1. Detail at least 2 reasons why your results turned out the way they did. This should be easy to do if your results are "unexpected", but even expected results can have multiple explanations. Really think about this, the answer "because I messed up in lab" (or any variation thereof) is not acceptable.
- The primary antibody wash was not left off for a significant amount of time to allow for binding to methylated cytosines. Evidence for this is based on the fact that a previous dot blot, which left the primary antibody was on for 12 hours, had dark stains while mine did not.
- The box I used to perform the washes in was larger than the previous used box, thus making the washes have less depth and possibly be less effective.-
emmatsMore likely the former reason than the latter.
2. What are two obstacles that you encountered during your lab work and experimental design? Did these obstacles affect your results? Why?
- The first obstacle was related to low number of individuals resulting in small amounts of duplication for 3 different treatments. By obtaining diploids we were able to three different treatments in addition to comparing diploids and triploids.
- A second obstacle was the high mortality rate in oysters in the low pH treatment after only 24 hours of exposure. This forced us to do premature sampling, and didn't allow for us to do direct comparisons between low and high pH treatments. However, it did give us some insight in the amount of stress an oyster may experience in regards to the two treatments.
3. Explain at least one aspect of your research and its results that have a greater impact outside of your own personal learning experience. What would you tell a non-scientist who challenged the importance of your research?
- The emerging understanding of epigenetics, specifically DNA methylation, is suggesting strong ties between the epigenome and gene expression ultimately resulting in physiological changes. Understanding these patterns in oysters is important due to their commercial and ecological importance. They are an important of the shellfish farming industry, and the play and important ecological role in providing habitat and filtering the water. Using molecular tehcniques to quanitify changes in physiology will result in a better understanding of response to stressful conditions such as ocean acidification.
4. What part of your research and analysis has completely stumped you? Is there anything you can do to find the answer or will it always remain a mystery?
- The most stumping moment so far has been the mystery as to why my dot blot did not work as planned. Ultimately I think it was a combination of different factors that could potentially be prevented with a second try, but most likely it will forever remain a mystery.
5. In about 3 sentences each, summarize 2 papers that you are going to cite in your own paper that give insight into the results that you found.
- Anthropogenic ocean acidifcation over the twenty-first century and its impacts on calcifying organims- The process of rising atmospheric CO2 levels is resulting in higher levels of acidity in the world's oceans. This is resulting in a shift in the carbonate buffer system resulting in a decline in the number of carbonate ions available for shell calcification. This may lead to a number of physiological changes that place unneeded stress on organisms, such as oysters, that utilize calcified shells as an important of their body structures.
- DNA methylation patterns provide insight into epigenetic regulation in the pacific oyster (Crassotrea gigas)- DNA methylation was detected in pacific oysters, and inferred to have regulatory abilities. It is likely that methylation patterns can alter the expression of stress genes such as HSP-70. More studies looking at comparison between stressed and non-stressed individuals is required to understand the importance of DNA methylation and stress response.
Date: 11/22/11
Summary:
Discussed results from last lab including the qPCR and methylation dot blot. Also we discussed the expectations for the paper draft due on Nov. 28th. And I had worked previously on processing samples to look at methylation patterns in all 33 oysters.
Methods:
See methods from previous lab to understand techniques used in qPCR and methylated cytosine dot blot.
Results:
Labeled methylated cytosine dot blot from previous lab. The fly DNA indicated by the label SC did not show up on the blot indicating low levels of methylation. The AET column indicated higher levels of methylation than any of the other samples. The columns on the left side did not have as high of levels of methylation of the other columns and may be due to them being different samples than the other Cg samples which do show methylation.

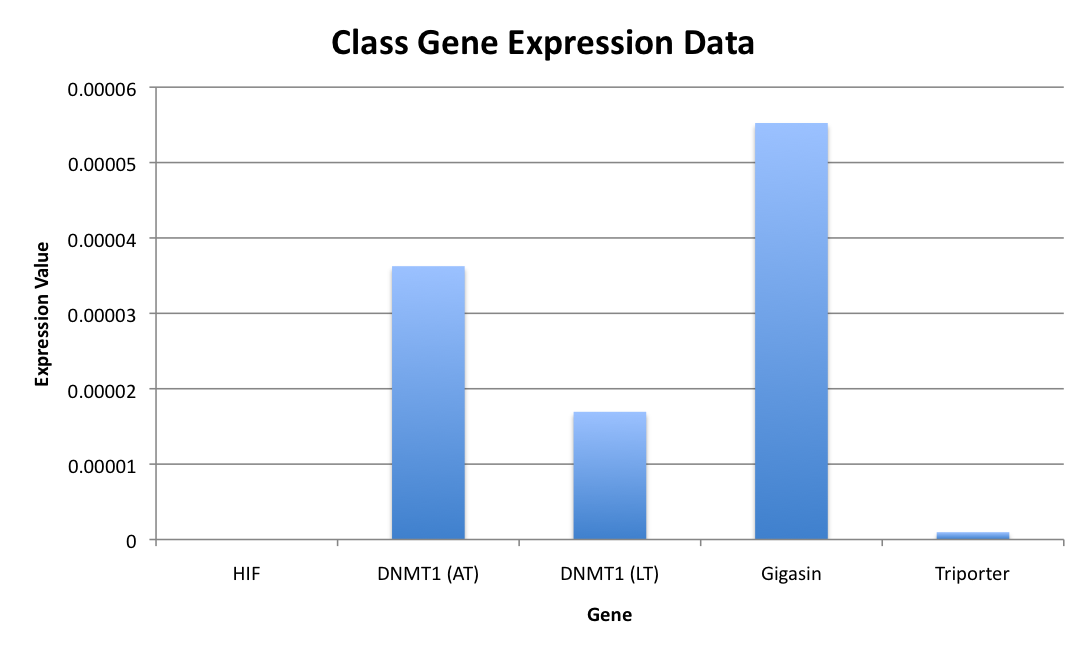
The above chart indicates the level of mRNA expression for various genes in various samples. The DNMT genes showed differing level of expressions in two different oyster samples. The gigasin showed much higher levels of expression than any of the other genes. And the triporter showed vary low levels of expression in sea urchin samples, while the HIF showed no level of mRNA expression.
Conclusions:
The dot blot can be used to make inferences about the level of methylation given that the concentration of the DNA used is known. A darker dot indicates increased methylation and comparisons can be made between similiar samples.
The level of mRNA expression varies between samples and genes, but it may be a result of the quality of the cDNA used in the qPCR process. A badly processed sample will register a signal that is not accurate the actual level of expression. In this case the gigasin sample was very clean and showed up well, as oposed to the triporter sample which did not amplify as well and had multiple peaks in the melt peak graph. This needs to be considered when interpeting the results.
The dot blot that I completed on 11/17 did not turn out as well as the other dot blot. This may have been due to not leaving the primary antibody on long enough or having a vessel that was too large to allow for the membrane to be adequetly washed in the solution. Additionally there appeared to be contamination on the membrane surface since there are large faint stain visible.
Reflection:
The qPCR seems like an especially useful way to quantify physiological changes in an organism. As the name implies the ability to quantify the amount of cDNA allows for more comparisons to be made. The qualitative nature of the dot blot limits it uses to being a simple indicator of whether DNA methylation is present or not. More complex techniques are required to get quantiative measurements for DNA methylation. In both cases the utmost care has to be taken in handling the samples since a slight mistake can result in inaccurate results.
Date: 11/15/11
Summary:
A methylation dot blot was performed to measure the expression of DNA methylation in oyster samples. DNA was diluted and put on to a membrane before being washed with antibodies. Additionally a qPCR was performed on cDNA samples. This involved creating a PCR mixture and running a gel for quantification.
Materials and Methods:
DNA dilution
- A DNA sample from a C.gigas triploid exposed to high pH conditions was chosen
- 41.32 uL of DNA was mixed with 39.68 uL of water to form 80 uL of 25ng solution
- This dilution was then further diluted to 5 dilutions containing 800, 400, 200, 100, and 50 ng of total DNA in 200ul of mixture. 60 ul of 20x SSC made up part of 200 ul with the rest being DNA and water for dilution.
qPCR
- First a master mix large enough for 10 reactions was formulated. This was 125ul of Master mix 2x, 10ul of syto-13 dye, 12.5 ul of upstream primer, 12.5ul of downstream primer, and 80ul of DI water.
- The primers used were for the DNMT1 gene and diluted to 10uM with TE buffer. And the two cDNA samples were triploid C.gigas.
- 2uL of cDNA were added to each of the positive reactions (2 x 2 samples =4) and 2uL of DI water were added to the 4 negative controls.
- Tubes were capped and spun down.
- Loaded into qPCR gel and run.
Dot blotting
- A trimmed nylon membrane was soaked in 6x SSC for 10 minutes.
- A similarly shaped piece of filter paper was soaked in 6x SSC until wet.
- We placed the membrane and filter paper in to the vacuum manifold.
- DNA dilutions were denatured by placing boiling water for 10 minutes and transfered to ice.
- 500 uL of 6x SSC were applied to each well and the vacuum was adjusted to take a few minutes for it to filter through
- DNA was spun down for 5 minutes.
- Entire volume of DNA samples (200 uL) was applied to each well.
- Samples filtered through by vacuum.
- Soaked a piece of filter paper in denaturation buffer.
- Membrane was transferred from manifold to denaturation soaked filter paper.
- Membrane was then placed on a neutralization soaked filter paper for 5 minutes.
- Membrane was placed on dry filter paper and dried.
- Membrane was put into the UV transluminator for 2 minutes at 120kJ
Antibody Washing
- 20 ml of blocking solution was formulated with 14 ml of water, 4 ml of part A blocker, and 2 ml of part B blocker.
- Placed the membrane in 10 ml of blocking solution and incubated for 30 minutes on the shaker tray.
- The blocking solution was removed and 20 ml of water was poured on and shaken for 5 minutes. This was repeated.
- 10 ml of antibody solution was prepared using 10 ml of blocking solution and 2 ul of antibody.
- The membrane was incubated with 10 ml of primary antibody solution for one hour.
- The membrane was then washed with 20 ml of TBS-T for 5 minutes four times.
- Incubated membrane in 10 ml of seconday antibody solution for 30 minutes.
- The membrane was rinsed for 2 minutes with 20 ml of water 3 times.
- Incubated the membrane with 5 ml of chromogenic substrate until color began showing.
Results:

The DNA dilutions loaded into column 11 showed up for all 5 dilutions. The strength does not appear to vary a lot with the amount of DNA present. The 100 and 50 ng samples may be fainter than the larger samples.
Conclusions:
Diploid C. gigas from a control pH demonstrates methylation. The fly DNA did not register a signal so it can be used a sort of control and we know that the oyster is demonstrating more methylation than the fly.
Reflection:
The DNA samples I used registered a positive signal for DNA methylation, which means not only that methylation is present, but that the experiment protocol was followed correctly. I can now perform the same procedure on my 30 other samples confident that I will get results. The purpose of this lab was to yet again introduce us to new molecular techniques that we can use in the lab. Quantification of cDNA is useful for measuring the amount of gene expression seen a tissue samples, and is more useful than performing a traditional PCR. A methylation dot blot is useful for measuring the amount of methylation in a tissue sample. This an indicator of the organisms epigenome and comparisons can be made between samples. Overall, these techniques would be useful for understanding environmental impacts on physiology.
Date:11/8/2011
Summary: Began working on independent research. Performed DNA extractions on 31 C. gigas tissue samples.
Materials and Methods:
DNA extraction
- Cut a sample of tissue that was .025-.05 g. Chopped tissue with razor blade to help with homogenization
- In a 1.5ml conical tube added .5ml of DNazol and 2.4uL of ProK.
- Incubated samples for 1 hour at room temperature.
- Spun samples at 10,000g for 10 minutes.
- Transfered supernatant to a new tube.
- Added .5 ml of 100% ethanol.
- Mixed sample by intverting
- Incubated for 1 minute at room temperature
- Spun samples for 5 minutes at 5000g.
- Removed all liquids and left DNA precipatate untouched at the bottom of the tube.
- Washed sample with 1 ml of 75% ethanol by pipetting in, inverting tube, and then pulling the ethanol back out.
- Repeat step 11.
- Add 300ul of .1% DEPC water and pipette up and down to mix.
- Place finished extractions in -20 C freezer.
Results
Hopefully some good extractions.
Conclusions
Performing so many extractions was difficult and if I had to do it again i would have spent more time homogenizing the tissue. It was hard to extract the supernatant efficiently without contamination. Similar difficulty in performing the washes.
Reflection
Processing samples takes time, and experience makes it go faster. I will do things differently next time.
Date:11/1/2011
Summary:
Using our cDNA PCR product from last week we placed samples into a gel and ran it for 45 minutes to determine presence and length of product. The -70 cDNA was present in both positive control and absent in the negative controls. The length was approximately 200 basepairs. Next we performed a protein extraction from a mantle tissue sample of C.gigas. This extraction was used to perform a western blot. In order to do so the extraction was processed and placed in a gel. The gel was then blotted onto a membrane that was stained through a process that invovled multiple antibody solutions.
Materials and Methods:
PCR gel electrophersis
- Placed 1.5% agrose gel in gel box and cover with 1xTAE buffer.
- loaded 7 uL of ladder to wells on both ends of both rows.
- loaded 25 uL of PCR product into each well.
- Ran the gel for 60 minutes at 100V
- Looked at gel in UV box to determine length and what was amplified.
- A 1.5ml snap cap tube was labeled with AD, 11/1.
- A .030 g triploid C. gigas mantle tissued sample was placed in the freshly labeled tube.
- 500 ul of CellLytic MT solution was added to the tube.
- A sterile pestle was used to homogenize the mixture. The mixture was inverted severa times.
- The tube was then spun at max speed in the centrifuge for 10 minutes.
- A fresh 1.5ml snap cap tube was labeled protein AD 11/1
- The original tube was removed from the centrifuge and the supernatant was pipetted into the new tube.
- In a 1.5 ml screw cap 15uL of protein extract and 15uL of 2x reducing sample buffer were added.
- Mixed and then centrifuged to pool liquid.
- Boiled sample for 5 minutes
- Centrifuged boiled sample for 1 minute
- Added 7 uL of later to one end of the gel
- Added 30uL of protein extract sample to each well of the gel
- Ran gel for 25 minutes at 150V
- Assembled the blotting sandwich with a filter paper, membrane, gel, more filter paper. This sandwich was placed on top of the positive anode and thoroughly soaked in tris-glycine transfer buffer.
- The cathode was placed on top and the blow was run for 30 minutes at 20V
- The gel was removed from the membrane and placed in a staining solution to make protein visible
- The membrane was rinsed twice in super pure water by placing in tray of water and placed on a shaker plate for 5 minutes.
- Next the membrane was placed in a new tray and covered with blocking solution.
- The tray was then incubated overnight on a shaker plate set to 1 revolution/minute.
- The membrane was rinsed with 20 ml of water for 5 minutes twice.
- Membrane washed with primary antibody solution (HSP-70) 3 times for 5 minutes at a time.
- Then incubate membrane with 10ml of secondary antibody solution for 30 minutes, twice.
- Incubate in chromogenic solution until purple bands appear.
- Dry membrane in open air.
- Look at membrane to see what proteins are present.
Results:

The above photo shows wells 1-9 from the second row of the gel run in lab. Wells 6 and 7 are the amplified HSP-70 cDNA product. The base pair length is approximately 200 basepairs. This is the expected length given the primers that were used. Wells 8 and 9 are negative controls and only a faint trace of primer dimer is present indicating uncontaminated samples.


Looking at the two above photos the ladder can be seen on the left most side of the SDS stained gel. Some extracted proteins may have had higher concentrations indicated darker bands on the gel.

This photo shows the western blot. There are no stains present indicating that no HSP-70 was present in any of the C.gigas or sea urchin samples.
Conclusions:
The results from the gel electrophoresis of the PCR products was as expected and encouraging. The HSP-70 mRNA was present in the original tissue samples as indicated by the singular bands seen on the gel. The length was as expected, and the negative controls remained uncontaminated. All of the steps were completed properly.
The western blot results were not as expected. HSP-70 protein was not expressed in any of the 12 different tissue samples. Protein was present as indicated by the stained gel, but the antibody wash shows that none of that protein was HSP-70. It is possible that a mistake occurred in the western blot process thus giving an all negative result.
Reflection:
Important methods were covered during the course of this lab. We were able to analyze our PCR products that we have now been working on for multiple weeks. Additionally we learned how to look at identifying if a specific protein is present by using a western blot. Both of these methods are useful for looking at expression of given proteins in a tissue sample. Ultimately the western blot is more indicative of what is actually present then the mRNA PCR product. Both can be used to answer questions regarding environmental stress and its molecular/cellular effects on an organism. Utilizing these methods will help us answer questions about ocean acidification stresses on oysters.
Date: 10/25/2011
Summary:
Using cDNA samples from last week we mixed primers and flourescent dye in to create a PCR. HSP70 for C.gigas forward and reverse primer that had been previously constructed were used. after mixing and formulation the tubes were added to a thermocycler where PCR could occur. The PCR will allow us to look at the samples with gel electrophroesis next week. Tissue was also samples from the 22 remaining oysters in our experiment. A single mantle sample and 3 gill tissue samples were taken from each individual. The samples were placed in the -80 C freezer for future analysis.
Materials and Methods:
PCR
- Spun primers (HSP70 F, HSP70 R) in centrifuge for 3 minutes to pool liquid.
- Added 204 uL of TE buffer to HSP70 F and 369 uL of TE buffer to HSP70 R.
- These mixtures were incubated at 45 C for 2 minutes.
- Primers were then vortexed, and spun down for 2 minutes to pool.
- A 9:1 dilution was performed for both primers. 10 uL of primer/TE buffer were added to a 1.5ml snap cap (labeled: AD,SC 10/25 R or F), and 90 uL of DI water was added to dilute.
- A mastermix was formulated in a 1.5ml tube (labeled: MM). 125 uL of GoTaq green master mix 2X, 5ul of both forward and reverse primers, and 105uL of DI water were added to this single tube. This mixture was vortexed.
- 48 uL of the mastermix were pippeted into one of four .5ml PCR tubes.
- 2uL of DI water were added to two of the tubes to act as negative controls (labeled: 1 NC, 2 NC)
- 2uL of cDNA sample were added to two of the tubes (labeled: 1 HS, 2 HS)
- Tubes were vortexed and spun down to pool liquids before being loaded into the thermocycler.
- The samples then went through a thermal cycle and were stored at -20 C when finished.
- Tubes were labeled Date, oyster number, diploid or triploid (2n/3n), dry or wet (D/W), and if not gill then tissue type(mantle).
- 3 gill tissue samples were taken from each oyster.
- 1 mantle tissue sample was taken from each oyster.
- After sampling tubes were placed in a -80 C freezer for storing.
- Sampled oysters were disposed of.
Results:
Amplfified cDNA that can be used for later analysis.
88 tissue samples that can be analyzed from DNA, mRNA, and protein.
Conclusions:
Science is fun.
Reflection:
The purpose of this lab is to further familiarize us with molecular techniques that can be used to answer questions. PCR is a powerful tool for analyzing tissue samples of all organisms. The steps performed today were not difficult and could be easily repeated. The second part of the lab allowed us to gather our tissue samples. The techniques we have learned in past labs will be useful fro analyzing the samples and answering the questions we orginally posed about stress and acidity.
Date: 10/18/2011
Summary:
Reverse transcriptase was added to a mixture of our previous C. gigas RNA sample to create cDNA that was put on ice for PCR analysis at a later date. The experimental design for the ocean acidifaction with oysters experiment was finalized and pH was measured from the water samples. Methods for constructing primers were reviewed.
Materals and Methods:
Reverse Transcription
- RNA sample (C. gigas gill) was removed from ice and mixed.
- .5ml PCR tube was labeled with AD, SC 10/18 cDNA.
- 5 uL of RNA sample,1 uL of oligo DT and 4 uL of DI water were added to the PCR tube
- The mixture was then incubated for 5 minutes at 70 C on a thermocycler.
- After incubation the tube was put on ice for 2 minutes.
- The mixture was spun down in a centrifuge for a few seconds.
- 5 ul of M-MLV 5x reaction buffer, 5 uL dNTPs, 1 uL of M-MLV RT, and 4 uL of DI water were added to the PCR tube.
- Mixture was vortexed then spun down in the centrifuge briefly.
- The mixture was then incubated for 60 minutes at 42 C and then heat inactive at 70 C for 3 minutes in a thermocycler
- Did not perform: spun down sample in centrifuge and stored on ice at -20 C
- Typed in keywords on ncbi database.
- located approriate genes
- Designed primers with the following parameters
- 20 base pairs in length
- melting points within 2 C of each other
- amplified length 80-200 base pairs long
- no peculiarities such as GC stretches of hairpins
Three treatment groups were placed in 2 trash cans. 12 diploid and 10 triploid oysters were placed in 7.24pH water, and 6 diploid and 5 triploid oysters were placed in 5.24pH water. 24 hours before sampling occurs 6 diploid and 5 triploid oysters will be removed from the 7.24pH water and placed out to dry. All oysters will be tissue sampled after 144 hours of exposure. 3 tissue samples will be taken from each for RNA, DNA and protein analysis. Lengths and weights of oysters will be recorded. pH and survival will be checked every 24 hours.
Results:
cDNA for pCR analysis
A good experimental design
primers
Conclusions:
Our results are what we expected.
Reflection:
The purpose of this lab was multi-faceted, and taught experimental design along with molecular lab techniques. Formulating primers is relatively easy and extremely useful for using cDNA techniques. The experimental design took a lot of refining and flexibility, but ultimately worked out in our favor to test multiple hypothesis about ploidy, acidity, and general stress. I understand the process of creating cDNA and why it is useful. Lessons learned during this lab will be useful for later labs.
Date: 10/11/2011
Summary: The purpose of this lab was to finish the process of RNA extraction of a C. gigas gill tissue sample for quantification. The homogenized mixture from the previous week was further processed by centrifuging and the addition of various chemicals including chloroform, isopropanol, and EtOH. After processing the tube was placed in a nanodrop machine that used the Beer-Lambert law to calculate the RNA extraction concentration level.
Materials and Methods:
RNA Extraction:
- Homogenized tissue (labeled: AD, SC, 10/4/11) from the previous lab was incubated at room temperature for 5 minutes.
- 200 uL of choloform were added. The mixture was then vortexed for 30 seconds until it was milky looking.
- The tube was then again incubated at room temperature for 5 minutes.
- The tube was then spun at max speed in the refrigerated centrifuge for 15 minutes.
- After carefully removing the tube from the centrifuge the aqueous layer was pippetted off and place in a new sterile microfuge tube. The interphase was disposed of.
- 500 ul of isopropanol were added to this new tube (labeled: AD, SC 10/11/11). The tube was inverted several times to mix.
- The was then incubated at room temperature for 10 minutes.
- The tube was then spun at max speed for 8 minutes in a centrifuge at room temperature.
- After centrifuging the supernatant was carefully removed leaving only the remains of a small white pellet.
- 1 mL of 75% EtOH was added to the tube with the pellet and vortex briefly to dislodge the pellet.
- The tube was then centrifuged at 7500g for 5 minutes at room temperature.
- The supernatant was then again removed leaving only the pellet.
- The tube was briefly spun in the centrifuge to pool the residual supernatant.
- After all supernatant was extracted the tube was left open to dry for 5 minutes at room temperature.
- 100 uL of .1% DEPC-H20 were mixed with the pellet by pippetting.
- The tube was then incubated at 55 degrees C for five minutes to solubilize RNA
- The tube was then removed from the incubating tray and flicked a few time before being placed on ice
RNA Quantification:
- 2ul of .1% DEPC-H20 were placed onto the nanodrop pedestal and the arm lowered.
- On the computer screen blanked was click to calibrate the device.
- Next 2 uL of RNA sample were placed onto the nanodrop pedestal and the arm lowered.
- The button measure was clicked on the screen. The Beer-Lambert law is used by the nanodrop spectrometer to calculate concentration
- The values of RNA concentration: 221.9 ng/uL, ratio A260/A280: 1.80, and ratio A260/A230: 2.36, were recorded.
- The pedestal was cleaned with a kimwipe and the next sample was placed on it.
- The RNA sample went back onto ice and eventually storage in a -80 degree C freezer.
Results:
RNA concentration: 221.9 ng/uL
A260/A280: 1.80
A260/A230: 2.36
Conclusions:
The ratio of A260/A280 is within the expected range of 1.8-2.0. The ratio of A260/A230 is outside the expected range of 1.5-2.0 at 2.36. This suggests that our sample may not contain only RNA.
The concentration of RNA is within the expected range and overall suggests a successful extraction. Based on these results we can compare our concentration levels to those found in other tissues, and to values found in literature.
Reflection:
The purpose of this lab was to introduce the methods that are involved with RNA extraction and ultimately quantification. These results are useful for measuring the amount of RNA in a given tissue. This is important because it quantifies the amount of transcription that is occurring in a tissue. Comparisons can be made between samples taken from specimens under different environmental regimes. Ultimately this kind of data is useful for understanding the mechanistic reactions of an organism to environmental change. One thing I hope to understand better is how these methods can be used to isolate RNA coding for a specific enzyme rather than just all of the RNA present in a random tissue sample.
Date: 10/4/2011
Summary: The lab was divided into two parts. First, the preliminary steps of RNA extraction were performed, which involved the homogenization of a C. gigas gill tissue sample with TriReagent. The homogenized mixture was than frozen in preparation for the next steps to be done in a week. The second half of the lab involved doing a protein extraction. An tissue sample from the digestive gland was homogenized in a similar way with addition of CellLyticMT solution. After being centrifuged the supernatant was pippetted off, a blank was created, and spectroscopy was performed to measure the absorbance of the sample. A standard curve was used to calculate the concentration from the absorbance.
Materials and Methods:
RNA extraction:
RNA Isolation:
- A 1.5 mL snap cap tube was labeled with AD,SC 10/4.
- A C. gigas gill tissue sample of .013g was taken off of the ice and added to the tube.
- 500uL of TriReagant were added to the tube with tissue.
- The tissue sample and TriReagant were then homogenizied by the use of a sterile pestle. And quickly vortexed.
- Another 500 uL of TriReagant was added to the tube.
- The tube was vortexed for 15 seconds then placed in a freezer a -80 degrees Celsius.
Observations: The sample was homogenized to the best of the pestles ability.
Protein extraction:
- A 1.5ml snap cap tube was labeled with AD, SC 10/4.
- A .019 g C. gigas digestive gland tissued sample was placed in the freshly labeled tube.
- 500 ul of CellLytic MT solution was added to the tube.
- A sterile pestle was used to homogenize the mixture. The mixture was inverted severa times.
- The tube was then spun at max speed in the centrifuge for 10 minutes.
- A fresh 1.5ml snap cap tube was labeled protein AD, SC 10/4
- The original tube was removed from the centrifuge and the supernatant was pipetted into the new tube.
- A 2ml snap cap tube was labeled BA, AD, SC 10/4 and 15uL of supernatant and 15uL of DI water were added.
- A second 2ml snap cap tube was labeled B, AD,SC 10/4 and 30uL of DI water were added. This is the blank.
- 1.5ml of bradford reagant were added to both 2ml tubes. The tubes were then inverted to mix
- The mixtures incubated at room temperature for 10 minutes.
- 1000 uL was removed from each 2ml tube and added to cuvette.
- The spectrometer was set to 595nm and the blank cuvette was mixed by pipetting, inserted into the spectrometer, and the "tare" button was pressed.
- Next the sample cuvette was added after mixing and a value of .331 was recorded. .
- The sample was removed, mixed again, and placed back into the spectrometer. A value of .330 was recorded.
- A standard curve was used with an equation of y=1013.9x
- The average absorbance reading of .3305 was substituted for x giving a value of 335.1ug/mL
- The dilution factor was calculated resulting in a final concentration of 670.2ug/mL
Observations: The two cuvettes were visibly different in color. The blank was blue, while the sample became brown during the incubation time.
Results:
Absorbance reading 1: .331
Absorbance reading 2: .330
Average: .3305
Standard curve y=1013.9x--> y=1013.9(.3305)--> y=335.1--> 2:1 dilution concentration = 670.2ug/mL
Conclusions:
The results are in the expected range and suggest that there is a fair level of protein in the digestive gland of C. gigas. Next step is to complete RNA extraction to determine its concentration.
Reflection:
The purpose of this lab was to learn new lab techniques and understand how to use methods like the bradford assay properly. These same steps could be applied to multiple samples to gain understanding of the level of protein in a variety of tissue samples from different parts of an organism and from different species. These results may also be particularly useful when looking at the same species exposed or grown in different conditions. One use may be in looking at protein concentration for fish feeding on different diets. Another use may be to look at how protein concentration varies with temperature. Protein is an important part of cellular and organism function. Quantifying it can be useful in many ways. I would like to look through the literature and see how our concentration reading compares to concentration readings in other tissues of other species. Also to further research exactly how such a value can be used. What specifically can be inferred from a protein concentration value? RNA extraction once complete will most likely be useful in a similar manner.