Tissue Extraction I: RNA and Protein extraction
Summary
Tissue was selected (Crassostrea gigas gill tissue at 37C) for RNA and protein extraction. RNA extraction was begun using Tri-Reagent. Protein was extracted and the concentration of protein was measured (913.75 μg/mL). Primers were then designed for heat shock protein 70 (HSP70).
Procedures
RNA Isolation
A piece of frozen tissue was placed in a 1.5 mL snap cap tube and then 500 μL of Tri-Reagent was added to the tissue. The tissue was then homogenized using a disposable pestle. Another 500 μL of Tri-Reagent was added to the tube and was then vortexed for 15 seconds. Tissue was then placed in storage at -80C.
Protein Extraction
EXTRACTION:
Frozen tissue was placed in a 1.5 mL snap tube and 0.5 mL of CelLytic MT solution was then added to the tube. The tissue was homogenized using a disposable pestle. The tube was then placed in a refrigerated microfuge for 10 minutes at maximum speed. After 10 minutes, the supernatant was extracted and put in a labeled tube.
QUANTIFICATION:
An aliquot of the sample was diluted in a 2 mL tube by mixing 15 μL of the protein sample and 15 μL of DI water. Then 1.5 mL of Bradford reagent was added. The solution was then incubated at room temperature for 10 minutes. From the sample, 1 mL was transferred into a cuvette. The absorbance was then measured at 595 nm in a spectrophotometer twice and the values averaged. Then the protein concentration was back-calculated using a provided standard curve. The remaining protein sample was stored at -20oC.
Measurements/Results
Spectrophotometer readings
- 0.462 A
- 0.441 A
Back-calculation of protein concentration
[protein] = 1011.9*0.4515*2 = 913.75 μg/mL
Conclusions/Next steps
A physiology of interest will be selected based on the tissue and primers will be designed for a related gene. The primers will then be used to analyze how much the gene is expressed.
Primer information
C. gigas - HSP70
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
GCCATCGGCATCGACCTGGG |
Plus |
20 |
149 |
168 |
59.90 |
70.00% |
| Reverse primer |
GAAGGCGACGTAGCTGGGCG |
Minus |
20 |
265 |
246 |
60.18 |
70.00% |
| Product length |
117 |
||||||
October 13, 2009
Tissue Extraction II: RNA extraction and protein gels Summary
The RNA extraction protocol was continued using the processed C. gigas tissue. Then an SDS-PAGE protein gel was run to analyze the proteins extracted from the C. gigas tissue in the previous lab.
Procedures
RNA isolation protocol
The tube containing the tissue with Tri-Reagent from the previous lab was thawed and allowed to incubate for 5 minutes at room temperature. Then 200 μL of chloroform was added to the sample. The tube was vortexed for 30 seconds. After allowing the tube to incubate for 5 minutes at room temperature, the tube was then microfuged in a refrigerator for 15 minutes at 16.1 rpm. When the tube was removed, the contents of the tube had not separated correctly. The tube was inverted several times, vortexed again (thoroughly), then put in the microfuge for another 10 minutes. After microfuging, the contents of the tube had separated properly. The aqueous phase was carefully removed into a new tube. Then 500 μL of isopropanol was added to the aqueous phase tube. The tube was inverted well then allowed to incubate at room temperature for 10 minutes. The tube was then microfuged in a refrigerator for 8 minutes at 14 rpm (still max. speed, just a different microfuge). The supernatant was removed and 1 mL of 75% EtOH was added to the tube. The tube was inverted slightly to dislodge the pellet then microfuged at 7500g for 5 minutes. Then the supernatant was removed and the pellet was allowed to dry at room temperature for 4 minutes. The pellet was then resuspended in 100 μL of 0.1% EDPC-H2O. The solution was pipetted up and down to dissolve the pellet. The tube was incubated at 55 degrees C for 5 minutes then placed on ice and stored.
RNA quantification
This procedure will be done in the next lab. We ran out of time during this lab.
Protein Gel protocol
The protein extract from the previous lab was thawed and mixed well through inverting. In a 1.5 mL screw cap tube, 15 μL of the protein sample and 15 μL of 2X Reducing Sample Buffer were combined. The sample was then boiled for 5 minutes. Then sample was centrifuged for 1 minute. The entire sample was loaded into the appropriate well using a gel loading tip. During the loading of my sample, an air bubble blew a significant amount of my sample out of the well, leaving less than 10 μL of protein sample left in the well (approximately 4.57μg of protein in that sample) . The electrodes were plugged in and the gel was run for 45 minutes. About 150 mL of Coomassie stain was added to a container. The gel was carefully removed after 25 minutes and was then placed in the Coomassie Stain. Then it was incubated on a shaker/rocker for 4 minutes. The stain was poured back into the original container. Then the gel was rinsed thoroughly with 10% acetic acid. Then about 250 mL of 10% acetic acid was added to the container with the gel and was allowed to incubate on shaker/rocker for about 15 minutes.
Measurements/Results

Picture 1. Final protein gel (My C. gigas sample is in well #7)

Picture 2. Molecular ladder weights. (Using the NuPAGE-MES column)
Although the amount of sample in well #7 was not enough to provide a detailed vision of the different proteins in the solution, a few distinct lines can be seen. Proteins with weights approximately 6, 32, 38, and 200 can be seen on the gel.
Conclusions/Next steps
During the next lab, the RNA quantification will be finished and we will begin working on running PCR with the genetic material.
October 20, 2009
Reverse Transcription and PCR
Summary
We quantified the RNA extracted from the tissue, reverse transcribed the RNA to complementary DNA and used the cDNA to run a PCR gel.
Procedures
RNA Quantification
The Nanodrop was zeroed using 2 μL of 0.1% DEPC-H2O. Then 2 μL of the RNA sample was pipetted onto the Nanodrop arm and measured. After quantification, the RNA was wiped off with a Kim Wipe.
Reverse Transcription
5 μL of RNA was transferred to a fresh PCR tube. The RNA was incubated at 75C for 5 minutes in the thermal cycler. The tube was then transferred immediately to ice and incubated for 5 minutes. The Master Mix was created using 4 μL of 5x AMV RT Buffer, 8 μL dNTPs, 1 μL AMV RTranscriptase, 1 μL Oligo dT Primer, and 1 μL of RNase free water.
Then, the Master Mix (15 μL total) was added to the RNA. The tube was vortexed, incubated at room temperature for 10 minutes, and then incubated at 37C for 1 hour in the thermocycler. After incubation, the tube was heat inactivated at 95C for 3 minutes. The cDNA was then left on ice to prevent breakdown.
PCR prep
Primers that were ordered were reconstituted by adding 327 μL of H2O to the forward primer and 265 μL of H2O to the reverse primer.
A master mix was created for the PCR samples. The ingredients were added in duplicate (for 5 reactions) to create enough mix for 4 samples (2 cDNA samples and 2 negative controls). For each reaction, 25 μL of GoTaq Green MM, 2.5 μL upstream primer, 2,5 μL downstream primer, and 18 μL of H2O were added. 48 μL of master mix was added to each PCR tube. Then 2 μL of DNA were added to the 2 sample tubes and 2 μL of H20 were added to 2 sample tubes. Then the reactions were put through the thermocycler.
Agarose gel prep
The gel box was put together and a small amount of 1x TAE was used to wash down the sides of the tray and the base. In a 1 L flask, 2 g of agarose and 150 mL of 1x TAE were combined and microwaved for about 3 minutes. The solution was allowed to cool and then 12 μL of ethidium bromide were added to the solution. The solution was mixed thoroughly and poured into the gel tray. Then the gel combs were added. After gel was set, it was placed in the refrigerator for use next week.
Measurements/Results
Nanodrop measurements
A260 absorbance: 23.322
[RNA]: 932.9 ng/μL
A260/280 ratio: 2.00
A260/230 ratio: 1.15
Since the A260/280 ratio is within the 1.8-2.0 range, the RNA seems clean. However, because the A260/230 ratio is below the 1.5-2.0 ratio, the RNA may have some phenol, ethanol or high salt in the solution still.
Conclusions/Next steps
Next week, we will use the gel we made and the PCR samples to run a PCR and analyze the samples.
October 27, 2009
Western Transfers and ImmunoblotsSummary
The PCR products from the last lab were run on gels and protein samples were also run out. Proteins were transferred from the gel to a nitrocellulose membrane and the membrane was probed with the HSP antibody.
Procedures
PCR gel
The agarose gel (made in the previous lab) was put into the gel box and the box was then filled with 1x TAE buffer until the wells were fully covered. The combs were then carefully removed from the wells. 7 μL of 100bp ladder was added to the far left lane. 25 μL of each PCR sample was added into the gel. Then the gel was run at 100V for about an hour. The finished gel was visualized on a UV transilluminator.
Protein Gel protocol
The protein extract from the previous lab was thawed and mixed well. In a 1.5 mL screw cap tube, 15 μL of the protein sample and 15 μL of 2X Reducing Sample Buffer were combined. The sample was then boiled for 5 minutes. Then sample was centrifuged for 1 minute. The entire sample was loaded into the appropriate well using a gel loading tip. During the loading of my sample, an air bubble blew a significant amount of my sample out of the well, leaving less than 10 μL of protein sample left in the well (approximately 4.57μg of protein in that sample) . The electrodes were plugged in and the gel was run for 45 minutes. About 150 mL of Coomassie stain was added to a container. The gel was carefully removed after 25 minutes and was then placed in the Coomassie Stain. Then it was incubated on a shaker/rocker for 4 minutes. The stain was poured back into the original container. Then the gel was rinsed thoroughly with 10% acetic acid. Then about 250 mL of 10% acetic acid was added to the container with the gel and was allowed to incubate on shaker/rocker for about 15 minutes.
Transferring proteins to membrane
The transfer buffer was cooled to 4°C. The filter paper, membrane, and gel were soaked in Transfer Buffer for 15 minutes. The blotting sandwich was assembled in this order: anode (+++), filter paper, nitrocellulose membrane, gel, filter paper, and cathode (- - -). The blot was transferred for 30 minutes at 20V. Then, the gel was removed from the sandwich and rinsed with transfer buffer.
Western blotting
Blocking Solution was prepared using 14 ml of ultra-filtered water, 4 ml of blocker/diluent (Part A), and 2 ml of blocker/diluent (Part B), for a total of 20 ml of solution. The membrane was placed in 10 ml of the appropriate Blocking Solution in a covered, plastic dish. It was then incubated for 30 minutes on a rotary shaker at 1 revolution/sec. The Blocking Solution was then decanted and the membrane was rinsed with 20 ml of water for 5 minutes, then decanted, then rinsed with 20 ml of water for 5 minutes again. 10 mL of Primary Antibody Solution was prepared (1:3000 dilution) using 10 ml of Blocking Solution and 3.3 μL of HSP 70 antibody. Then, the membrane was incubated with 10 ml of Primary Antibody Solution overnight.
The next day, the Primary Antibody Solution was decanted and the membrane was washed with 20 ml of prepared Antibody Wash for 5 minutes then decanted. This was repeated 3 times. The membrane was incubated in 10 mL of Secondary Antibody Solution for 30 minutes, then decanted. The membrane was again washed with 20 ml of prepared Antibody Wash for 5 minutes then decanted, and repeated 3 times. The membrane was then rinsed in 20 ml of water for 2 minutes and decanted. This was repeated twice. The membrane was incubated in 5 ml of Chromogenic substrate until purple bands developed (about an hour). Then, the membrane was again rinsed in 20 ml of water for 2 minutes and decanted, and repeated twice. Then the membrane was dried.
Measurements/Results
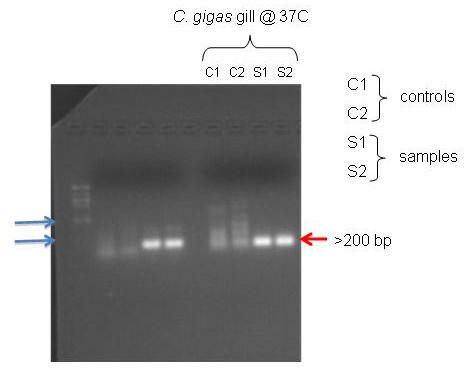
Figure 1. PCR gel results
According to NCBI, the primers I designed for the C. gigas gill should have amplified a 117 bp section. The PCR gel shows bands that are just below the 200 bp marker on the ladder. It seems that the primers worked.
Figure 2. Western blot image. (My C. gigas tissue is in the farthest right well.)
The antibodies from the Western blot did not bind to the proteins from my C. gigas tissue, even though the oyster had been through heat shock.
Conclusions/Next steps
There may have been some contamination in the controls of the PCR gel. The antibody may not have binded to the DNA during the Western blot because it may not be able to bind to bivalve DNA, since the antibody is probably derived from a mammal. Since it is mammal-derived, but for the broad HSP70, it's hit or miss whether or not it's going to bind to non-mammal DNA.
In the next lab, we will use quantitative PCR to further analyze the tissue samples.
November 3, 2009
Quantitative PCRSummary
cDNA samples from previous weeks were run through a qPCR cycle.
Procedures
A master mix was prepared for 7 reactions (2 negative controls, 2 RNA controls, 2 cDNA samples, and 1 for volume recovery). For each reaction, 25 μL of 2X Immomix master mix (175 μL total), 2 μL of Syto-13 dye at 50 μM concentration (14 μL total), 1 μL of upstream primer at 10 μM concentration (7 μL total), 1 μL of downstream primer at 10 μM (7 μL total), and 19 μL of ultra pure water (133 μL total) were combined.
48 μL of this master mix was added to each of 6 PCR tubes. Then (after thawing) 2 μL of cDNA were added to 2 tubes, 2 μL of ultra pure water were added to 2 tubes, and 2 μL of diluted RNA were added to the remaining 2 tubes. (RNA was diluted by adding 1 μL of RNA to 3 μL of ultra pure water per reaction). The wells were capped and loaded on to the PCR plate.
The qPCR was run as follows:
- Incubate at 95C for 10 min
- Incubate at 95C for 15 sec
- Incubate at 55C for 15 sec
- Plate read
- Incubate at 72C for 30 sec
- Plate read
- Repeat steps 2-6, 39 times
- Incubate at 95C for 1 min
- Incubate at 55C for 1 sec
- Melting curve from 55C-95C, read every 0.5 sec, hold for 30 secs
- Incubate at 21C for 10 min
Measurements/Results
Fig 1. qPCR results (turquoise and violet are cDNA, orange and blue are negative control, green and red are RNA control)
The cDNA amplified, but so did the RNA control tubes. There was no amplification in the negative control (water) tubes.
Conclusions/Next steps
Since the RNA control tubes amplified, that means there was genomic DNA carryover in the RNA solution. Since the genomic DNA bumps are similar to the cDNA bumps, it shows that the primers didn't go over any introns in the genomic DNA. Therefore, the genomic DNA and cDNA products were about the same size. This shows that the HSP70 primers used on the genetic material found the HSP70 gene expressed.
If I were to continue with this tissue, I would want to use DNAse in the RNA solution to get rid of any genomic DNA carryover and run the sample again.
Project timeline
11/10 - Experimentation, gathering samples, isolating RNA and extracting protein
- 12 crabs will be used for experiment (6 control, 6 stressed).
- Control crabs will each be placed gently in a 25-50 mL beaker, with just enough water to cover the animal, and allowed to remain there, without disturbance for an hour.
- Experimental crabs will be placed in a salad spinner and spun at random for a couple minutes. Then each crab will be placed in a 25-50 mL beaker, with just enough water to cover the animal, and allowed to remain there, without disturbance for an hour.
a. RNA isolation
i. About 0.5 g of (gill) tissue from each animal will be placed in a 1.5 mL snap cap tube and 500 μL of Tri-Reagent will be added.
ii. The tissue will be homogenized using a disposable pestle.
iii. Add another 500 μL of Tri-Reagent to tube and vortex for 15 seconds.
iv. Store at -80C.
b. Protein extraction
i. About 0.5g of (gill) tissue from each animal will be placed in a 1.5 mL snap cap tube and 0.5 mL of CelLytic MT solution will be added.
ii. The tissue will be homogenized using a disposable pestle.
iii. Put in a refrigerated microfuge for 10 minutes at maximum speed.
iv. After 10 minutes, extract supernatant and put in labeled tube. Store at -20C.
5. At the end of the hour, the water from the beakers will be placed in 15 mL conical tubes and frozen and stored at -20C.
Materials needed:
- 12 beakers (25-50 mL)
- 12 conical tubes (15 mL)
- 36 snap cap tubes (1.5 mL)
- microfuge
- vortex
- salad spinner
- Tri-Reagent
- Disposable pestles
- CelLytic MT solution
- dissection tools
- dissection scope
- filtered sea water
11/17 - Protein quantification, water sample analysis
Water sample analysis
1. Thaw water samples.
2. Using sampling kits, test and record ammonia, nitrite, nitrate, pH, and phosphate levels in each water sample.
3. Put water samples back in storage at -20C.
Protein Quantification:
8. In a fresh 2 mL tube labeled as 'sample', dilute an aliquot of the sample 1:2 by pipetting 15uL of your protein sample and 15uL of DI water and mixing well.
9. In a second 2 mL tube pipette 30uL of DI water (this tube will serve as your blank). Label tube as 'blank'
10. To both tubes add 1.5mL of Bradford reagent.
11. Invert the tubes several times and then incubate at RT for 10mins.
12. Mix the 'blank' tube and transfer 1mL to a plastic, disposable cuvette.
13. Zero the spectrophotometer using your blank sample
14. Mix the 'sample' tube and transfer 1 mL to a plastic, disposable cuvette
15. Measure the absorbance at 595nm and record the value.
16. Remove the cuvette from the spectrophotometer. Using a P1000 set to 1mL, carefully pipette the solution in the cuvette up and down a couple of times to mix.
17. Measure the absorbance at 595nm and record the value.
18. Average the two absorbance values you recorded.
19. Back-calculate your protein concentration using the standard curve
Protein Gel
1. Begin boiling water on hot plate.
2. Thaw your protein extract from last week. Mix well by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
Supply list
- pH/nitrate/nitrite/phosphate/ammonia water test kits
- cuvettes
- Coomassie Protein Assay Reagent
- Protease inhibitor cocktail
- spectrophotometer
- protein gel box (SR provided)
- SDS/PAGE gels
- trays for staining gels
- protein ladder marker
- digital camera
- power supply
- microfuge tube racks
- running buffer (SR provided)
- coomassie stain (SR)
- platform rocker/shaker
- plastic wrap
- sandwich bags
- gel loading tips
- pipettors (1ul-1000ul)
- sterile, 1.5mL screw cap tubes
- pipet tips (filter - RNAse free)
- RNase free water
- hot plate
- tube "floatie" (8 tube capacity)
- glass container for boiling water that can accommodate "floatie"
- 2X SDS reducing sample buffer (SR provided)
- heating block/water bath
- acetic acid
- chloroform
- isopropanol
- ethanol
- ice buckets
- phenol/chloroform waste containers (liquid/solid)
- filter tips
- methanol
- 50 ml Falcon tubes / holders
- DEPC treated water (SR provided)
- Normal light box (SR provided)
11/24 - Protein gel, cont.
Protein Gel
1. Begin boiling water on hot plate.
2. Thaw your protein extract from last week. Mix well by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly as demonstrated.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well using a gel loading tip.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL (does not have to be measured - just need enough to cover the gel) of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Be careful not to dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible. This may need to incubate O/N. If so, cover container with plastic wrap and leave on shaker/rocker.
12/1-RNA isolation
RNA isolation
1. Turn on heating block to 55C
2. Incubate tube at room temperature (RT) for 5 mins.
3. In the fume hood, add 200uL of chloroform to your sample and close the tube.
NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
4. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
5. Incubate tube at RT for 5 mins.
6. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
7. Gently remove tube from microfuge. Be sure not to disturb the tube.
8. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
9. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
10. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
11. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
12. Incubate at RT for 10 mins.
13. Spin in refrigerated microfuge at max speed for 8 mins.
14. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
15. Remove supernatant.
16. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
17. Spin in refrigerated microfuge at 7500g for 5mins.
18. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
19. Briefly spin tube (~15s) to pool residual EtOH.
20. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
21. Leave tube open and allow pellet to dry at RT for no more than 5mins.
22. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
23. Incubated tube at 55C for 5mins. to help solubilize RNA.
24. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample
RNA quantification
p1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedistal and lower the arm.
2. Click "Blank", to zero the instrument
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your A260 absorbance, RNA concentration (ng/µL), A260/280 ratio and A260/320 ratio.
NOTE: The Nanodrop uses the Beer-Lambert Law to calculate RNA concentration for you. See Lab 1 notes on RNA extraction for more information on the calculation and how to evaluate RNA purity using A260/280 and A260/A320 ratios.
5. Raise the arm and wipe off you sample with a Kim Wiple
6. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
7. Store at -80C.
12/3-DNAse, RNA quant
12/4- Reverse transcription, prep qPCR plates
November 10, 2009
Experimentation, gathering samples, isolating RNA and extracting protein Summary
A stress experiment was conducted using 12 Hemigrapsus spp. crabs (6 stressed, 6 control). Water samples from live crab excretions were collected and then gill tissue was removed and fixed to isolate RNA and extract protein.
Procedures
Experimentation
12 crabs of similar size were selected from the collected individuals (6 control, 6 stressed). The control crabs were placed in individual 25 mL beakers which had 13 mL of filtered sea water in them. The beakers were placed in a tray with ice to keep the water cool so as to minimize heat stress. The experimental crabs were placed in a salad spinner and vigorously spun for 2.5 minutes. Then, the experimental crabs were removed and put into individual 25 mL beakers with 13 mL of filtered sea water. Both groups were allowed to sit for an hour to allow for crabs to excrete any compounds. After an hour, the water samples were put in 15 mL conical tubes and frozen at -20C.
After the hour, the crabs were killed to halt metabolism and any other biological processes. The crabs were dissected and the gill tissue removed. One gill from each animal was used for RNA isolation and one gill was used for protein extraction.
RNA isolation
About 0.015g of gill tissue per animal was placed in a 1.5 mL snap cap tube and 500 μL of Tri-Reagent was added to each tube. The tissue was then homogenized with a disposable pestle. Another 500 μL of Tri-Reagent was added to each tube and the tube was vortexed for 15 seconds. The tubes were then stored at -80C.
Protein extraction
About 0.015g of gill tissue per animal was added to a 1.5 mL snap cap tube with 0.5 mL of CelLytic MT solution. The tissue was homogenized with a disposable pestle. Then the tubes were put in a refrigerated microfuge for 10 minutes at maximum speed (14 rpm). After 10 minutes, the tubes were removed and the supernatant was extracted. The tubes were then stored at -20C.
Measurements/Results
None for this stage.
Conclusions/Next steps
Next week I will quantify the protein samples using a Bradford Assay, analyze the chemical content of the water samples, and continue with RNA isolation and quantification. I will also try to run an SDS-PAGE gel, if time allows.
November 17, 2009
Protein quantification, preparation for protein gel Summary
The protein extractions were quantified using a Bradford Assay and calculations and dilutions were done in preparation to run an SDS-PAGE gel.
Procedures
Protein quantification
Protein samples were diluted 1:2 by pipetting 15 μL of sample into a tube and adding 15 μL of DI water. A blank was made by adding 30 μL of DI water to another tube. Then 1.5 mL of Bradford reagent was added to all tubes. The tubes were then inverted and incubated at RT for 10 minutes. 1 mL of the "blank" tube was transferred to a cuvette. The spectrophotometer was zeroed using the blank sample. The sample tubes were mixed and 1 mL was added of each tube to 12 different cuvettes. The absorbance of the samples were measured at 595nm twice and averaged. The protein concentrations were back-calculated using the standard curve provided. (NOTE: Some lab error caused the protein samples to sit out for more than 10 minutes before analysis.)
Protein sample dilution (in preparation for protein gel)
After the protein sample concentrations were calculated, the samples were standardized in preparation for the protein gel. The lowest protein concentration was used as the standard. Using the equation C1*V1=C2*V2, the amount of the protein sample to be transferred and diluted was calculated. Then, 15 μL of the sample dilutions were transferred to tubes with 15 μL of 2X Reducing Sample Buffer.
Measurements/Results
Spectrophotometer readings (@ 595nm)
| Reading #1 |
Reading #2 |
Avg |
[protein] |
|
| S1 |
0.062 |
0.060 |
0.061 |
123.5 |
| S2 |
0.058 |
0.055 |
0.0565 |
114.4 |
| S3 |
0.040 |
0.040 |
0.04 |
81.0 |
| S4 |
0.047 |
0.047 |
0.047 |
95.1 |
| S5 |
0.065 |
0.066 |
0.0655 |
132.6 |
| S6 |
0.050 |
0.052 |
0.051 |
103.21 |
| C1 |
0.013 |
0.014 |
0.0135 |
27.3 |
| C2 |
0.060 |
0.059 |
0.0595 |
120.4 |
| C3 |
0.010 |
0.013 |
0.0115 |
23.3 |
| C4 |
0.029 |
0.027 |
0.028 |
56.7 |
| C5 |
0.045 |
0.046 |
0.0455 |
92.08 |
| C6 |
0.043 |
0.041 |
0.042 |
85.0 |
Dilution measurments (using C1V1=C2V2 where C1= conc., C2=23.3, and V1=20 uL)
| Conc. |
Volume |
H20 |
|
| S1 |
123.5 |
4 |
16 |
| S2 |
114.4 |
4 |
16 |
| S3 |
81.0 |
6 |
14 |
| S4 |
95.1 |
5 |
15 |
| S5 |
132.6 |
3.5 |
16.5 |
| S6 |
103.2 |
4.5 |
15.5 |
| C1 |
27.3 |
20 |
- |
| C2 |
120.4 |
4 |
16 |
| C3 |
23.3 |
20 |
- |
| C4 |
56.7 |
8 |
12 |
| C5 |
92.08 |
5 |
15 |
| C6 |
85.0 |
5.5 |
14.5 |
Next week I will run the SDS-PAGE gel and finish RNA isolation and quantification.
November 24, 2009
SDS-PAGE gel
Summary
A protein gel was run using the protein samples from last week.
Procedures
In screw cap tubes, 15 uL of the undiluted protein sample and 15 uL of 2X Reducing buffer were combined for each sample. Then the samples were boiled for 5 minutes. The SDS-PAGE gel box was put together while the samples were boiling and the wells were rinsed thoroughly. Immediately after 5 mins of boiling, the samples were centrifuged for 1 minute to pool the liquid. Then 10 uL of a molecular ladder was added to the first well. Then all 30 uL of each sample was added to each well. The lid was put on and the gel was run for 30 minutes at 150 V. After 30 min, the gel was removed from the box and added to a container containing Coomassie Stain. The gel was allowed to sit in the stain for 5 min on a shaker/rocker. Then, the stain was poured back into the original container and the gel was rinsed briefly (twice) with 10% acetic acid. Then it was immersed in acetic acid and allowed to sit overnight on a shaker/rocker. The next day, pictures were taken of the gel.
Measurements/Results
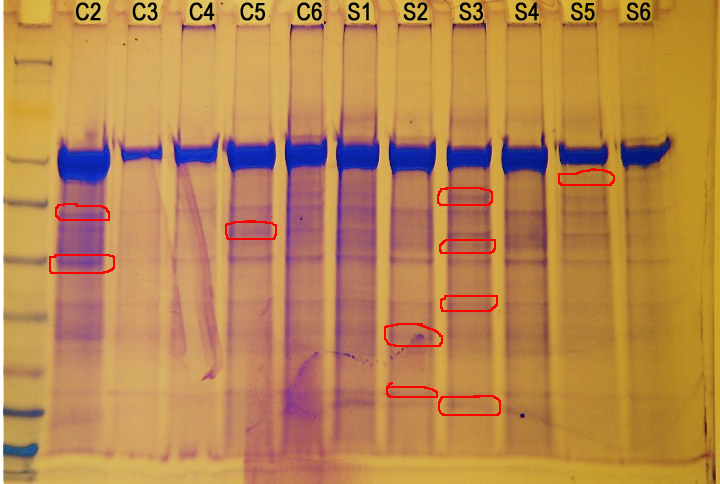
Figure 1. Protein gel results (not normalized). Circled in red are proteins of interest.

Conclusions/Next steps
There doesn't seem to be a correlation between protein expression and amount of stress. However, there are several different proteins being expressed in different animals.
Next week, I will attempt to finish RNA isolation and quantification and run another protein gel after filtration.
December 1, 3, 4, 2009
RNA isolation, DNAse, RNA quantification, filtered SDS-PAGE
Summary
RNA was isolated from the tissue fixated at the beginning of the procedure. Then 20.5 uL of the sample was DNAsed. Then I attempted to quantify the RNA concentration, but there was severe contamination and I had to abandon the rest of the gene analysis. I filtered protein samples to filter out the large protein found in all the samples and ran another SDS-PAGE.
Procedure
RNA isolation
The tubes containing the tissues with Tri-Reagent from the previous lab were thawed and allowed to incubate for 5 minutes at room temperature. Then 200 μL of chloroform were added to the samples. The tubes were vortexed for 30 seconds. After allowing the tubes to incubate for 5 minutes at room temperature, the tubes were then microfuged in a refrigerator for 15 minutes at 14 rpm (max). The aqueous phase was carefully removed into a new tube. Then 500 μL of isopropanol was added to the aqueous phase tubes. The tubes were inverted well then allowed to incubate at room temperature for 10 minutes. The tubes were then microfuged in a refrigerator for 8 minutes at 14 rpm (max). The supernatant was removed and 1 mL of 75% EtOH was added to the tubes. The tubes were inverted slightly to dislodge the pellets then microfuged at 7500g for 5 minutes. Then the supernatant was removed and the pellets were allowed to dry at room temperature for 4 minutes. The pellets were then resuspended in 100 μL of 0.1% EDPC-H2O. The solution was pipetted up and down to dissolve the pellets. The tubes were incubated at 55 degrees C for 5 minutes then placed on ice and stored.
DNAse
In a 0.5 mL tube, 2.5 uL of DNAse buffer, 1 uL of Turbo DNAse, and 20.5 uL of the RNA sample were combined and then incubated for 30 minutes at 37C. Then another 1 uL of Turbo DNAse was added and the solution was again incubated for 30 minutes at 37C. then 2.5 uL of inactivation reagent was added and the tubes were mixed then allowed to incubate for 2 minutes at room temperature. The tubes were centrifuged at 10,000 rcf for 1.5 minutes. The supernatant was then transferred to new tubes.
RNA quantification
The Nanodrop was zeroed using 2 μL of 0.1% DEPC-H2O. Then 2 μL of the RNA sample was pipetted onto the Nanodrop arm and measured. After quantification, the RNA was wiped off with a Kim Wipe. After measuring 4 samples, it was realized that there were peaks presenting themselves that indicated either ethanol contamination or contamination from the DNAse procedure. Since time restrictions didn't allow for another attempt at DNAsing a sample, the gene analysis branch of lab procedure was abandoned.
Filtrated SDS-PAGE
Filtration was done using a Microcon model YM-50 to filter out proteins larger than 50 kD. 250 uL of protein sample was put into the the filter tube. 250 uL of PCR water was also added to the filter tube to increase volume. The tubes were microfuged at 14,000 g for 12 minutes. Then the filtered protein was used in the SDS-PAGE.
In screw cap tubes, 15 uL of the filtered protein sample and 15 uL of 2X Reducing buffer were combined for each sample. Then the samples were boiled for 5 minutes. The SDS-PAGE gel box was put together while the samples were boiling and the wells were rinsed thoroughly. Immediately after 5 mins of boiling, the samples were centrifuged for 1 minute to pool the liquid. Then 10 uL of a molecular ladder was added to the first well. Then 30 uL of each sample was added to each well. The lid was put on and the gel was run for 30 minutes at 150 V. After 30 min, the gel was removed from the box and added to a container containing Coomassie Stain. The gel was allowed to sit in the stain for 5 min on a shaker/rocker. Then, the stain was poured back into the original container and the gel was rinsed briefly (twice) with 10% acetic acid. Then it was immersed in acetic acid and allowed to sit for 15 minutes on the shaker/rocker. Then it was briefly rinsed again then immersed in acetic acid and allowed to sit overnight on a shaker/rocker. The next day, pictures were taken of the gel.
Conclusions/Results
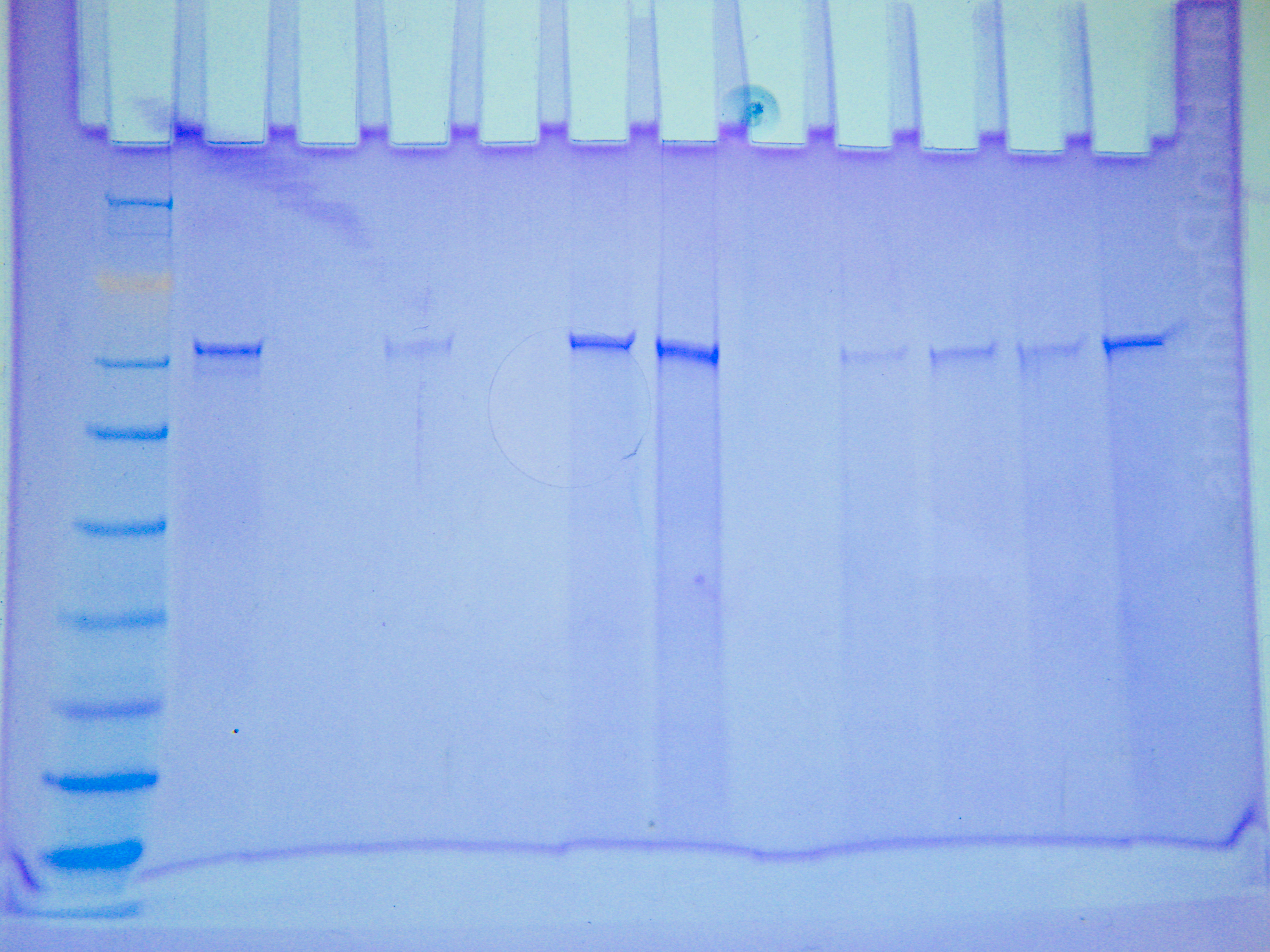 |
| Figure 1. Filtered protein sample SDS-PAGE gel |
The filter seemed to have filtered out some of the large protein around 62 kD, but it did not filter it all out. Additionally, it seemed to have filtered out everything smaller than 50 kD. The lack of smaller bands could also be attributed to the dilution of the sample that occurred during filtration.
Next steps
The protein sizes from the first gel will be investigated and identified using a database containing protein information about crabs.