12/3/12
Capstone paper working draft: HereData analysis: Here
- qPCR raw data formatted from CFX96 output to PCR miner format
- run on PCR miner
- R0 calculated (R0 = 1/(avg. efficiency of gene +1)^ avg. CT of replicate sample)
- R0 normalied with actin to get normalized R0 numbers (normalized R0 = R0 gene/ R0 actin)
- Statistics and graphs made from normalized R0
10/12/12
Today: 1. Run qPCR with normalizing gene primers that worked on check gelqPCR procedure:
Single reaction volume:
2X Sso Fast Evagreen – 10ul
Forward primer (10uM) – 0.5ul
Reverse primer (10uM) – 0.5ul
H2O – 8ul
Template – 1ul
Master mix (for 3 volumes: 1 sample mix, 1 neg. qPCR control, and 1 neg. cDNA control)
-made 4 reaction volumes
-made two seperate master mixes: a. actin primers ; b. acidic ribosomal protein primers
2X Sso Fast Evagreen – 10ul x 4 = 40ul
Forward primer (10uM) – 0.5ul x 4 = 2ul
Reverse primer (10uM) – 0.5ul x 4 = 2ul
H2O – 8ul x 4 = 32ul
1. Use white opaque well plate - 6 wells (Row A= actin master mix; B= acidic ribosomal protein master mix;; Column 1= sample mix; 2= -qPCR; 3= -cDNA)
- For -cDNA used column 5 well
- For sample mix used 1uL from each well in column 1 (8uL total)
10/2/12
Today: 1. Repeat procedure from 10/1/12 because gel did not workPrimer 3 and 4 worked on gel. Will use one of them as my normalizing gene.
10/1/12
Today: 1. Run PCR of normalizing genes 2. Check gel on PCR productLabels: "#, letter"
1= elongation factor; 2= g3p dehydrogenase; 3= actin; 4= acidic ribosomal protein
S= sample mixture; C= cDNA negative control; P= PCR negative control
Sample mixture - mix 1uL of samples
Procedure
Make 1.3% agarose gel
1. 110uL TAE buffer with 1.4g agarose - after cool add 11uL EtBr -- note: gel partially solidified when mixing EtBr, so re-heated in microwave until fully dissolved
2. Gel poured into mold with 20 well comb
a.
3. Run for about minutes 30 at 107V
Ran gel for too long. Samples came off of the gel.
Next time: Need to redo and run sample on check gel for shorter time.
6/19/12
Today: 1. Re-suspend primer pellets 2. Make 10mM working stock primers 3. PCR subset cDNA with normalizing gene primersProcedure:
Re-suspend primers:
1. Look at label on tube for nmol of primer in tube.
2. Spin down tubes to gather pellet on bottom of tubes.
3.. Add TE Buffer pH 8.5 (ETS 10.20.11) to primer tube
- Amount of buffer added to make 100mM solution. (uL added is 10*#nmol of primer)
Example: Ol_Arp_F - 28.6nm, add 286uL TE buffer.
4. Primer stock stored in -20C
Working stock primers:
1. Dilute from 100mM to 10mM.
2. 10uL of primer stock with 90uL of PCR water
3. Mix and spindown
-Labeled "primer name, 10mm, 6/19/12"
PCR tubes labeled - "sample/-cDNA/-PCR , PCR, gene name"
Next time: Make agarose gel and run PCR to see if normalizing gene primers are working.
6/18/12
Today: 1. Interpret qPCR results 2. create cDNA from Dnase treated RNA samples 3. Picked up normalizing gene primersqPCR results: CFX96 file
- Look at melting curve - see multiple products for one sample, all other samples negative as expected. See primer dimers.
primers were not resuspended. stored in -20C.
Next time: Run a check gel with normalizing gene primers.
6/13/12
Today: 1. Spec all Dnase treated RNA samples 2. Design primers for O. lurida normalizing genes 3. qPCR subset of RNA samples to check if Dnase treatment was successfulProcedure:
Spec sample in two groups. a. 1-23 b. 24-44,X,0
Spec results: Sheet "6_13_12_SpecData_DnaseSamples.t" - Spec results
Normalizing genes primers: Primer information
qPCR done with subset of samples: 3G, 6G, 15G, 17G, 28G, 30G, 40G, XG
Single reaction volume:
2X Sso Fast Evagreen – 10ulkk
Forward primer (10uM) – 0.5ul
Reverse primer (10uM) – 0.5ul
H2O – 8ul
Template – 1ul
Master mix (for 11 volumes: 8 samples, 2 negative control (H2O), one positive control (pooled cDNA))
-made 12 reaction volumes
2X Sso Fast Evagreen – 10ul x 12 = 120ul
Forward primer (10uM) – 0.5ul x 12 = 6ul
Reverse primer (10uM) – 0.5ul x 12 = 6ul
H2O – 8ul x 12 = 96ul
- Use white opaque well plate (12 wells, 6X2) – samples arranged as follows:
| 7 |
8 |
9 |
10 |
11 |
12 |
|
| A |
3G |
6G |
15G |
17G |
28G |
30G |
| B |
40G |
XG |
Neg. Con. |
Neg. Con. |
Pos. Con. |
Blank |
- add 19ul of master mix to each tube
- add 1ul template to each tube: PCR H2O for negative control, pooled cDNA for positive control
- template samples were diluted to same mass of sample as would have been added after cDNA step. Calculations here:
- pooled cDNA created by adding 1ul of each cDNA sample from 5/11/12 subset (8ul total volume): mixed and spun down
- Cap tubes with qPCR specific caps
- Centrifuge plate in Sorvall for 2min at 3000g
- Sign up for qPCR machine use
- Well added to top left corner of thermal cycler [95C, 95C, 65C; 40 cycles]
- File saved to dropbox folder: “Roberts Lab CFX96 Data”
Samples diluted: diluted samples stored in -20C
Pooled cDNA from 5/11 subset stored in -20C
Next time: interpret qPCR results
6/12/12
Today: Treat RNA samples with Dnase to remove residual DNA.Procedure:
Manufacturer protocol: tools.invitrogen.com/content/sfs/manuals/cms_055740.pdf
Use 0.5ml tubes.
50ul reaction volume:
44ul of 5ug RNA + H2O - calculations here (spreadsheet called "Dnase calculations"): Dnase calculations
5ul of 10X Dnase buffer - want to add 0.1 volumes
1ul of Dnase
Split into 4 groups (all gill tissue):
a. X,0, 24-33
b. 34-44
c. 1-12
d. 13-23
Same procedure followed for each group:
1. Make reaction volume following calculations from spreadsheet - mix by tapping tubes, spin down.
2. Incubated in hot water bath at 37C for about 1 hour
3. Add 5ul of Dnase inactivation reagent - use 0.1 volumes
4. Inactivate at room temp for about 2 min- make sure to mix (tap) tubes periodically during this time
5. Centrifuge at 10,000g for 1.5min - used 4C centrifuge
6. Use pipette to extract supernatant from tubes and transfer to a clean 0.5ml tube.
---Tubes labeled "# G pRNA"
7. Stored in -80C.
6/7/12
Today: 1. RNA extraction of all samples except subset from 5/12/12. 2. Spec extracted RNAProcedure:
Extraction in two batches: a. 1-24 b.25-44,0,X
RNA samples stored in -80C. Box labeled "Bradley -80C Tri-R"
Spec data: Spec data
Next time: Setup for cDNA.
6/1/12
Today: Transfer tissue to tri-reagent. Completed all gill samples except for those already done during subset test.Procedure:
Homogenized samples in groups of 10, 10, 10 and 12. Total of 42 samples today. 4 samples were previously done for subset (6G, 20G, 29G, 35G)
Samples stored in -80C.
Next time: Start RNA extraction of samples. Max 24 at a time.
5/29/12
Today: Re-make 1.3% agarose gel. Run samples on gel.Gel made 5/23/12 had wells that had bottom pulled out by comb. Need to remake gel.
Procedure:
Made gel with same procedure from 5/23/12
Used 105mL TAE buffer with 1.4g agarose.
Modification in procedure- took out the comb very slowly while submerged in buffer in electrophoresis box. Helps slide out better so bottom of well does not get pulled out.
Run gel:
Fill electrophoresis box with TAE buffer, with just enough buffer to thinly cover over entire gel.
Run at 100V for 3 min. then to speed up process ran at 115V for rest of the time. Took about 25 minutes total.
Photo of gel. Lane assignment of gel here:
Note that all primers worked except for crim (lane 17). For negative control, we see primer dimers for many of them.
Picture of gel saved to Bradley sites folder.
Gel disposed in trash.
Next time: Now confident that primers work. Will start RNA extraction for all samples.
5/23/12
Today: 1. Use designed primers to PCR cDNA from 5/11/12. 2. Pour 1.3% agarose gelProcedure:
PCR
Note: make sure to use PCR specific tubes (in drawer below thermal cycler)- have thinner walls for more efficient heat transfer
Set-up:
Primer dilutions:
-Ordered primers are 100mM
-For PCR want 10mM primers
-Dilute by adding 10uL of 100mM primer to 90uL PCR H2O ::: total of 100uL of 10mM primers
-Did this procedure for primer ordered
- Primers 10mM stored in -20C Bradley's Box
Master mix prep:
-Want separate master mix for each primer pair. Have 7 primer pairs so need 7 master mixes.
-Master mix: (25uL for one reaction volume) :: make 4 reaction volumes for 3 treatments (samples, cDNA neg. control, PCR neg. control) (vortex)
- 12.5uL 2X Apex red x 4 = 50uL
- 0.5uL F primer x 4 = 2.0uL
- 0.5uL R primer x 4 = 2.0uL
- 6.5uL H2O x 4 = 42uL
- added to "samples" treatment- 1uL of sample mix separate from master mix
- added to -cDNA treatment- 1uL of -cDNA from 5/11/12
- added to -PCR treatment- 1uL of PCR H2O
1. Use PCR well plate (28 samples total)
- table key:
- rows: 4=sample mix; 5=-cDNA; 6=-PCR; 7=-PCR (used left over master mix- maybe slightly less than 24uL)
- columns: (Each letter has unique master mix because unique primer) A=Pygm; B=U2a; C=Igfr; D=Hsp; E=Prkg; F=Crim; G=Bag
3. Thermal cycler conditions:
- 95C 10min
- 40X: 95C 15s :: 55C 15s :: 72C 15s
- Annealing temp= 55C (5C lower then melting temp)
Agarose Gel preparation:
1. Because PCR product is fairly small (100-120bp) want a higher resolution gel. Make 1.33% gel.
2. Use large mold so can fit 28 well comb
3. 105mL TAE buffer with 1.4g agarose
- do not stick anything into agarose bottle to contaminate: just tap out of bottle into weight boat
5. Microwave- with swirling every one minute: microwave until completely clear
6. Re-weight flask and add DI water to original weight if needed
7. Let cool to approx. 55C- test by being able to hold flask by bottom with bare hand comfortably for 30sec
8. Once cool, added 10.5uL EtBr [1:10,000 dilution of EtBr at 10mg/mL]- final concentration of EtBr= 1ug/mL --- gently swirl, do not introduce bubbles
9. Pour gel into mold, and let solidify
10. Comb pulled out, wrapped in saran wrap, stored in 4C fridge.
Next time:
Run PCR product on agarose gel.
5/11/12
Today: RNA extraction from sample stored in tri-reagent (5/1/12), spec extracted RNA, and reverse transcription of RNA to cDNA.Procedure:
RNA extraction: 8 samples [4M, 6G, 12M, 20G, 25M, 29G, 35G, 41M]
1. Extraction
- 1mL sample and 0.2mL chloroform (add under hood)
- centrifuge: 12,000rpm for 15min @ 4C
- 0.5mL isopropanol (poured new isopropanol (labeled 5/11/11, isopropanol, BC), wait 10 min for precipitation
- centrifuge: 12,000rpm for 8min @ 4C
- samples stored at -80 after centrifuge for about 2 hours (11:30am to 1:30pm)
- took samples out of -80C, let thaw at room temp for about 5 minutes
- 1mL 75% ethanol (10/11/11)
- centrifuge: 7,200rpm for 5min @4C
- EtOH pipetted out --> spot spun --> pipette out remaining liquid --> in fume hood to completely evaporate EtOH
- 50uL DEPC H2O (from 10/6/09 SJW) for samples 25M and 29G, all other samples 50uL nuclease free water
- stored on ice before spec
- Note: for solubilizaiton, make sure to use DEPC H2O, it can degrade nucleases. When pouring DEPC H2O, do not pour into beaker, use treated containers like 15mL or 50mL conical vials.
Spec procedure: (samples stored on ice)
Login on computer: Open "ND-1000" program
In program:
1. select "nucleic acid"
2. program will prompt for "initialize"- put 2uL nanopure water on pedal, close, open, and wipe lever and pedal with kim wipe
-note: make sure RNA-40 constant is selected
3. add 2uL of solvent (RNA pellet solvent is DEPC H2O) to pedal, close lever, hit "blank"
4. lift lever and wipe lever and pedal with kim wipe
5. add 2uL of sample, close lever, label sample on computer, hit "measure"
6. repeat steps 4 and 5 for all samples
7. save data by going to reports: save report --> save to excel file
Spec data: with calculations for amount of RNA for cDNA - Spec data
cDNA procedure:
5/9/12
Re-designed primers because originally done off of wrong sequence data.New Primers designed: Primer information
5/1/12
Today- Test a subset of samples for compatibility to primers. Today, will transfer frozen tissue into tri-reagent.Procedure: Randomly selected oysters to use for testing qPCR primers
- Put 0.5mL Tri-Reagent in microfuge tubes (1.5mL- fit the homogenizer)
- these tubes were labeled # of oyster, M/G for mantle or gill tissue
- Frozen tissue samples stored on dry ice to keep frozen, while tri-reagent stored on normal ice
- Quickly to avoid melting of tissue, cut piece of tissue and place in tri-reagent tube.
- Put leftover tissue back in frozen tube and place on dry ice.
- Tissue is tri-reagent tube is homogenized using blue plunger
- Once homogenized, place tube back on ice
- Tools for cutting (forcep and scissors) were cleaned using treatment of 10% bleach, water, ethanol, and flame
- Same procedure for all sample
- Sample done today: 4M, 6G, 12M, 20G, 25M, 29G, 35G, 41M
10. vortex each tube and stored tubes in -80C
Note: Primers created from before were done using the sequence of incorrect species. Olympia oyster consensus sequence is on Dr. Roberts notebook. Need to re-design primers using the sequences from this file.
Plan- Continue with RNA extraction and making cDNA.
4/20/12
Today- mechanical stress and sample tissueProcedure: randomly selected which oysters got the stress treatment
1. Mechanical stress
- Sorvall centrifuge- set at 1000rpm, 15C, 5 minutes
- spun 4 oysters at a time- 2 in each barrel
- if spun 3 oysters, used a shucked oyster to balance weight
- note- make sure do hold down top/lid when starting
- shuck oyster- insert instrument into back of oyster shell, use twisting motion to pry open
- using forceps and scissors, but section of mantle and gills
- process repeated for all oysters
- tissue samples stored in 1.5mL tubes - labeling system here:
- Notes on specific oysters:
- #17- Sam noticed that it may have been dead at time of sampling
- #2- normal sized oyster but had #X settled on its shell
- #X- much small oyster (1/3 size) that settled on shell of #2
- Notes on specific oysters:
- note: for oysters that were spun were sampled as soon as possible after stress treatment - at most 5 minutes between centrifuge and sample
- used bleach and sand to clean instruments between oysters
- tubes stored in -80C freezer - boxes labeled with name and date
- The rest of oyster shell and body were stored in a plastic bag in the -20C freezer (on the right side "door" shelf") : labeled "Olympia Oyster 4/20/12"
Other notes: transferred oysters from treatment room into lab using roll cart. Was not very smooth and there was noticeable physical movement during transport.
Plan: possibly extract RNA. Also need to get qPCR primers of genes. Get information on oysters.
4/17/12
4:30pm added about 1mL of algae feed to both containersre-adjusted the aeration device- no air-flow when I first came in.
4/16/12
3:00pm changed water in both containersAlso fed oysters about 2mL of algae feed
Notes:
-water level at about 2.5 inches
-scrubbed both tanks clean before adding water
-tanks looked noticeably dirtier (lots of pseudofeces) since last check on Friday (4/13)
-having some trouble with getting enough flow from aeration stones- will need to monitor daily to make sure there is consistent flow
-make sure to monitor temp inside room- was at 15C
4/13/12
12:30pm added about 1.5mL of algae feed to both containersSlight adjustment of aeration stones to get same air flow in both containers
No observable differences in oysters
Plan- Keep monitoring oysters. Change water and feed again on Monday (4/16).
Start to prepare/label tissue biopsy tubes for collection- will need separate tubes for gill, mantle, and whole body
Plan to perform centrifuge and biopsy collection on Friday (4/20)
4/9/12
10:30am changed the water in both containers.- Used water from black can (same source as original water)
- Added about 2mL of algae feed to both containers
- Left all other conditions the same
No apparent deaths of oysters. Moved oysters gently while changing water, and avoided excess movement.
Water level at about 2.5 inches
Will continue monitoring condition of oysters. Not sure if we need to change water again.
Plan to talk with Dr. Roberts about details on tissue sampling and centrifuge treatment.
4/6/12
About 12:30pm added approx. 2mL of algae feed to both containers with disposable pipette.Adjusted aeration stones to same airflow and centered them closer to oysters.
Plan:
Keep monitoring oysters.
Change the water on Monday (4/9/12).
-Need information on oysters
Info on feed:
http://www.reedmariculture.com/pdf/product_shellfish_diet_1800.pdf
4/4/12
Olympia oysters came in sometime in the morning. 44 oysters split into two groups, separate containers.22 per container.Came in around 2:00pm and set the timer for photoperiod treatment. Tarps are not completely draped around 12D:12N container, secure on one side. Left one side open to allow adding the food. Will close everything up once added.
Need to ask for information on the oysters (age, etc.)
Plan to run treatment for about one week. Current holding conditions are 12C, aeration stone. Group A- 12 hours light ; 12 hours dark, Group B- 24 hours light.
*For group A- Light from 6am to 6pm
qPCR Primers:
1. Found the mRNA sequence on NCBI2. Put into primer3plus- parameters= product: 80-130bp; primer: 18-23bp; Tm: 58-62C; CG content: 40-65%; left the rest at default. Check primers for self-pairing, hairpins, and consecutive G/C's
3. Run primers through BLAST to check for specificity
Xenopus laevis insulin-like growth factor 1 receptor (igf1r)
e-value: 2E-121
NCBI Reference Sequence: NM_001088265.1
Primers: product: 115bp
Left: ATGCTTCTGTGAACCCGGAG – 20bp, Tm: 60C
Right: CCAAACGAACCTTGCCCAAG – 20bp, Tm: 60C

Bos taurus U2 small nuclear RNA auxiliary factor 1-like 4 (U2AF1L4)
e-value: 6.00E-84
NCBI Reference Sequence: NM_001034778.1
Primers: product: 130bp
Left: GCAGACGGATCTCACTGTCA – 20bp, Tm: 60C
Right: GGTTGTCGCACACATTCATC – 20bp, Tm: 60C
Bos taurus protein kinase, cGMP-dependent, type I (PRKG1)
e-value: 0
NCBI Reference Sequence: NM_174436.2
Primers: product: 109bp
Left: CGAGGTTATGCCAAACTGGT – 20bp, Tm: 60C
Right: GGATGATCTCTGGGGCTACA – 20bp, Tm: 60C – but 4 G’s in a row??

Danio rerio cysteine rich transmembrane BMP regulator 1 (chordin like) (crim1)
e-value: 3.00E-38
NCBI Reference Sequence: NM_212821.1
Primers: product: 130bp
Left: AAAACTGGGATGACGACCAG – 20bp, Tm: 60C
Right: GCGAACTCAAATGGGTTGTT – 20bp, Tm: 60C

Bos taurus BCL2-associated athanogene 2 (BAG2)
e-value: 6.00E-18
NCBI Reference Sequence: NM_001034264.1
Primers: product: 83bp
Left: TTGCCTCCCATTCCACTTAC – 20bp, Tm: 59.9C
Right: AGCTCGGTGTGCGTAAATCT – 20bp, Tm: 59.9C

Homo sapiens heat shock protein 90kDa alpha (cytosolic), class B member 1 (HSP90AB1)
e-value: 0
NCBI Reference Sequence: NM_007355.2
Primers: product: 114bp
Left: ATGGAAGAGAGCAAGGCAAA – 20bp, Tm: 60C
Right: GCAGCAAGGTGAAGACACAA – 20bp, Tm: 60C

Capstone Notes:
Experimental temp: http://www.nodc.noaa.gov/dsdt/cwtg/npac.html
Photoperiod: http://aa.usno.navy.mil/cgi-bin/aa_rstablew2.pl
9/29/11
Today: 1. Make gel and run PCR product from 9/1/11
Methods:
-Make gel
1. Made approx. 1% agarose gel (75mL TAE buffer w/ 8g agarose)- use slightly larger flask (500mL)
2. Microwave ~3min with swirling every 1min: check for agarose to be completely soluble
3. Added 6uL EtBr and swirled
4. Wait to cool down a bit and pour into mold
Notes:
- can take up sides of mold if it does not have rubber sides
- EtBr stored in 4C
1. Loaded samples (from left to right)
- 10kb ladder- loaded 5uL (http://genefish.fish.washington.edu/~srlab/Steven/Commercial%20Protocols:Manuals/HyperLadderI.jpg)
- 6E PCR - 10uL
- 8E PCR - 10uL
- 1L PCR - 10uL
- 4L PCR - 10uL
- -PCR control from 9/1/11 - 10uL
- -PCR control from 9/1/11 - 10uL
- -PCR control from 9/1/11 - 10uL
2. Add 1x TAE buffer to fill the entire electrophoresis box (buffer should be covering gel)
3. Hook up electricity cables- DNA should run towards +charge (voltage in power box to 107V)
4. Check for bubbles on + end to make sure it is working
5. Run time dependent of purpose of gel (check gel vs. size identification)
Results:
See expected bands for all PCR products and no bands for neg. controls
Next time: PCR all samples of cDNA from (8/31/11) to check for differences between early and late groups. Expect to see difference in band intensity.
9/1/2011
Today: Select a few cDNA samples to PCR. Test to see if assay works.
Methods: Primer info at 6/24/11
cDNA from 8/31/11. Used samples 6E, 8E, 1L, and 4L. Also made 3 neg. controls with H2O.
Reaction volume: 25ul (2X Apex red= 12.5uL, F primer= 0.5uL, R primer= 0.5uL, template= 1.0uL, H2O= 10.5uL)
1. Master Mix (20uL per tube; made 7x for 7 tubes ):
- 12.5uL 2X Apex red x 7 = 87.5uL
- 0.5uL F primer x 7 = 3.5uL
- 0.5uL R primer x 7 = 3.5uL
- 6.5uL H2O x 7 = 45.5uL
3. Template (5uL) added after master mix
4. Thermocyling parameters (folder BC :: file APEXBC)
- 95C 10min
- 40X: 95C 15s :: 55C 15s :: 72C 20s
- Annealing temp= 55C (5C lower then melting temp)
- Extend time= 20s per cycle (max 300kb amplicon) [1000kb/min]
Note: make sure to use PCR tubes (at drawer under the thermocycler- tubes have thinner walls, heat transfers more quickly)
Next time: make agarose gel and run PCR product on gel
8/31/2011
Today: Make cDNA from 8/5/11 and 8/11/11 RNA extractions
Methods:
Reverse transcription
-Assume less than 1ug RNA from extraction
1. Want 25 reaction volumes (10 early, 14 late, and 1 - control). Made 26 reaction volume master mix.
2. 17.75uL RNA extract with 0.5uL oligo dT primer. H2O used for - control.
3. Break secondary structure and anneal primer: thermocycler 70C for 5min
4. Master Mix (for 26 tubes) [6.75uL per tube]
- 5uL buffer x 26 = 130uL
- 1.25uL dNTPs x 26 = 32.5uL
- 0.5uL reverse trans x 26 = 13uL
- Master mix total= 130 + 32.5 + 13= 175.5uL
6. Heat inactivate: thermocycler 95C for 3min
7. Store -20C
Next time: Decide how to proceed with cDNA. Could compare telomerase expression in early and late senescence.
8/29/2011
Today: Spec the extracted samples from 8/5/11 and 8/11/11 to check for nucleics
Methods: Use nanodrop
Results:
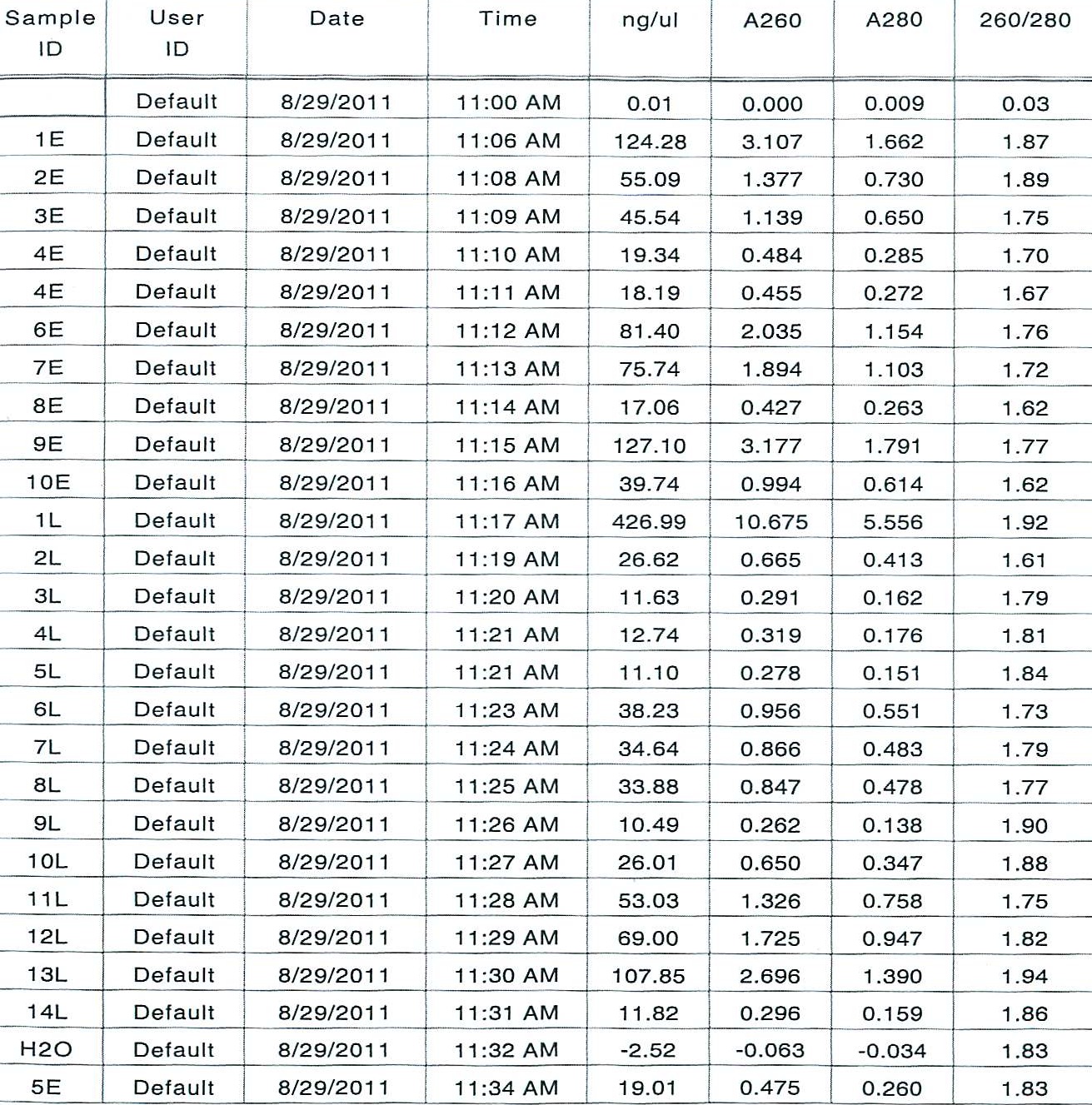
Next time: Start making cDNA
8/11/2011
Today: RNA extraction of 8/2/2011 sample stored in Tri-Reagent (2nd half)
Methods: Extracted 3-14 late (12 samples total): Check 8/2/2011 for labeling
1. Extraction
- 1mL sample and 0.2mL chloroform
- centrifuge: 12,000rpm for 15min @ 4C
- 0.5mL isopropanol (5/3/11)
- centrifuge: 12,000rpm for 8min @ 4C
- 1mL 75% ethanol (9/15/09)
- centrifuge: 7,600rpm for 5min @4C
- 50uL DEPC H2O
- stored in -80C
Spilled a bit of sample 9L during extraction step
Next time: spec samples
8/5/2011
Today: RNA extraction of 8/2/2011 sample stored in Tri-Reagent
Methods: Extracted 1-10 early and 1-2 late (12 samples total): Check 8/2/2011 for labeling
1. Extraction
- 1mL sample and 0.2mL chloroform
- centrifuge: 12,000rpm for 15min @ 4C
- 0.5mL isopropanol
- centrifuge: 12,000rpm for 8min @ 4C
- 1mL 75% ethanol
- centrifuge: 7,600rpm for 5min @4C
- 50uL DEPC H2O
- stored in -80C
Next time: Extract other 12 samples from sample set
8/2/2011
Today: 1. Prepare samples for extraction by putting tissue in Tri-Reagent
2. Results from re-spec of 7/12/2011
Methods:
Sample information and labeling information:
https://spreadsheets.google.com/spreadsheet/ccc?key=0ArvO3xgNTmCidFc4TkdYcWQ3dGxYcGs3Z3lpRVJkOWc&hl=en_US#gid=0
1. add 500uL tri-r to tube
2. add small piece of tissue (1/4 size of pinky nail)
3. homogenize in tube
4. add 500uL tri-r to tube
5. stored in -80C
Notes:
- new homogenizer for each tube
- sterilize forceps with isoprop and flame before each tissue
- vortex after homogenize
Next time: Begin RNA extraction of samples
7/12/2011
Today: 1. RNA extraction of 44,46,47,62,63 and 64 samples of senescent sockeye salmon males
from: https://spreadsheets.google.com/spreadsheet/ccc?key=0AtV_gF766XZAdDNmV09UX1M0UlFUUkloQ3gtWlByTHc#gid=0
2. Spec product of RNA extraction from today (7/12/11) and 7/5/11
Methods:
-RNA extraction
Sample were homogenized and stored in Tri-Reagent at an earlier date
Labels:
RNA 11 = 44 ; RNA 12 = 46 ; RNA 13 = 47 ; RNA 14 = 62 ; RNA 15 = 63 ; RNA 16 = 64
1. Extraction
- 1mL sample and 0.2mL chloroform
- centrifuge: 12,000rpm for 15min @ 4C
- 0.5mL isopropanol (5/3/11)
- centrifuge: 12,000rpm for 8min @ 4C
- 1mL 75% ethanol (7/5/11)
- centrifuge: 7,600rpm for 5min @4C
- 50uL DEPC H2O (5/3/11)
- stored in -80C
1. follow instructions in program
2. clean with kimwipe
3. add 2uL sample- tap/mix sample before adding
-- Had some samples not fully dissolved leading to inconsistent spec readings. Will slightly heat samples next time to make sure they are dissolved before do spec.
Notes:
- ideally 260/280= 2.0 and 260/230= 2.0
- nucleic acids absorb at 260nm
Next time: Redo do spec and look for consistent readings
7/5/2011
Today: RNA extraction of 9,10,11,12, 13 and 14 samples of pre-senescent sockeye salmon males
from: https://spreadsheets.google.com/spreadsheet/ccc?key=0AtV_gF766XZAdDNmV09UX1M0UlFUUkloQ3gtWlByTHc#gid=0
Methods:
Sample were homogenized and stored in Tri-Reagent at an earlier date
Labels:
RNA 1 = 9 ; RNA 2 = 10 ; RNA 3 = 11 ; RNA 4 = 12 ; RNA 5 = 13 ; RNA 6 = 14
1. Extraction
- 1mL sample and 0.2mL chloroform
- centrifuge: 12,000rpm for 15min @ 4C
- 0.5mL isopropanol (5/3/11)
- centrifuge: 12,000rpm for 8min @ 4C
- 1mL 75% ethanol (7/5/11)
- centrifuge: 7,600rpm for 5min @4C
- note: may have lost pellet for RNA 1
- 50uL DEPC H2O (5/3/11)
- stored in -80C
Next time:
Could extract RNA from senescent males
7/1/2011
Today: Run PCR product on gel
Methods:
Samples loaded in wells:
- 10kb ladder- loaded 5uL (http://genefish.fish.washington.edu/~srlab/Steven/Commercial%20Protocols:Manuals/HyperLadderI.jpg)
- 56A cDNA from 6/17/11 - 10uL
- 58A cDNA from 6/17/11 - 10uL
- -cDNA control from 6/17/11 - 10uL
- -PCR control from 6/24/11 - 10uL
- -PCR control from 6/24/11 - 10uL
- blank
- blank
2. load wells with samples (don't need to go far into well, loading dye helps sample fall down into well)
3. quickly attach current to mold (black at wells and red at bottom: "runs to red")
4. voltage set at 100V and run for 50min
5. UV machine to view bands
Results:
| left to right: 10kb ladder, 56A, 58A, -cDNA, -PCR, -PCR |
Gel supports conclusion that PCR was successful. All negative controls were as expected.
Notes:
1. make sure no bubbles when looking at gel
2. can add EtBr to buffer to increase clarity of bands
3. reasons bands don't show up: a. did procedure wrong b. no mRNA expressed
Next time: RNA extraction of more 2009 senescent male brain tissue samples
Want to determine what kind of gene expression information be can get out of these tissue samples
6/24/2011
Today: 1. PCR 2. Make gel
Full technique: http://genefish.wikispaces.com/protocols#Protocols-Conventional%20PCR%20%282x%20Apex%20Red%29
Methods:
Primer Info (from Caroline):
Forward Primer: AAAGGCTTTGTGATGGCACT
20bp tm 59.7 GC 45%
Reverse Primer: CGTTTTCTTCCCAAACTGGA
20bp tm 60.1 GC 45%
Amplicon size 150bp
Primer designing method: http://www.evernote.com/pub/cgstorer/carolinesnotebook#b=c5e05cfb-8b7e-42dd-abd1-490de048dfa5&n=79961e44-55c4-444b-a486-6297defa405b
--PCR
Reaction volume: 25ul (2X Apex red= 12.5uL, F primer= 0.5uL, R primer= 0.5uL, template= 1.0uL, H2O= 10.5uL)
1. Master Mix (20uL per tube; made 7x for 5 tubes ):
- 12.5uL 2X Apex red x 7 = 87.5uL
- 0.5uL F primer x 7 = 3.5uL
- 0.5uL R primer x 7 = 3.5uL
- 6.5uL H2O x 7 = 45.5uL
3. Template (5uL) added after master mix
4. Thermocyling parameters (folder BC :: file APEXBC)
- 95C 10min
- 40X: 95C 15s :: 55C 15s :: 72C 20s
- Annealing temp= 55C (5C lower then melting temp)
- Extend time= 20s per cycle (max 300kb amplicon) [1000kb/min]
--Making gel
1. 1% agarose gel (1g agarose per 100mL 1x TAE buffer): used small mold- 0.75g agarose and 75mL buffer
2. put into flask but do not mix, microwave until it starts to boil and stop- look for all of agarose to be dissolved.. keep microwaving if not
3. take out and swirl, let sit until lukewarm (able to hold flask without mit and little steam coming out of flask)
4. add 7.5uL EtBr- swirl to mix [10uL EtBr per 100mL buffer]
4. pour into mold (make sure there are no bubbles- get them out with pipette tip) and add comb- used 8 well comb
5. let cool until solid
6. stored in 4C fridge
Notes:
1. Pipette small volumes directed into the liquid, not above, to help precision and accuracy
2. Careful with agarose- do not contaminate, expensive
3. Master mix into general waste
Next time: Run PCR product on gel
6/17/2011
Today: 1. Reverse transcription- RNA => cDNA 2. Determine primers and reagents for PCR next time
Full technique: http://genefish.wikispaces.com/protocols#Protocols-Reverse%20Transcription%20%28Promega%20M-MLV%20Protocol%29
Manufacturer protocol: http://genefish.fish.washington.edu/%7Esrlab/Steven/Commercial%20Protocols:Manuals/Promega%20-%20MMLV%20RT.pdf
Methods:
Reverse transcription
-Assume less than 1ug RNA from extraction
1. Made 3 reaction volumes (56A, 58A and negative control [w/ RNase free H2O])
2. 17.75uL RNA extract or H20 with 0.5uL oligo dT primer
3. Break secondary structure and anneal primer: thermocycler 70C for 5min
4. Master Mix (for 3 tubes) [6.75uL per tube]
- 5uL buffer x 3 = 15uL
- 1.25uL dNTPs x 3 = 3.75uL
- 0.5uL reverse trans x 3 = 1.5uL
- Master mix total= 15 + 3.75 + 1.5= 20.25uL
6. Heat inactivate: thermocycler 95C for 3min
7. Store -20C
Notes:
Reverse Transcriptase expensive- keep in freezer and don't contaminate
Gene identification: GeneBank on NCBI
- decided to look at telomerase gene of O. nerka
-forward and backwards primers bought from IDT (20-21bp, annealing temp 53-55C)
- primers from 100mM stock solution diluted to 10mM working solution
Use 2X Red Apex master mix: contains Taq polymerase, dNTPs, buffer, dye
PCR H2O = RNase-free H2O
Master mix- make 2 tubes extra for negative PCR control.
Next time: Complete PCR of cDNA
6/13/2011
Today: Learning lab techniques- RNA extraction => Do this at RNA extraction bench/ station
Full technique: http://genefish.wikispaces.com/protocols#Protocols-RNA%20Extraction
Manufacturer protocol: http://www.mrcgene.com/tri.htm
Methods:
Two samples- replicated extraction (labeled 56A, 56B and 58A, 58B)
Notes:
1. Homogenize
- check tissue weight to determine Tri volume
- homogenized frozen sample- let thaw and vortex before extraction
- under hood and hazardous waste tips- chloroform and Tri
- vortex after chloroform added
- transfer aqueous (clear) only- do not get bottom layer
- invert tube 2-3 times after isoprop- no vortex
- isopropanol- normal waste
- place "hinge" part of tube towards outside when centrifuge
- spun for 8 min @ 12,000rpm 4C
- large pellet- 1. pour out liquid 2. add 75% ethanol 3. centrifuge 5 min @ 7,600rpm 4C 4. pour out liquid 5. spot spin (up to 1000rpm and stop)
- small pellet- 1. pipette out liquid 2./3. same
- flick tube to dislodge pellet before wash if large pellet
- after spot spin- 1. pipette out excess liquid 2. put tube under hood to evaporate alcohol // spot spin again if still alcohol
- volume of H2O w/ DEPC dependent on pellet size- today added 50uL H2O
- can store at -80C
General lab info:
Shared room:
- Dishwasher- settings for glass or plastic, add detergent
- Autoclave
Sample information:
O. nerka brain tissue- females in senescence: used sample # 56 and 58
Sampling data set: https://spreadsheets.google.com/spreadsheet/ccc?key=0AtV_gF766XZAdDNmV09UX1M0UlFUUkloQ3gtWlByTHc#gid=0
Next time: See if extraction was successful. See how much RNA was extracted.IGF1R
