PCR Results:
I retrieved the results from my PCR. The Cytochrome P450 results ended up being too inconsistent with low fluorescences so I decided to throw out that data and not include it in my analysis. The HSP70 and Settlement Inducing Factor primers resulted in good amplification so I set a threshold above my waters and recorded the efficiencies and C(t) values. From the C(t) values I was able to calculate and arbitrary expression value using the equation: Arbitrary expression value =10^(-(0.3012*Ct)+11.434). By comparing the expression values, I can see relatively how much cDNA is being amplified| Well / Set |
Dye |
Content |
Description |
Efficiency |
C(t) |
Expression Value |
| A3 |
SBG1 |
Sample |
N/A |
N/A |
||
| A4 |
SBG1 |
Sample |
79.12% |
37.59 |
1.293874042 |
|
| B3 |
SBG1 |
Sample |
45.72% |
38.12 |
0.895892704 |
|
| B4 |
SBG1 |
Sample |
44.44% |
38.56 |
0.660279783 |
|
| C3 |
SBG1 |
Sample |
N/A |
N/A |
||
| C4 |
SBG1 |
Sample |
N/A |
N/A |
||
| D3 |
SBG1 |
Sample |
N/A |
N/A |
||
| D4 |
SBG1 |
Sample |
86.69% |
36.63 |
2.517932016 |
|
| E3 |
SBG1 |
Sample |
103.52% |
37.58 |
1.302878748 |
|
| E4 |
SBG1 |
Sample |
N/A |
N/A |
||
| F3 |
SBG1 |
Sample |
89.20% |
32.37 |
48.32323495 |
|
| F4 |
SBG1 |
Sample |
N/A |
N/A |
||
| G3 |
SBG1 |
Sample |
67.84% |
33.34 |
24.65993912 |
|
| H3 |
SBG1 |
Sample |
48.71% |
38.31 |
0.785286262 |
| Well / Set |
Dye |
Content |
Description |
Efficiency |
C(t) |
Expression Value |
| A1 |
SBG1 |
Sample |
N/A |
N/A |
||
| A2 |
SBG1 |
Sample |
N/A |
N/A |
||
| B1 |
SBG1 |
Sample |
29.76% |
35.61 |
5.108201262 |
|
| B2 |
SBG1 |
Sample |
N/A |
N/A |
||
| C1 |
SBG1 |
Sample |
22.37% |
23.46 |
23326.41511 |
|
| C2 |
SBG1 |
Sample |
N/A |
N/A |
||
| D1 |
SBG1 |
Sample |
24.03% |
26.77 |
2348.962048 |
|
| D2 |
SBG1 |
Sample |
38.78% |
36.61 |
2.553101025 |
|
| E1 |
SBG1 |
Sample |
N/A |
N/A |
||
| E2 |
SBG1 |
Sample |
N/A |
N/A |
||
| F1 |
SBG1 |
Sample |
77.37% |
36.63 |
2.517932016 |
|
| F2 |
SBG1 |
Sample |
N/A |
N/A |
||
| G1 |
SBG1 |
Sample |
24.25% |
29.11 |
463.51949 |
|
| H1 |
SBG1 |
Sample |
19.58% |
30.97 |
127.5956886 |
Conclusion: The Settlement Inducing factor showed high amplification in the soap and bleach experimental groups while relatively low amplification in the control group. This indicates that more settlement inducing factor gene was transcribed when the tissue was removed. The HSP70 showed a slightly higher amplification in the bleach group but still not significant enough to make a definite conclusion. My next steps, if I were to continue my research would be to try out different primers or actually isolate the chemicals to see if I can pinpoint the cause of the increase in settlement inducing factor protein.
Wednesday, December 2, 2009
PCR:
- Make three master mixes for 15 reactions (14 running reactions of PCR + 1 extra reaction). One master mix for each primer pair: Indu, HSP70, and HC
- 2x Immunomix: 12.5uL x 15 rxns = 187.5 uL
- Syto-13 Dye: 1.0uL x 15 rxns = 15 uL
- Upstream Primer: 0.5uL x 15 rxns = 7.5 uL
- Downstream Primer: 0.5uL x 15 rxns = 7.5 uL
- Ultra Pure Water: 9.5uL x 15 rxns = 142.5 uL
- Put 24uL of master mix in each PCR well.
- Add 1uL of cDNA or PCR water to each PCR well.
- Fill out PCR sheet.
- Place in refrigerator for Mac to run PCR.
Observations/Conclusions:
I made the three different master mixes using a total of 24uL of MM per well/reaction (1 Master Mix per primer pair). Hopefully this will give me better amplification and better absorbances. My final step is to analyze the results of my PCR.
Tuesday, December 1, 2009
PCR Results:
From my results I have concluded to go forward with the HSP70, Settlement Inducing Factor Complex, and the HC Cytochrome P450 Primers. My next step will be to run all my cDNA samples using those primers to see and compare the amplification.Monday, November 30, 2009
Reconstituted Primers:
- Add nm (different for each primer) x 10uL of water to each primer.
- Make a working stock of primer by adding 90uL of water to 10uL of newly diluted primer solution.
PCR (testing primers):
- Place in each well:
- 2x Immunomix: 25uL
- Syto-13 Dye: 2.0uL
- Upstream Primer: 1.0uL
- Downstream Primer: 1.0uL
- Ultra Pure Water: 19uL
- cDNA or H2O: 2uL
- Fill out PCR sheet.
- Place in refrigerator for Mac to run PCR.
Observations/Conclusions:
I did my wells individually, which was wrong. I should have made a master mix for each primer pair (4). By doing so, will create less variability and chance of error/contamination. Next time, when I do my actual PCR with the primer that I know will amplify my cDNA, I will make a master mix for each primer pair and pipet into the according wells.
Tuesday, November 24, 2009
Normalize RNA:
- Add calculated amount of RNA to new tube according to table below
- Add calculated amount of 0.1% DEPC H2O according to table below
- Invert several times
- Spin tubes briefly to pool liquid
- Incubate tube at 75C for 5 mins in thermocycler
- Transfer tube immediately to ice and incubate for at least 5 mins
| Sample |
ng/uL |
uL of RNA added |
uL of H20 added |
| S1 |
815.5 |
1.9 |
8.1 |
| S2 |
190.7 |
8.2 |
1.8 |
| S3 |
181.9 |
8.6 |
1.4 |
| S4 |
448.4 |
3.5 |
6.5 |
| B1 |
292.8 |
5.3 |
4.7 |
| B2 |
1087 |
1.4 |
8.6 |
| B3 |
615.1 |
2.5 |
7.5 |
| B4 |
198.1 |
7.9 |
2.1 |
| C1 |
376 |
4.16 |
5.84 |
| C2 |
320.6 |
4.9 |
5.1 |
| C3 |
156.4 |
10 |
--- |
| C4 |
144.2 |
10 |
--- |
Observations:
By normalizing all my RNA concentrations ensures that I have equal amounts of RNA in each solution. That way, when I make cDNA through reverse transcription, I will have equal concentrations of cDNA per unit of volume. Then, when I PCR, I will be able to correlate the amount of amplification with each solution since they all started out with the same amount of cDNA available to amplify. Normalizing the RNA solution was relatively simple. I calculated the amount of water needed to be added by using the equation: C1(concentration that was quantified)*V1(10uL)=C2(desired concentration)*V2(amount of water to be added)
Reverse Transcription:
MASTER MIX PER RXN:
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
- Make Master mix for 13 reactions
- Add 15 uL of Master Mix to 5 uL of Normalized RNA
- Vortex
- Spot spin
- Place tubes in thermocycler (Main -> RT)
- Incubate at RT for 10mins (in thermocycler)
- Incubate at 37C for 1 hour (in thermocycler)
- Heat inactivate at 95C for 3 mins (in thermocycler)
- Remove from thermocycler
- Spot spin
- Store at -20C
Observations:
I completed the protocol for reverse transcription with any problems. The reverse transcription was hopefully able to convert all my RNA to cDNA using the primers and dNTPs.
Conclusions: I normalized my RNA solutions so that they all have equal concentrations. I also made cDNA through reverse transcription so my next steps would be to test that my primers will lay down on my cDNA through PCR. I will test my four primer pairs and hope for amplification.
Friday, November 20, 2009
DNAase:
- Add to a 0.5mL tube: 2.5 uL of DNAase Buffer, 1 uL Turbo DNAase, 20.5 uL RNA sample.
- Incubate at 37C for 30 minutes
- Add 1uL of Turbo DNAase
- Incubate at 37C for another 30 minutes
- Add 2.5uL of inactivation reagent
- Vortex at room temperature for 2 minutes
- Transfer supernatant to new tube
RNA Quantification:
- Pipette 2uL of 0.1% DEPC-H2O onto the Nanodrop pedestal and lower the arm.
- Click "Blank" to zero the instrument.
- Pipette 2 uL of RNA sample onto the Nanodrop pedestal and lower the arm.
- Click "Measure" and record your A260 absorbance, RNA concentration (ng/uL), A260/280 ratio and A260/320 ratio.
- Raise the arm and wipe off your sample with a Kim wipe.
- Repeat for all samples.
- Store samples at -80C
| Sample |
ng/uL |
A260 Abs |
A260/280 |
A260/230 |
| S1 |
815.5 |
20.388 |
1.99 |
1.16 |
| S2 |
190.7 |
4.769 |
1.65 |
0.67 |
| S3 |
181.9 |
4.548 |
1.97 |
1.01 |
| S4 |
448.4 |
11.209 |
1.94 |
1.07 |
| B1 |
292.8 |
7.321 |
1.9 |
0.71 |
| B2 |
1087 |
27.176 |
2.01 |
1.58 |
| B3 |
615.1 |
15.378 |
1.99 |
1.18 |
| B4 |
198.1 |
4.952 |
1.88 |
0.75 |
| C1 |
376 |
9.4 |
1.95 |
1.04 |
| C2 |
320.6 |
8.015 |
1.93 |
0.93 |
| C3 |
156.4 |
3.91 |
1.87 |
0.72 |
| C4 |
144.2 |
3.606 |
1.9 |
0.82 |
Observations:
The DNAase process was a very simple and easy way to ensure no DNA is present in my RNA solution. After adding the inactivation reagent, the solution had a clear top layer and a white sandy lower layer. The upper layer contained the RNA, while the lower one contained all the DNAase and inactivation reagent. I removed the top layer slowly in order to ensure none of the lower layer got into my solution. This would be extremely bad further down the process when I start making my cDNA because it could possibly degrade or mess up my cDNA product.
Conclusions:
After quantifying the RNA I need to normalize all my solutions to the same concentrations of RNA. This will ensure that all solutions will have the same amount of starting material or RNA before I create cDNA to the specific genes of interest. If one solution started with more RNA than another solution then it would be more likely to have more cDNA amplification relative to the other because more RNA was available. By normalizing all solutions ensures that all solutions start at the same level with a known amount of RNA.
Tuesday, November 17, 2009
RNA Isolation (cont.):
- Thaw tube at room temperature for 5-10 mins.
- Add 200 uL of chloroform and close tube.
- Vortex for 30 seconds.
- Incubate tube at room temperature for 5 minutes.
- Spin tube in refrigerated microfuge at max speed for 15 minutes.
- Transfer aqueous, clear top portion to a fresh microfuge tube and close tube.
- Add 500 uL of isopropanol to the new tube.
- Close tube and invert several times until the solution does not appear viscous or lumpy.
- Incubate at room temperature for 10 minutes.
- Spin in refrigerated microfuge at max speed for 8 minutes.
- Remove supernatant but not white pellet.
- Add 1 mL of 75% EtOH to the pellet and votex to dislodge pellet.
- Spin in refrigerated microfuge for 5 mins.
- Carefully remove supernatant but not the pellet.
- Spin tube to pool residual EtOH and remove it using P20 tips.
- Leave tube open to dry at room temperature for about 5 minutes.
- Resuspend pellet in 100uL of 0.1% DEPC-H2O by pipetting up and down until pellet is dissolved.
- Incubate tube at 55 C for 5 minutes
- Remove and flick a few times to mix sample.
- Place on ice.
After vortexing for the first time, the solution became a milky emulsion, which was good. After microfuging again, the solution was separated into three layers. The top layer was clear, then there was a milk emulsion layer, and at the bottom there was a pink layer. After adding the isopropanol, the solution was clear and homogenous. After spinning the solution in step 9, a small white pellet was present in each solution, which is supposedly the RNA. This was definitely a good sign that indicates there is a good possibility that my solutions contain RNA. The pellet was then resuspended and then soluble in teh DEPC-H2O. This is my RNA stock solution that I must now DNAase and quantify.
Conclusion:
Now that I have a stock RNA solution, I have to DNAase the solution in order to ensure no DNA impurities and to quantify my RNA solutions. The DNA could cause amplification during PCR, giving unclear results of amplification. We then need to quantify the solution in order to normalize all solutions with the same concentrations of RNA.
Tuesday, Novemeber 10, 2009
Lab #6: Mollusc Xenobiotic Response
TIMELINE:
11/10: Run Experiment, Order Primers, Start RNA Isolation11/17: Continue RNA Isolation
11/20: DNAase and RNA Quantification
11/24: Normalize and Reverse Transcription
11/30: PCR
12/8:
12/9: Presentations
Experiment:
1. Collect barnacle specimens from the Fisheries Lab.2. Prepare experimental environments as follows:
100 ppm experimental variable
- Control: 2L seawater + 2mL seawater
- Bleach: 2L seawater + 2mL 6% Sodium Hypochlorite Bleach
- Soap: 2L seawater + 2mL Ultra Concentrated DAWN Dishwashing Soap
4. Run experiment for 2 hours.
Observations:
Tank Temperature: 16 degrees C
Start Time: 12:50 pm
End Time: 2:50 pm
At 30 mins, I noticed 3 barnacles are open and feeding in the control tank, while only 1 barnacle is open and feeding each in the bleach and soap tank.
At 60 mins, I noticed none of the barnacles are open and feeding in any of the three tanks.
At 90 mins, I noticed 1 barnacle is open and feeding in the control tank, while none of the barnacles are open and feeding in the bleach and soap tanks.
At 120 mins, I noticed, again, none of the barnacles are open and feeding in any of the three tanks.
RNA Isolation:
- Add 500 uL of TriReagent to 12 1.5mL snap cap tubes. Store on ice.
- Remove 4 barnacles from each of the variable treatments. Remove barnacle from shell using tools and add to tube containing TriReagent.
- Carefully homogenize the tissue using disposable pestle.
- Add another 500 uL of TriReagent to the tube and close cap.
- Store at -80 degrees C.
I isolated the RNA and stored it without any trouble. The next step would be to continue my RNA isolation.
-
Tuesday November 3, 2009
Lab #5: Quantitative PCR
QUANTITATIVE PCR:
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).1. Prepare master mix for seven reactions: Prepare enough reactions to run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template). Add 1 additional reaction to the total to ensure sufficient volume recovery.
For a 50μl reaction volume: -
| Component |
Volume |
Final Conc. |
| Master Mix, 2X (Immomix) |
25µL |
1x |
| Syto-13 dye (50uM) |
2-5µL |
2 - 5µM |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| Ultra Pure Water |
to 48uL |
NA |
3. Thaw cDNA samples.
4. Add 2uL cDNA template to each reaction. Repeat.
5. Add 2uL of ultra pure water to the negative control wells. Repeat.
6. Add 2uL of RNA to each reaction. Repeat.
7. Cap the wells securely.
8. Load the plate, verify the PCR conditions and start the run.
9. Analyze results.
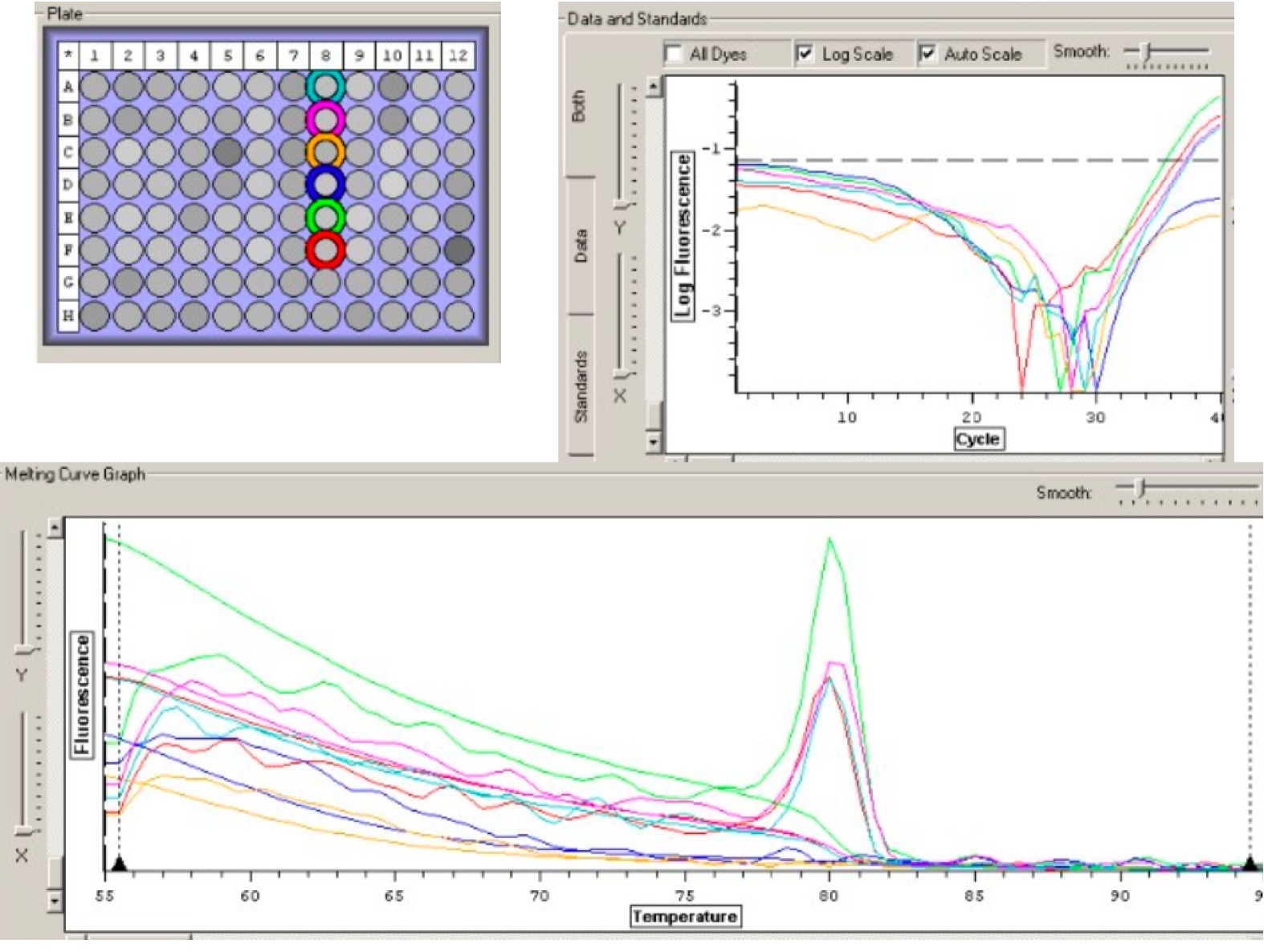
Figure 1: My PCR results. Wells (from top to bottom): cDNA, cDNA, negative control, negative control, RNA, RNA
Observations:
From my PCR results, I can conclude that my cDNA was present in my cDNA sample because wells 1 and 2 showed amplification. The negative controls (wells 3 and 4) did not show amplification, indicating no contamination. Finally, because wells 5 and 6 showed amplification, my RNA sample contains cDNA (-
Conclusion:
My next steps will be to DNAase my RNA solution to get rid of the cDNA crossover. I will also start my barnacle-xenobiotic study.
Tuesday October 27, 2009
Lab #4: Western Transfer Immunoblots
-RUN PCR PRODUCTS ON GEL
1. Place gel in gel box and fill with 1x TAE buffer to fully cover wells.
2. Remove combs from wells.
3. Load 7uL 100bp ladder in far left lane
4. Load 25uL of your PCR sample into the gel
5. Run gel at ~ 100V for ~ 1hr
6. Visualize the gel on the UV transilluminator
Observations:
The 25 microliters of cDNA and blank were loaded onto gels. After running for an hour, the wells loaded with my cDNA showed a light band at molecular weight 200-250. There was also another light band that was present on both the cDNA and the blank wells. However, it was concluded that they were the result of primer dimers. This is consistent with the expected length, according to NCBI, of 273 bp.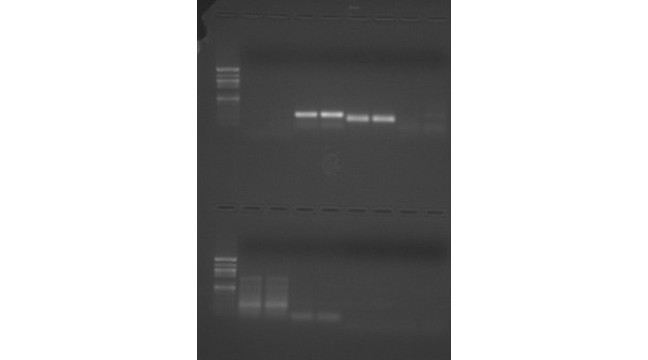
Figure 1: PCR with sample in bottom half in lanes 6 and 7 loaded with cDNA and 8 and 9 loaded with blank.
RUN PROTEIN GEL
1. Begin boiling water on hot plate.
2. Thaw you protein extract from last week. Mix by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge for 10 seconds.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well using a gel loading tip.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Do not dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible.
Observations:
The protein gel was run for us prior to lab by Mac. The results is pictured in Figure 2. My sample was loaded into well #5 and the results indicated that there is indeed protein in my sample.
Figure 2: Protein gel after soaked in blue dye with sample loaded in well #5.
TRANSFER MEMBRANES TO PROTEINS
1. Cool the transfer buffer to 4°C.
2. Soak the filter paper, membrane and gel in Transfer Buffer for 15 minutes.
3. Assemble the blotting sandwich in a semi-dry blotting apparatus as follows:
• Anode (+++)
• Filter paper
• Nitrocellulose Membrane
• Gel
• Filter paper
• Cathode (– – –)
4. Transfer the blot for 30 minutes at 20V.
5. Remove the gel from the sandwich and rinse with transfer buffer.
6. Use a cotton swab to remove any adhering gel from the membrane.
Observations:
We carried out a semi-dry blotting apparatus with the anode on top, then filter paper, Nitrocellulose Membrane, gel, filter paper, and, finally cathode. It was important to have no air bubble. After it ran for 30 mins at 20V, we carried out the western blotting procedure.WESTERN BLOTTING PROTOCOL
1. Prepare 20 mL of Blocking Solution
Ultra filtered Water 14 ml
Blocker/Diluent (Part A) 4 ml
Blocker/Diluent (Part B) 2 ml
Total Volume 20 ml
2. Place the membrane in 10 ml of the appropriate Blocking Solution in a covered, plastic dish provided in the kit. Incubate for 30 minutes on a rotary shaker set at 1 revolution/sec.
3. Decant the Blocking Solution.
4. Rinse the membrane with 20 ml of water for 5 minutes, then decant. Repeat once.
5. Prepare 10 mL of Primary Antibody Solution (1:3000 dilution)
Blocking Solution 10 ml
HSP 70 antibody 3.3 µl
Total Volume 10 ml
6. Incubate the membrane with 10 ml of Primary Antibody Solution OVERNIGHT
-
Figure 3: Western blotting result with my sample loaded in lane 5.
Observations:
We incubated the membrane in the Blocking Solution and incubated for 30 minutes on a rotary shaker. Afterwards, we rinsed the membrane and incubated the membrane with Primary Antibody Solution overnight. The result is pictured in Figure 3. Because we used an antibody for HSP 70, I expected to see a positive result. However, I do not see a good indication of a positive result. The possible reasons for this could have resulted from the antibody not binding to the HSP70, or the seondary antibody not binding to the first antibody.Conclusion:
We have just performed qualitative PCR. Our next step is to perform quantitative PCR, which is used to amplify and simultaneously quantify a targeted DNA molecule. It enables both detection and quantification of a specific sequence in a DNA sample.
-
Tuesday, October 20, 2009
Lab #3: Reverse Transcription and PCR
-RNA QUANTIFICATION
NOTE: Always keep your RNA samples on ice!
1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedistal and lower the arm.
2. Click "Blank", to zero the instrument
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your A260 absorbance, RNA concentration (ng/µL), A260/280 ratio and A260/320 ratio.
5. Raise the arm and wipe off you sample with a Kim Wiple
6. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
7. Give your samples to Mac for storage at -80C.
Observations:
We basically thawed our solution and loaded one small drop onto the RNA Quantification machine. We recorded the absorbances and here were my results:
A 260 Abs: 1.587
[RNA] (ng/microliters): 63.5
A 260/280: 1.74
A 260/230: 1.02
From the data, I could tell that my RNA sample was not very concentrated, with only 63.5 nanograms per microliter.
REVERSE TRANSCRIPTION PROTOCOL
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 5 ul of your RNA to a fresh PCR tube. Bring the volume up to 5uL with PCR water.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix (MM)
PER RXN
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave cDNA on ice or store at –20C
PCR
Prepare your samples in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| GoTaq®Green Master Mix, 2X |
25 |
1x |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| DNA template |
1–5μl |
<250ng |
Observations:
A master mix was made for 5 reactions instead of 4. This master mix was made of 125 microliters of GoTaq Green Master mix, 12.5 microliters of upstream primer, 12.5 microliters of downstream primer, and 90 microliters of water. To this master mix, 2 microliters of cDNA was added to two of the capsules and 2 microliters of water was added to the blank capsules. The cDNA and blank solutions resulted in having a green color to them. These PCR solutions were then subjected to the following thermal profile:
90C 10mins
20 cycles
95C 30 sec
55C 30 sec
72C 90sec
72 C 3 mins
4C forever
The final product will be used in the next lab to be run on gels. The gels will indicate if the correct cDNA was amplified by the PCR process.
PREPARE AGAROSE GELS (groups of 4)
1. weigh 2g of agarose and mix with 150mL 1x TAE in a 1L flask
2. microwave solution for ~ 3 minutes
3. cool solution (you should be able to touch the flask for a few seconds), then add 12uL ethidium bromide.
4. mix thoroughly by swirling, then pour into gel tray.
5. add gel combs
6. after gel is set, wrap in plastic wrap and place gel in the fridge for next week.
Observations:
We weighed out the 2g of agarose, which was a white powdery substance, and mixed it with 150mL of TAE in a flask. The white, opaque solution was then microwaved until the solution turned clear. Ethidium bromide, which is very toxic, was then added to the clear solution and poured into gel tray. We added the combs, let it set and then stored it.
Conclusion:
In this lab we have quantified our RNA sample, which will tell us the quantity and quality of our RNA solutions. We then used our known primers to purify and amplify a specific cDNA sequence through the process of PCR. From this, we will be able to observe the expression of our specific gene of interest. Later we will quantify this product. We have also prepared agarose gels for the next lab where we will run our PCR products on these gels. The gels will allow us to see the purity of our PCR products and see if the correct cDNA was amplified by the PCR cycles.
Tuesday, October 13, 2009
Lab #2: Tissue Extraction II
RNA Isolation Procedure (cont.):
1. Turn on heating block to 55C .2. Incubate tube at room temperature (RT) for 5 mins.
3. In the fume hood, add 200uL of chloroform to your sample and close the tube. Pipet carefully and quickly.
4. Vortex vigorously for 30s.
5. Incubate tube at RT for 5 mins.
6. Spin tube in refrigerated microfuge for 15 mins. at max speed.
7. Gently remove tube from microfuge.
8. Slowly and carefully transfer the aqueous phase to a fresh microfuge tube. Do NOT transfer ANY of the interphase.
9. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
10. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
11. Mix by inverting the tube numerous times until the solution appears uniform.
12. Incubate at RT for 10 mins.
13. Spin in refrigerated microfuge at max speed for 8 mins.
15. Remove supernatant.
16. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube.
17. Spin in refrigerated microfuge at 7500g for 5mins.
18. Carefully remove supernatant. Do not remove pellet.
19. Briefly spin tube (~15s) to pool residual EtOH.
20. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
21. Leave tube open and allow pellet to dry at RT for no more than 5mins.
22. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
23. Incubated tube at 55C for 5mins. to help solubilize RNA.
24. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
25. Quantitate RNA yield using spectrophotometer.
Observations:
After spinning the tube in the refrigerated microfuge for 15 mins (step 11), the solution was separated into three layers. The top layer was the clear, RNA-containing interphase. The second layer consisted of a milky emulsion and the the bottom layer was a clear pink solution. The clear top layer was then extracted for further purification. After adding the isopropanol, incubating, and microfuging a very small, white-opaque pellet was formed at the bottom of the tube. The isolated RNA will be used in an RNA Quantification procedure to measure the quantity and quality or our RNA solution.
RNA Quantification Procedure:
NOTE: Always keep your RNA samples on ice!1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedistal and lower the arm.
2. Click "Blank", to zero the instrument
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your A260 absorbance, RNA concentration (ng/µL), A260/280 ratio and A260/320 ratio.
5. Raise the arm and wipe off you sample with a Kim Wiple
6. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
7. Give your samples to Mac for storage at -80C.
Observations:
Time did not allow use to proceed on to the RNA Quantification. We will continue this procedure during the next lab period.
Protein Gel Procedure:
1. Begin boiling water on hot plate.
2. Thaw you protein extract from last week. Mix by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge for 10 seconds.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well using a gel loading tip.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Do not dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible.
Observations:
The product was a clear blue solution, which we loaded into the wells. The ladder used was SeeBlue Ladder and a 4-20% Tris-HEPES gel. However, we used the molecular weights for NuPAGE MES gel, which had the closest values to the gel used. The final solutions consisted of glycerol, betaME, and SES. The glycerol allowed the solution to sink into the wells. The betaME acted as a reducing agent to break down the disulfide bonds. Boiling the solution was also used to break down the proteins' tertiary and quaternary structures. Finally, the SES coats the proteins, causing them to have similar charges. This way, when the proteins are moving along the gel ladder, they will be separated by size and the charges will not affect the way in which certain proteins move. The result of the ladder for the sample protein was very light, which can be explained by the fact that my sample in the well only contained 10.31 micrograms of proteins. Other ladders with a higher concentration of proteins in their solution had darker, more visible results.
Results:

Figure 1: Group 2 Gel wells. A SeeBlue Ladder was used to run our protein samples. The result is shown above. From left to right, the wells contain the reference protein sample, herring heart, salmon brain, hard clam foot, sea scallop, barnacle, gigas, and oyster gill.

Figure 2: This is the reference key of known proteins and their molecular weights. For our purposes we used molecular weights for NuPAGE MES gel (closest to our 4-20% Tris-HEPES gel). Proteins of smaller molecular weights travel the furthest from the well. By comparing our unknown proteins to proteins of known molecular weights, we can estimate the number of different sized proteins in our unknown protein sample.
My sample, the salmon brain, was loaded into the third well from the left. Although the bands were very light, due to the fact that my sample only contained 10.31 micrograms of protein, four distinct bands stood out. Their molecular weights of those bands of proteins were about 49, 44, 38, and 17 kDa. There were also some non distinct bands between molecular weights 49-62 kDa, which may or may not be significant amounts.
Tuesday, October 6, 2009
Lab #1: Tissue Extraction
Sample Tissue Used: Salmon Brain
RNA Extraction:
Procedure:
- Add 500 microliters of TriReagent to a 1.5 mL snap cap tube and store it on ice.
- Add a piece of frozen tissue to the tube containing TriReagent.
- Carefully homogenize the tissue using a disposable pestle.
- Add 500 microliters of TriReagent to teh tube and close the tube.
- Vortex the tube for 15 seconds.
Observations:
The isolation of the RNA from the salmon brain was difficult to homogenize. The result of my efforts was a large piece of tissue stuck at the bottom of the tube. Hopefully, a sufficient amount of tissue was homogenized in order to extract RNA.
Protein Extraction:
Procedure:
Extraction:
- Add 0.5 mL of CellLytic MT solution to a 1.5 mL snap cap tube.
- Add a piece of frozen tissue to the tube containing CellLytic MT solution.
- Homogenize the tissue with a disposable pestle.
- Close the tube and invert it several times.
- Spin the tube in a refrigerated microfuge for 10 minutes at maximum speed.
- While spinning, label fresh tube with "Protein", source organism/tissue, initials, and date.
- Carefully transfer supernatant to labeled tube and store tube on ice.
Quantification:
- In a fresh 2 mL tube labeled as "sample", dilute 15 microliters of protein sample with 15 micorliters of DI water to make a 1:2 dilution.
- In a second 2 mL tube pipette 30 microliters of DI water, which will serve as your blank. Label tube a "blank."
- Add 1.5 mL of Bradford reagent to both tubes.
- Invert the tubes several times and then incubate at room temperature for 10 minutes.
- Mix the 'blank' tube and transfer 1 mL to a plastic, diposable cuvette.
- Zero the spectrophotometer using the blank sample.
- Mix the 'sample' tube and transfer 1 mL to a plastic, disposable cuvette
- Measure the absorbance at 595nm and record the value.
- Remove the cuvette from the spectrophotometer. Using a P1000 set to 1mL, carefully pipette the solution in the cuvette up and down a couple of times to mix.
- Measure the absorbance at 595nm and record the value.
- Average the two absorbance values you recorded.
- Calculate your protein concentration using the standard curve below.
- Store protein sample at -20 degrees Celsius.
Observations:
I found that, again, the salmon brain was difficult to homogenize, but decided to continue with a piece of tissue stuck at the bottom of the tube. After the microfuge, in order to obtain a viable absorption reading, the solution containing the sample had to be diluted 1:2. This means the we had to add equal amounts of the protein sample with DI water. The resulting solution would contain 1 part sample solution to every 2 parts resulting solution. This dilution factor must be accounted for when calculating the total amount of protein in the original solution sample. After adding Bradford reagent in step 10, the blank sample resulted in a clear, dark-gray solution, while the protein sample resulted in a clear, dark-blue solution. The resulting absorption readings for the sample solution was 0.338A and 0.341A, which has an average absorption of 0.3395A. Using the Bradford Assay Protein Kit curve, with an equation of y = 1011.9x, one can plug in the average absorption value for x and solve for y. Doing so, I obtained a value of 343.54. Accounting for the dilution factor of 1:2, one can then multiply the obtained value by 2. The result of my protein extraction was a protein concentration of 687.08 micrograms/mL.
Results:
Absorption 1: 0.338A
Absorption 2: 0.341A
Ave Absorption: 0.3395A
Total Protein Concentration: 687.08 micrograms/mL