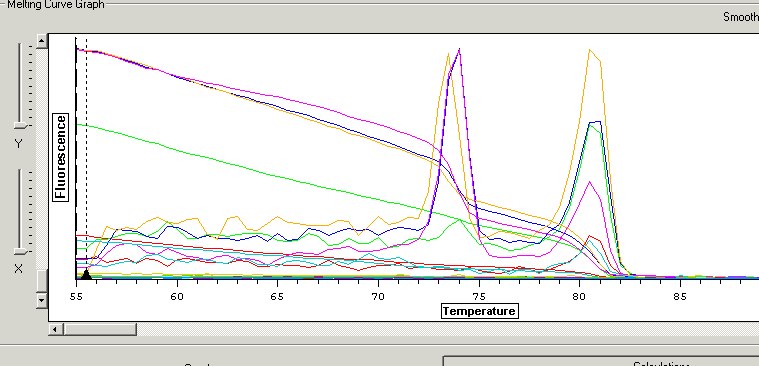
1. Red= control 2. Green=RBT(1) 3. Blue=RBT(2) 4. Orange=RBT+Herring(1) 5. Pink=RBT+Herring(2) 6. Lt. Blue= control
Key- my samples are in row G.
There is definitely something strange going on. All of my samples have 2 peaks, indicating contamination or carryover. Since my controls are also peaked at the higher temperature and my previous cDNA analyses (nanodrop and PCR) did not raise any big flags, I am inclined to think that these results are indicative of contamination at some point in the process of sample prep. My first RBT rep looks the cleanest but still has a small peak at 74 degrees where the rest of the brain samples have large peaks. There are no peaks at the beginning, so I don't have any indication of primer dimers. Since I suspect the water, I could test my results by running the samples again using water from 2 different tubes.
11 February 2009
Worked on tracking down a genotype approach to gender determination for the coho I am using. Found the following references that show a sex-linked growth hormone sequence (GH-Y):
Du SJ, Devlin RH, Hew CL. 1993. GENOMIC STRUCTURE OF GROWTH-HORMONE GENES IN CHINOOK SALMON (ONCORHYNCHUS-TSHAWYTSCHA) - PRESENCE OF 2 FUNCTIONAL GENES, GH-I AND GH-II, AND A MALE-SPECIFIC PSEUDOGENE, GH-PSI. DNA Cell Biol 12:739-751.
Devlin, R. H., Biagi, C. A., Smailus, D. E. 2001. Genetic mapping of Y-chromosomal DNA markers in Pacific salmon. Genetica 111(1):43-58.
The authors used primers GH5 and GH6 in PCR to identify males. Sent request to library for e-copy of Du article.
Also worked on primer designing with a couple of successes and a few failures. Found the following in the Coho nucleotide database:
1.Oncorhynchus kisutch (coho salmon) growth hormone mRNA, complete cds
2.Coho salmon growth hormone (GH) mRNA, 3' end
3.Oncorhynchus kisutch growth hormone 2 (GH-2-b) gene, partial cds
4.Oncorhynchus kisutch growth hormone 2 (GH-2-a) gene, partial cds
5.Oncorhynchus kisutch growth hormone 1 gene, intron C, complete sequence
6.Oncorhynchus kisutch growth hormone type-2 (GH2) gene, partial cds
7.Oncorhynchus kisutch growth hormone 1 (GH1) gene, intron d
8.Oncorhynchus kisutch growth hormone 2 (GH2) gene, intron d
9.Oncorhynchus kisutch vitellogenin A (VtgA) gene, exons 1 through 7 and partial cds
10.Oncorhynchus kisutch estrogen receptor alpha 1 (ERa1) mRNA, partial cds
Only 1 Vtg record and one estrogen receptor

LOCUS DQ248228 1615 bp mRNA linear VRT 30-MAR-2007
DEFINITION Oncorhynchus kisutch estrogen receptor alpha 1 (ERa1) mRNA, partial
cds.
ACCESSION DQ248228
VERSION DQ248228.1 GI:82409099
KEYWORDS .
SOURCE Oncorhynchus kisutch (coho salmon)
ORGANISM Oncorhynchus kisutch
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Actinopterygii; Neopterygii; Teleostei; Euteleostei;
Protacanthopterygii; Salmoniformes; Salmonidae; Salmoninae;
Oncorhynchus.
REFERENCE 1 (bases 1 to 1615)
AUTHORS Nagler,J.J., Cavileer,T., Sullivan,J., Cyr,D.G. and Rexroad,C. III.
TITLE The complete nuclear estrogen receptor family in the rainbow trout:
Discovery of the novel ERalpha2 and both ERbeta isoforms
JOURNAL Gene 392 (1-2), 164-173 (2007)
REFERENCE 2 (bases 1 to 1615)
AUTHORS Nagler,J.J., Cavileer,T.D., Cyr,D.G. and Rexroad,C. III.
TITLE Direct Submission
JOURNAL Submitted (17-OCT-2005) Biological Sciences and Center for
Reproductive Biology, University of Idaho, Life Science South, Rm
252, Moscow, ID 83844-3051, USA
FEATURES Location/Qualifiers
source 1..1615
/organism="Oncorhynchus kisutch"
/mol_type="mRNA"
/db_xref="taxon:8019"
gene <1..1615
/gene="ERa1"
/note="csERa1"
CDS <1..1501
/gene="ERa1"
/note="nuclear steroid receptor"
/codon_start=2
/product="estrogen receptor alpha 1"
/protein_id="ABB73308.1"
/db_xref="GI:82409100"
/translation="FLHPPSHHGLPSQSYYLETSSTPLYRSSVVTNQLSASEEKLCIA
SDRQQSYSAAGSGVRVFEMANETRYCAVCSDFASGYHYGVWSCEGCKAFFKRSIQGHN
DYMCPATNQCTMDRNRRKSCQACRLRKCYEVGMVKGGLRKDRGGRVLRKDKRYCGPAG
DREKPYGDLEHRTAPPQDGGRNSSSSLNGGGGCRGPRITMPPEQVLFLLQGAEPPALC
SRQKVARPYTEVTMMTLLTSMADKELVHMIAWAKKVPGFQELSLHDQVQLLESSWLEV
LMIGLIWRSIHCPGKLIFAQDLILDRSEGDCVEGMAEIFDMLLATVSRFRMLKLKPEE
FVCLKAIILLNSGAFSFCSNSVESLHNSSAVESMLDNITDALIHHISHSGASVQQQPR
RQAQLLLLLSHIRHMSNKGMEHLYSIKCKNKVPLYDLLLEMLDGHRLQSPGKVAQAGK
QTEGPSTTTTTSTGSSIGPMRGSQDTHIRSPGSGVLQYGSPSSDQMPIP"
ORIGIN
1 cttcctccac cccccaagcc accatggtct ccccagccag tcatactacc tggagacctc
61 gtccacaccc ttatacaggt cgagtgtggt aaccaatcag ctgtcagcgt cagaggagaa
121 gctctgcatc gcctccgata ggcagcagtc gtacagtgca gcagggtcag gggtcagggt
181 gtttgagatg gccaacgaga cgaggtactg tgcggtctgc agcgactttg cctccgggta
241 ccactacgga gtttggtcct gcgagggctg caaagccttc ttcaaaagga gcatccaagg
301 tcacaatgac tacatgtgcc ctgcgactaa ccagtgtaca atggacagga atcgtaggaa
361 gagctgccag gcatgccgcc tcagaaagtg ttatgaagtg gggatggtga aaggaggctt
421 gcgtaaggac cgcggtgggc gggttctcag gaaggataag cggtattgtg gccctgctgg
481 tgacagagag aaaccctacg gtgacctgga gcacaggaca gcgccccctc aggacggggg
541 taggaacagc agcagcagtc tcaatggtgg tggaggatgt cgtgggccca gaatcaccat
601 gcctcctgaa caggtgctgt tcctgctgca gggggcagag cctccggccc tgtgttctcg
661 tcagaaggtg gcccgcccct acacagaggt caccatgatg accctgctca ccagcatggc
721 tgacaaggag ctggtgcaca tgatcgcttg ggctaagaaa gtaccaggtt tccaggagct
781 gtctctccat gaccaggtgc agctgctgga gagttcctgg ctggaggtgc tgatgatcgg
841 actcatatgg cggtccatcc actgccccgg gaaactcatc ttcgcccagg acctcatact
901 ggacaggagt gaaggggact gtgtggaggg tatggctgag atcttcgaca tgctcctggc
961 cactgtgtct cgcttccgca tgcttaaact gaagcctgag gagtttgtgt gcctcaaagc
1021 catcatcttg ctcaactctg gtgccttctc cttctgttct aactctgtgg aatccctcca
1081 caacagctcg gcagtggaaa gcatgctgga caacatcacc gacgccctca tccaccacat
1141 cagccattca ggagcctctg tgcagcagca gcccagacgg caggcccagc tcctgctctt
1201 gctctcacac atcagacata tgagcaacaa aggcatggag cacctttaca gcataaaatg
1261 taagaacaaa gttcctctgt acgacctgct cctggagatg ctggacggtc accggctcca
1321 atccccaggc aaagtggccc aagctgggaa acagaccgag ggcccctcta ccaccactac
1381 cacctccaca ggctccagca tagggccgat gcgaggcagc caggataccc acatcagaag
1441 ccctggttcg ggggtactcc agtatggctc ccccagctca gaccagatgc ccattccgtg
1501 agatacagaa ataggtattt gtagatacgg aagatgtaaa agtcctatta catacagtat
1561 ggtatagaga gagatattta gaaatgtatg gaggtaagaa tggattaaaa atatg
FASTA:
>gi|82409099|gb|DQ248228.1| Oncorhynchus kisutch estrogen receptor alpha 1 (ERa1) mRNA, partial cds CTTCCTCCACCCCCCAAGCCACCATGGTCTCCCCAGCCAGTCATACTACCTGGAGACCTCGTCCACACCC TTATACAGGTCGAGTGTGGTAACCAATCAGCTGTCAGCGTCAGAGGAGAAGCTCTGCATCGCCTCCGATA GGCAGCAGTCGTACAGTGCAGCAGGGTCAGGGGTCAGGGTGTTTGAGATGGCCAACGAGACGAGGTACTG TGCGGTCTGCAGCGACTTTGCCTCCGGGTACCACTACGGAGTTTGGTCCTGCGAGGGCTGCAAAGCCTTC TTCAAAAGGAGCATCCAAGGTCACAATGACTACATGTGCCCTGCGACTAACCAGTGTACAATGGACAGGA ATCGTAGGAAGAGCTGCCAGGCATGCCGCCTCAGAAAGTGTTATGAAGTGGGGATGGTGAAAGGAGGCTT GCGTAAGGACCGCGGTGGGCGGGTTCTCAGGAAGGATAAGCGGTATTGTGGCCCTGCTGGTGACAGAGAG AAACCCTACGGTGACCTGGAGCACAGGACAGCGCCCCCTCAGGACGGGGGTAGGAACAGCAGCAGCAGTC TCAATGGTGGTGGAGGATGTCGTGGGCCCAGAATCACCATGCCTCCTGAACAGGTGCTGTTCCTGCTGCA GGGGGCAGAGCCTCCGGCCCTGTGTTCTCGTCAGAAGGTGGCCCGCCCCTACACAGAGGTCACCATGATG ACCCTGCTCACCAGCATGGCTGACAAGGAGCTGGTGCACATGATCGCTTGGGCTAAGAAAGTACCAGGTT TCCAGGAGCTGTCTCTCCATGACCAGGTGCAGCTGCTGGAGAGTTCCTGGCTGGAGGTGCTGATGATCGG ACTCATATGGCGGTCCATCCACTGCCCCGGGAAACTCATCTTCGCCCAGGACCTCATACTGGACAGGAGT GAAGGGGACTGTGTGGAGGGTATGGCTGAGATCTTCGACATGCTCCTGGCCACTGTGTCTCGCTTCCGCA TGCTTAAACTGAAGCCTGAGGAGTTTGTGTGCCTCAAAGCCATCATCTTGCTCAACTCTGGTGCCTTCTC CTTCTGTTCTAACTCTGTGGAATCCCTCCACAACAGCTCGGCAGTGGAAAGCATGCTGGACAACATCACC GACGCCCTCATCCACCACATCAGCCATTCAGGAGCCTCTGTGCAGCAGCAGCCCAGACGGCAGGCCCAGC TCCTGCTCTTGCTCTCACACATCAGACATATGAGCAACAAAGGCATGGAGCACCTTTACAGCATAAAATG TAAGAACAAAGTTCCTCTGTACGACCTGCTCCTGGAGATGCTGGACGGTCACCGGCTCCAATCCCCAGGC AAAGTGGCCCAAGCTGGGAAACAGACCGAGGGCCCCTCTACCACCACTACCACCTCCACAGGCTCCAGCA TAGGGCCGATGCGAGGCAGCCAGGATACCCACATCAGAAGCCCTGGTTCGGGGGTACTCCAGTATGGCTC CCCCAGCTCAGACCAGATGCCCATTCCGTGAGATACAGAAATAGGTATTTGTAGATACGGAAGATGTAAA AGTCCTATTACATACAGTATGGTATAGAGAGAGATATTTAGAAATGTATGGAGGTAAGAATGGATTAAAA ATATGPrimers for ERa1:
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
CGATAGGCAGCAGTCGTACA |
Plus |
20 |
136 |
155 |
60.03 |
55.00% |
| Reverse primer |
TGGCAGCTCTTCCTACGATT |
Minus |
20 |
369 |
350 |
59.98 |
50.00% |
| Internal oligo |
Plus |
||||||
| Product length |
234 |
||||||
| Product Tm |
|||||||
| Product Tm - min(OLIGO Tm) |
|||||||
----
Products on allowed transcript variants
----
Products on potentially unintended templates
----
Products on target templates
----
Primer pair 3
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
AATCGTAGGAAGAGCTGCCA |
Plus |
20 |
350 |
369 |
59.98 |
50.00% |
| Reverse primer |
CCTCCACCACCATTGAGACT |
Minus |
20 |
576 |
557 |
59.96 |
55.00% |
| Internal oligo |
Plus |
||||||
| Product length |
227 |
||||||
LOCUS AF454747 1477 bp DNA linear VRT 09-APR-2003
DEFINITION Oncorhynchus kisutch vitellogenin A (VtgA) gene, exons 1 through 7
and partial cds.
ACCESSION AF454747
VERSION AF454747.1 GI:29150215
KEYWORDS .
SOURCE Oncorhynchus kisutch (coho salmon)
ORGANISM [[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=8019|Oncorhynchus kisutch]]
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Actinopterygii; Neopterygii; Teleostei; Euteleostei;
Protacanthopterygii; Salmoniformes; Salmonidae; Salmoninae;
Oncorhynchus.
REFERENCE 1 (bases 1 to 1477)
AUTHORS Buisine,N., Trichet,V. and Wolff,J.
TITLE Complex evolution of vitellogenin genes in salmonid fishes
JOURNAL Mol. Genet. Genomics 268 (4), 535-542 (2002)
PUBMED [[http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&list_uids=12471451|12471451]]
REFERENCE 2 (bases 1 to 1477)
AUTHORS Buisine,N., Trichet,V. and Wolff,J.
TITLE Direct Submission
JOURNAL Submitted (03-DEC-2001) Laboratoire de Biologie et Chimie
Moleculaires, Universite de Bretagne Sud, Campus Universitaire Yves
Coppens, Tohannic, Vannes 56017, France
[[#comment_29150215]][[#feature_29150215]]FEATURES Location/Qualifiers
source 1..1477
/organism="Oncorhynchus kisutch"
/mol_type="genomic DNA"
/db_xref="taxon:[[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=8019|8019]]"
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=1&to=1477&view=gbwithparts|gene]] <1..>1477
/gene="VtgA"
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&itemID=2&view=gbwithparts|mRNA]] join(<1..43,209..229,371..522,614..865,953..1114,
1205..1353,1445..>1477)
/gene="VtgA"
/product="vitellogenin A"
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=1&to=43&view=gbwithparts|exon]] <1..43
/gene="VtgA"
/number=1
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&itemID=1&view=gbwithparts|CDS]] join(4..43,209..229,371..522,614..865,953..1114,
1205..1353,1445..>1477)
/gene="VtgA"
/codon_start=1
/product="vitellogenin A"
/protein_id="[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=AAO72348.1|AAO72348.1]]"
/db_xref="GI:29150216"
/translation="MRAVVLALTLALVASQSVNFAPDFEASKTYVYKYEALLLGGLPE
EGLARAGVKVISKVLISAVAENTYLLKLVNPEIFEYSGVWPNDPFVPAAKLTSALAAQ
FSIPIKFEYAKGVVGKVLAPTAVSETVLNVHRGILNILQLNIKKTQNVYELQEAGAQG
VCKTHYVIREDAKAERIHLTKSKDLNNCQQRIMKDFGLAYTEKCVECRQRGEALMGAA
TYNYLMKPADNGALILEATVTELHQFTPFNEMSGAAQMEAKQMLTFVEIKKA"
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=209&to=229&view=gbwithparts|exon]] 209..229
/gene="VtgA"
/number=2
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=371&to=522&view=gbwithparts|exon]] 371..522
/gene="VtgA"
/number=3
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=614&to=865&view=gbwithparts|exon]] 614..865
/gene="VtgA"
/number=4
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=953&to=1114&view=gbwithparts|exon]] 953..1114
/gene="VtgA"
/number=5
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=1205&to=1353&view=gbwithparts|exon]] 1205..1353
/gene="VtgA"
/number=6
[[http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=29150215&from=1445&to=1477&view=gbwithparts|exon]] 1445..>1477
/gene="VtgA"
/number=7
ORIGIN
[[#sequence_29150215]] 1 gccatgagag cagtagtact tgcactgact ctagcccttg tgggtaagta cagtttttct
61 gtcttatact agtgcagata ttataactaa cttcatgtaa atataactaa gttatgtaaa
121 tatagaaatg tacaattctg tatctatgac attttttgaa aaacattcag taacatattg
181 tgacaataat ttttgtttta tcttgtagcg agtcaatctg ttaactttgg taagtacata
241 ttatttttat ggatttttac ttatgtttat taggtagttt cacttctgca tttttttcga
301 gttgaataaa ttaagtaaaa ccagttctca gatcacaaga tcatcaaata tgttttgatt
361 gttattacag cccctgattt tgaagccagt aagacctatg tgtacaagta tgaggcactg
421 ctcctgggtg gtctgccaga ggagggtctg gctagagctg gagtaaaagt aatcagcaaa
481 gttcttatca gtgcagttgc agagaatacc tacttgctga aggtatttat tatacatgaa
541 gttcccacac caacaatatc ttaatgtaat gtccattaca ttgactaata catcaattga
601 tatcttcttg tagcttgtga accctgagat ctttgagtac agtggtgtgt ggcccaacga
661 tcctttcgtc ccagctgcaa aactcacttc agccctggct gctcagttct cgattcccat
721 caagtttgag tatgccaagg gtgttgtggg taaggtactt gcccccactg ctgtctctga
781 aacagtgctg aatgtccata gaggtatcct gaacattctt cagctcaaca tcaagaagac
841 acaaaatgtc tatgagttgc aggaggtaaa acttgtatat gtacatatag atgcaaaaca
901 tgtttcagta agcaatgttt gaattcctca ctcatgctct cttttgtgtt aggctggagc
961 tcagggagtg tgcaagaccc actatgtgat cagggaagat gccaaggcag aacgcatcca
1021 tttgaccaag agcaaggatc tcaataactg ccagcagaga atcatgaagg actttggtct
1081 ggcttacaca gagaagtgtg tagagtgccg gcaggtatga gcataagtat ctattttggg
1141 ggttggaaaa acataaccag aaacacttca catgacaata caactctgtg ttttgtctaa
1201 acagagaggg gaggccctga tgggagctgc cacttacaac tacctcatga agcccgctga
1261 caatggtgct ctgatcttgg aggccactgt tactgagctc catcagttca caccattcaa
1321 tgagatgtca ggagctgccc aaatggaagc aaagtatgtt gactgatttg tggtataaaa
1381 ggaaaacatt ttcattttat taagcttata ggtttaacag tttcccatga acatcctttc
1441 acagacaaat gttgactttc gttgagatca agaaggc
//
FASTA:>gi|29150215|gb|AF454747.1| Oncorhynchus kisutch vitellogenin A (VtgA) gene, exons 1 through 7 and partial cds GCCATGAGAGCAGTAGTACTTGCACTGACTCTAGCCCTTGTGGGTAAGTACAGTTTTTCTGTCTTATACT AGTGCAGATATTATAACTAACTTCATGTAAATATAACTAAGTTATGTAAATATAGAAATGTACAATTCTG TATCTATGACATTTTTTGAAAAACATTCAGTAACATATTGTGACAATAATTTTTGTTTTATCTTGTAGCG AGTCAATCTGTTAACTTTGGTAAGTACATATTATTTTTATGGATTTTTACTTATGTTTATTAGGTAGTTT CACTTCTGCATTTTTTTCGAGTTGAATAAATTAAGTAAAACCAGTTCTCAGATCACAAGATCATCAAATA TGTTTTGATTGTTATTACAGCCCCTGATTTTGAAGCCAGTAAGACCTATGTGTACAAGTATGAGGCACTG CTCCTGGGTGGTCTGCCAGAGGAGGGTCTGGCTAGAGCTGGAGTAAAAGTAATCAGCAAAGTTCTTATCA GTGCAGTTGCAGAGAATACCTACTTGCTGAAGGTATTTATTATACATGAAGTTCCCACACCAACAATATC TTAATGTAATGTCCATTACATTGACTAATACATCAATTGATATCTTCTTGTAGCTTGTGAACCCTGAGAT CTTTGAGTACAGTGGTGTGTGGCCCAACGATCCTTTCGTCCCAGCTGCAAAACTCACTTCAGCCCTGGCT GCTCAGTTCTCGATTCCCATCAAGTTTGAGTATGCCAAGGGTGTTGTGGGTAAGGTACTTGCCCCCACTG CTGTCTCTGAAACAGTGCTGAATGTCCATAGAGGTATCCTGAACATTCTTCAGCTCAACATCAAGAAGAC ACAAAATGTCTATGAGTTGCAGGAGGTAAAACTTGTATATGTACATATAGATGCAAAACATGTTTCAGTA AGCAATGTTTGAATTCCTCACTCATGCTCTCTTTTGTGTTAGGCTGGAGCTCAGGGAGTGTGCAAGACCC ACTATGTGATCAGGGAAGATGCCAAGGCAGAACGCATCCATTTGACCAAGAGCAAGGATCTCAATAACTG CCAGCAGAGAATCATGAAGGACTTTGGTCTGGCTTACACAGAGAAGTGTGTAGAGTGCCGGCAGGTATGA GCATAAGTATCTATTTTGGGGGTTGGAAAAACATAACCAGAAACACTTCACATGACAATACAACTCTGTG TTTTGTCTAAACAGAGAGGGGAGGCCCTGATGGGAGCTGCCACTTACAACTACCTCATGAAGCCCGCTGA CAATGGTGCTCTGATCTTGGAGGCCACTGTTACTGAGCTCCATCAGTTCACACCATTCAATGAGATGTCA GGAGCTGCCCAAATGGAAGCAAAGTATGTTGACTGATTTGTGGTATAAAAGGAAAACATTTTCATTTTAT TAAGCTTATAGGTTTAACAGTTTCCCATGAACATCCTTTCACAGACAAATGTTGACTTTCGTTGAGATCA AGAAGGC
Primers for Vtg:
Primer pair 1
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
TAAGGTACTTGCCCCCACTG |
Plus |
20 |
751 |
770 |
59.99 |
55.00% |
| Reverse primer |
CCTGAGCTCCAGCCTAACAC |
Minus |
20 |
965 |
946 |
60.01 |
60.00% |
| Internal oligo |
Plus |
||||||
| Product length |
215 |
||||||
Primer pair 3
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
TGAAGTTCCCACACCAACAA |
Plus |
20 |
537 |
556 |
59.98 |
45.00% |
| Reverse primer |
CAGTGGGGGCAAGTACCTTA |
Minus |
20 |
770 |
751 |
59.99 |
55.00% |
| Internal oligo |
Plus |
||||||
| Product length |
234 |
||||||
25 February 2009
Pulled livers and sexed coho for my project today. 10 coho smolts each from a control and a pesticide cocktail treatment group were used. The X is carryover notation from the main project and denotes the location where these fish were reared. C is control, T is pesticide treatment (pulsed cocktail exposure from fertilization to smoltification).
| Group |
Phenotype gender |
Whole liver weight (mg) |
RNA liver sample weight (mg) |
Protein liver wt (mg) |
Comments |
| XC-1 |
M |
129 |
64 |
65 |
|
| XC-2 |
M |
120 |
55 |
65 |
|
| XC-3 |
M |
125 |
67 |
58 |
|
| XC-4 |
M |
127 |
57 |
70 |
|
| XC-5 |
M |
113 |
62 |
51 |
|
| XC-6 |
M |
170 (91) |
47 |
44 |
1/2 dropped on floor when split- final weight of tissue used was 91mg |
| XC-7 |
M |
80 |
42 |
38 |
|
| XC-8 |
M |
101 |
52 |
49 |
|
| XC-9 |
F |
116 |
58 |
58 |
|
| XC-10 |
F |
95 |
50 |
45 |
|
| XT-1 |
F |
55 |
35 |
20 |
Only used 500ul of Tri Reagent |
| XT-2 |
F |
88 |
60 |
28 |
|
| XT-3 |
F |
87 |
45 |
42 |
|
| XT-4 |
F |
65 |
36 |
29 |
Only used 500 ul of Tri Reagent |
| XT-5 |
M |
74 |
46 |
28 |
|
| XT-6 |
F |
80 |
~40 |
40 |
Forgot to weight before putting in reagent (use for sample run set) |
| XT-7 |
F |
107 |
61 |
46 |
|
| XT-8 |
M |
129 |
79 |
50 |
|
| XT-9 |
F |
64 |
43 |
21 |
|
| XT-10 |
F |
123 |
73 |
50 |
I intend to run 4 samples through the procedures to work out the kinks, then run 8 from each group for the project.
The phenotypic sex is opposite between groups. Christine indicated that females are more rare, noting that she needed 2 for an experiment once and went through 20 fish before meeting her quota. She was not surprised when the 6 control group fish we brought to practice on were all males. My control group ratio meets those anecdotal expectations. The treatment group fish I labelled female clearly had ova, though they were less developed (shorter) than the control group ova.
1 March 2009
Zr-protein
Primer pair 6
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
CATCACTGTGGGACCCTCTT |
Plus |
20 |
938 |
957 |
59.96 |
55.00% |
| Reverse primer |
TGAACTGCAGCAGGAATGTC |
Minus |
20 |
1150 |
1131 |
59.99 |
50.00% |
| Internal oligo |
Plus |
||||||
| Product length |
213 |
||||||
| Product Tm |
|||||||
| Product Tm - min(OLIGO Tm) |
|||||||
Products on allowed transcript variants----
Products on potentially unintended templates----
Products on target templates----
Primer pair 7
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
CAGTACCATTGTGGCTGTGG |
Plus |
20 |
688 |
707 |
60.02 |
55.00% |
| Reverse primer |
AAGAGGGTCCCACAGTGATG |
Minus |
20 |
957 |
938 |
59.96 |
55.00% |
| Internal oligo |
Plus |
||||||
| Product length |
270 |
||||||
Vtg
Primer pair 6
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
CCAGTTGTCGCTCTCATCAA |
Plus |
20 |
221 |
240 |
59.98 |
50.00% |
| Reverse primer |
AAGCCACCTCCAATGTCATC |
Minus |
20 |
450 |
431 |
59.93 |
50.00% |
| Internal oligo |
Plus |
||||||
| Product length |
230 |
||||||
ER-b
Primer pair 4
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
ACACGGTGACCATAAGCACA |
Plus |
20 |
440 |
459 |
60.03 |
50.00% |
| Reverse primer |
GTGCCCGATGTCAGTGTATG |
Minus |
20 |
824 |
805 |
59.99 |
55.00% |
| Internal oligo |
Plus |
||||||
| Product length |
385 |
||||||
ER-a
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
CCTCTGGGTACCACTACGGA |
Plus |
20 |
651 |
670 |
59.98 |
60.00% |
| Reverse primer |
CGCTTGTCCTTCCTGAGAAC |
Minus |
20 |
882 |
863 |
59.99 |
55.00% |
| Internal oligo |
Plus |
||||||
| Product length |
232 |
||||||
2 March 2009
Isolated RNA from all samples today. NanoDrop spec'd the first 8 but samples XT_2, XT_5, XT_6, XT_7, XT_8 were over 2000 ng/ul so I diluted them with another 100 ul of .1% DEPC H2O. Will spec all again tomorrow. Just realized I should have set this notebook up to enter newest entries on top...
3 March 2009
NonoDrop spec'd the remaining RNA and diluted as above anything over 2000 ng/ul.
4 March 2009
DNased all samples. RVTranscriptase too low to run reverse transcription. Nanodropped all samples for post DNase RNA values.
5 March 2009
Isolated DNA with 10% Chelax. Did not know to shake chelax until I had added it to half my samples, so I sucked it out and added new, spun chelax to them all. .5 ml Chelax added to liver samples, homogenized, vortexed, heated at 95 C for 20 min, then put in the freezer (-20C). Forgot to put a block on top of samples while they were heating and 3 lids popped open during the process.
Reverse transcribed RNA using 1/2 the listed amounts from lab 3 above to make a total of 10 ml cDNA due to low RVTranscriptase supply. Placed samples in thermocycler, set program and walked away for an hour. Returned to find the thermocoupler in query mode, noted that my samples were at room temp, reset program and actually turned it on this time. During post-thermocycler centrifugation (7 secs) one of the microtube lids broke (treatment sample #8) and the tube split. It was sitting in a 1.5 ml tube that I prepped for the centrifuge yesterday so I sucked the microtube and cDNA out and put it all in a fresh 1.5 ml tube with a lid and put it with the rest of the cDNA in the freezer. Checked phenotype data and following the trajectory of my evening the broken sample was one of 2 boys out of the 10 treatment samples... Then I forgot that I drove in and took the bus home.
6 March 2009
Centrifuged DNA samples at max speed in refrigerated centrifuge for 5 min. PCR thermocycled the DNA samples as per lab 3 (95-55-72 C cycling- protocol 441 on thermocycler) and ran them out on agarose gel.
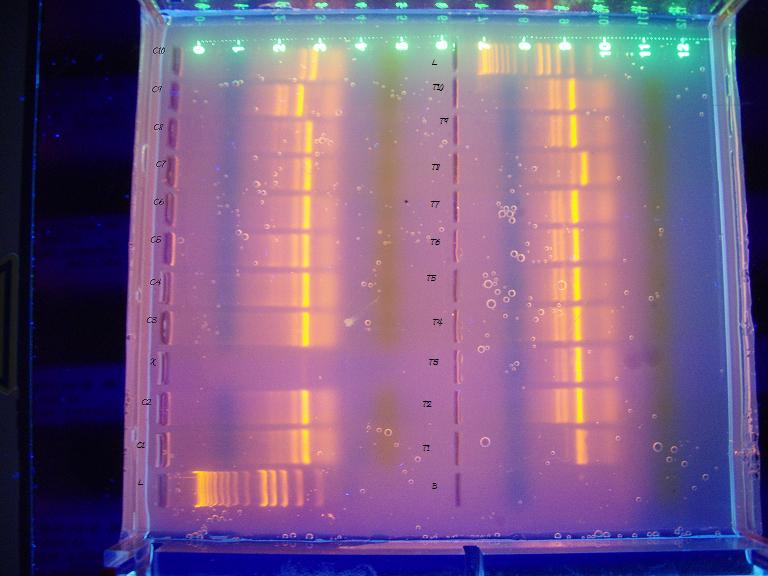
Left side are controls, right are treatments. T2 & T3 are mislabelled, the samples run blank, T1,T3,T2 from the bottom. I am a little concerned that I didn't run the gel long enough, though I ran it for over an hour and 30-45 min is the recommended time. The bands I am looking for should be 290 bp and the ladder looks a bit squished at the bottom, further fueling my duration concerns. According to the phenotype analysis, the last two controls (C9 & C10, top left 2 columns) were female. The columns that differ from the majority also differ from eachother, leaving me with no conclusive evidence of gender determination.
9 March 2009
After reviewing the PCR gel with Steven and comparing it to the ladder, it turns out I had something after all.
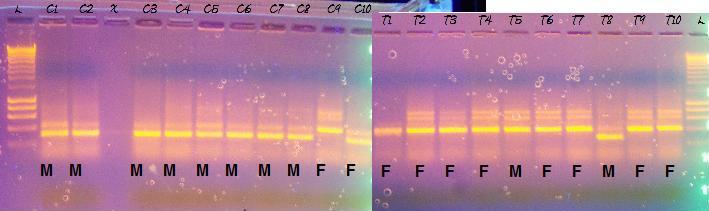
The black text is the phenotype result for each sample. According to this gel, I was 1 off in my phenotyping of each of the control and treatment categories (C10 and T5) and the gender ratio between treatment and controls are flipped in a 9/10 ratio; males are king of controls and females rule the treatments. I ran a Chi-square test on a 2x2 contingency table with the gender results and the two sets are statistically different from eachother. The Gh5/GH6 primer pair should have resulted in males with a band at 290bp. The band on the ladder corresponding to most of the bands on the right side of the above image is at 400 bp; the rest of the bands are ~290. Kerensa thought that gender determination occurred at 40 days. Devlin et al (2001) feminized chinook salmon by treating hatched embryos with estradiol 1x/wk throughout hatch to first feeding period followed by dietary estradiol for another 10 weeks. Given the potential for sex reversal with early exposure to estrogenic compounds, I expected to find a lot of phenotypic females in the treatment group, and I expected the genomic DNA to reveal that they were, in fact, males.
10 March 2009
Started QPCR with Vtg primer today following methods from lab 5. Thought I might start another primer set to run overnight, but I didn't want to wait for John's to finish and I wanted to make sure the first set was ok before proceeding.
11 March 2009
Started the ER-alpha and ER-Beta QPCR this morning. Reviewed Vtg results with Steven and there were double peaks in all of the data though the waters were clean, indicating that perhaps my annealing temps were off or I have contaminated cDNA. Will not be able to determine cDNA contamination from the ER set because the computer crashed in the middle of it. Steven offered to have someone in his lab run my samples after John's set this afternoon.
12 March 2009
ER results: ER-alpha was not detected but ER-beta did have some non-significant trend of upregulation in the treated fish. Vtg results may also have been multiple Vtg products and a similar non-significant trend was seen in that data. ER results interesting in that ER alpha is thought to be more correlative with Vtg and ER b less so.
Abstract from presentation:
Effects of a Pesticide Cocktail on Coho Reproductive Physiology
Amy Yahnke
Fish 441
13 March 2009
Multiple pesticide residues detected in urban streams during storm events show that anthropogenic contaminants rarely occur as single compounds in the environment, and therefore should not be treated as such when evaluating their toxicological effects. “Pesticide cocktails” contain endocrine disrupting compounds that can disrupt sexual differentiation and reproductive physiology of aquatic organisms. This study evaluates 3 endpoints of Coho (Onchorhynchus kisutch) exposure to endocrine disrupting compounds in a pesticide cocktail designed from August-December maximums reported in urban streams. Coho treated from egg to smolt received 4-day pulsed exposure to a mixture of 8 herbicides (2,4-D, Dichlobenil, MCPP, Prometon, Triclopyr, MCPA, Simazine, Dicamba), 1 fungicide (Pentachlorophenol), 2 insecticides (Diazinon and Carbaryl), and 1 transformation product (4-nitrophenol). Pentachlorophenol is estrogenic and Carbaryl is thyroidogenic, and 4 other compounds from the mixture are identified as initial screening priority compounds for the Environmental Protection Agency and EU’s endocrine disruptor investigations. Coho smolts were dissected and phenotypic sex was determined by presence of testes or ova. Genotypic sex was determined using PCR on genomic DNA isolated from livers (GH-5 and GH-6 primers), with the hypothesis that treated fish would show differential phenotypic and genotypic genders due to xenoestrogen-induced feminization. Three genes associated with estrogen response in teleost reproductive systems, hepatic vitellogenin (Vtg), estrogen receptor alpha (ER-α), and estrogen receptor beta (ER-β), were analyzed using QPCR. Vtg and ER-α are shown to inductively respond to xenoestrogens in fish. Phenotypic and genotypic results were similar, but showed a significant (X2, p<0.005) flipped sex ratio favoring females in the treated fish (controls 9:1, exposed 1:9). No ER- α was detected, but ER- β and Vtg showed a non-significant trend of higher expression levels in treated fish. Vtg expression was also higher in the two phenotypic female (1 genotypic) controls. Further investigations should include pesticide cocktail effects on gender determination and sex ratios and increased sample sizes for reproductive gene expression analysis.