Summary: This week, I analyzed the results from last weeks qPCR. The expression values varied between 0.08 to 69. The Vtg normalized expression value group averages for the groups were as follows: Mock control = 8.95, Mock treatment = 29.32, LPS control = 5.82, and the LPS treatment = 12.23. These differences aren't substantial, but there is a slight increase in average Vtg expression in the treatment groups, and a lower Vtg expression in the LPS groups.
Reflection: The purpose of this week was to analyzed the results of qPCR and determine how the results apply to the project. The increase in Vtg in treatment groups is expected because of the increase exposure to EDCs, which mimic estrogen-like hormones. The reduced expression in LPS treated groups is interesting, because these groups were injected with LPS bacteria, indicating that there is less expression during a immune response than there is to chemical stressors.
12/5/10 - Lab 9: qPCR
Summary: The results from the RNA test qPCR showed contamination of a few samples. These samples were DNased. This week, we ran a test qPCR for all the primers, and found some interesting results. Although some primers didn't work (were not expressed or didn't fit with coho DNA), many of the primers had some expression. I ran qPCR on the Mock Treatment, Mock Control, LPS Treatment, and LPS control using the Vtg primer. This gene was especially interesting since during the test run, this was the only sample with two peaks in the melting curve. When we ran the samples in qPCR for all the treatment groups, we changed the temperature to 60ºC rather than the normal 55ºC.
Reflection: The purpose of this weeks lab was to test the expression of Vtg gene on the Mock Treatment, Mock Control, LPS Treatment, and LPS Control groups to determine if there is a difference in expression between the groups. My results were not as clean as I would have liked them to be. The melting point had two peaks, which could mean that the primer coded for more than one gene in coho salmon. The gene was found on NCBI, and originally was designed for rainbow trout. Also, some of the duplicate samples did not have a C(t) value within a point of each other. Could this be due to operator error? What else would account for this difference?
11/28/10 - Lab 8: More samples?!?
Summary: When looking over the samples that we had and comparing it to the treatment groups, we noticed a lot of inconsistencies and gaps and concluded that there must be another box with more samples. When we found the remaining samples, we quantified the RNA with the nanodrop mass spectrophotometer, normalized the samples to 200ng/uL, and ran the RNA samples on qPCR to test for contamination. Because of the snow storm, we haven't yet seen the results to determine if there is DNA contamination/carry-over, or completed any other intended procedure or tests.
Reflection: The purpose of this weeks lab was to bring all the remaining samples up to the same step as the original samples. We had other goals to complete this week, but because of the lovely snow and the extended holiday weekend, everything had to be postponed and will be completed next week.
11/20/10 - Lab 7: Quantify RNA, Normalize RNA, Test RNA, DNase, and Reverse Transcription
Summary: To determine if we have a sufficient amount of RNA to run qPCR on each of our 14 primers, we quantified RNA using the Nanodrop and measured the A260 absorbance, the A260/280 ratio, and the A260/230 ratio. We found the results of the A260 absorbance to vary between less than 100 ng/uL to over 3,000 ng/uL, with the majority of the samples having sufficient RNA for reverse transcription. To make all the samples of the same concentration, we normalized the RNA samples by creating 10uL of 200ng/uL. Each of the RNA samples were analyzed for DNA contamination and carry-over using qPCR. The qPCR results showed only 12 samples required decontamination using DNase: 11, 25, 28, 32, 41, 42, 45, 58, 59, 61, 76, 77. DNasing is required for contaminated samples to remove DNA present in the RNA samples so when we run qPCR on our primers later, the results reflect only the DNA being expressed and nothing carried over from RNA extraction. We performed reverse transcription to create cDNA which will be used later to run qPCR with our primers.
Reflection: We completed several procedures this week in preparation for measuring gene expression of 14 primers. We measured RNA concentration to later normalize and dilute all RNA samples to be the same concentration. We ran qPCR to test for DNA contamination, and DNased samples that had carry-over DNA so when we later measure gene expression, we are certain that all DNA present in the samples corresponds to the gene we are measuring, and not carry-over from RNA extraction. We reverse transcribed RNA to cDNA which will be used to run qPCR for each of the primers. Because we have very little RNA available for reverse transcription, and will be running qPCR for all 14 primers, we will need to dilute the cDNA. When the primers arrive, we will first test each of the primers to make sure they work (and/or are expressed) in our samples, and then will begin to measure gene expression using qPCR. It was a bit unclear how the procedures that I wasn't involved in were completed, so I wish people would update the group Wiki-page with each of the steps they completed and how exactly it was done, but other than that, I think our group and our project is running smoothly.
11/12/10 - Lab 6: Reading papers and choosing primers
Summary: We all spent the week reading papers and designing primers, which will be used later in determining gene expression in qPCR.
Purpose: to design primers that will be used in determining gene expression in qPCR.
Materials/Methods: I designed primers found in literature in the NCBI website.
Results: Primers I chose for the project include:
| TargetGene |
AssessionNumber |
Forward Sequence |
Reverse Sequence |
Reference |
| Vtg |
AJ417877 |
GCGCAGAACGACGAGGCCA |
GGGATGCAGCGACCGTTCCC |
Palstra et al. 2010 |
| IL 12B |
AJ548830 |
CGAGGCCGAGCAGACTGCC |
CCCAGCCCAAGACGGACAGC |
Djordjevic et al. 2009 |
Djordjevic, B. S. Skugor, S.M Jorgensen, M. Overland, L.T. Mydland, and A. Krasnov. 2009. Modulation sf splenic immune responses to bacterial lipopolysaccharide in rainbow trout (Onchorhynchus mykiss) fed lentinan, a beta-glucan from mushroom Lentinula edodes. Fish and Shellfish Immunology 26: 201-209
Palstra, A.P., D. Crespo, G. van den Thillart, and J.V. Planas. 2010. Saving energy to fuel exercise: swimming suppresses oocyte development and downregulates ovarian transcriptomic response of rainbow trout. Am J Physiol Regul Integr Comp Physiol 299: R486-R499.
Conclusion: I chose those specific primers based on literature I read during my research. I chose to look at Vitellogenin because we are examining the liver tissue and I thought it would be an interesting to see how a reproductive gene was affected by the pesticide exposure and/or pathogen exposure. Looking back, it probably isn't the best choice for this project, since we are specifically looking at how the pesticide exposure affects immune response, but if there was time and enough RNA sample, it would be an interesting side project. I chose to look at Interleukin-12 subunit beta because it is a cytokine involved in the immune response. The gene acts on Th1 cells, cells involved with cell memory that mediates long-term protections to an intracellular pathogen. It would be interesting to see if there is a difference in expression between the 24 and 48 hour treatments.
Reflection: We need to work on communication within the group. We had a specific plan to choose primers by Friday; however, only four primers had been designed before class began on Friday. There were no plans to meet after class or work on choosing the primers together. We also need to decide on the next steps in the project and delegate roles to individual people.
11/8/10 - Research Proposal
Background
Habitat destruction and degradation have contributed to the widespread decline and extirpation of Pacific salmonid populations (Oncorhynchus sp.) throughout the Pacific Northwest (Nehlsen et al. 1991). Several salmonid species, including chinook (O. tshawytscha), coho (O. kisutch), sockeye (O. nerka), chum salmon (O. keta), and steelhead (O. mykiss) have been listed as threatened or endangered under the Endangered Species Act (ESA). Among the leading factors contributing to their decline is a decrease in water quality, which is partially due to an increase in contaminants such as pesticides.
Nationwide, more than 95 percent of river and stream samples contained at least one pesticide, with over half the streams sampled containing five or more pesticides (Gilliom et al. 1999). Although they are typically sub-lethal, pesticides in high concentrations have been found to be toxic to fish and still affect fish with low concentrations (Engelhaupt 2008). Pesticide exposure in salmon is linked to changes in feeding behavior, growth, and survival (Baldwin et al. 2009), and a loss of olfactory abilities (Engelhaupt 2008). Results from studies show that toxins can limit salmonid populations and have delayed impacts that occur when the fish migrate to the ocean (Baldwin et al. 2009). Early toxic exposure can also lead to impairment in fish immune response, increasing an organism’s susceptibility to disease (Eder et al. 2008).
Research Objective
In this study, we propose to measure how pesticide exposure in early developmental stages can affect immune response in later life stages by examining gene expression in liver samples.
Approach/Methods
Samples will be collected from specimens that have undergone the following treatments during developmental stages. As eggs and alevins. the sampled Coho salmon were exposed to a pesticide cocktail until they grew into fry. Once they were able to feed, the fry were raised for another two months without pesticide contamination. Then as fry, they were injected with Lipopolysaccharide (LPS) and Polyinosinic-polycytidylic acid (Poly I:C). Samples were collected from the liver 24 and 48 hours following the injection and used for RNA extraction.
Primers designed for the project will include heat shock proteins (hsp) to measure the stress reaction to pesticide exposure and/or pathogen exposure, and cytokines to measure the immune response to the pathogen injection. Complementary DNA (cDNA) is synthesized using 5µL of M-MLV 5X Reaction Buff, 5µL of dNTPs, 1µL of M-MLV RT, and 4µL of water for every 5µL of RNA sample. The reverse transcription reaction proceeds for 60 min at 42ºC and then heat inactivated at 70º for 3 min on the thermocycler. Quantitative PCR (qPCR) is performed with 2µL of cDNA combined with the Master Mix, containing 25µL 2X Immomix, 2µL Syto-13 dye (50µM), 2.5µL upstream primer (10µM), 2.5 downstream primer (10µM), and 16µL of ultra pure water for a total volume of 50µL per reaction. Amplification conditions are 5 min at 95ºC, 40 cycles of 30 s at 95ºC, 30 s at 55ºC, and 90 sec at 72ºC, followed by 3 min at 72ºC.
Time Schedule
Collection of samples: already completed
Primer design: November 9-15, 2010
Reverse Transcription: November 9-15, 2010
qPCR: November 16-22, 2010
Analysis: November 23-29, 2010
Completion of project report: December 6, 2010
Presentations: December 6-10, 2010
Anticipated Results
There are several groups from the experimental portion of the project: half of the samples were exposed to a pesticide cocktail, and half were a control group. From those, a third were injected with a viral Poly I:C, a third with bacterial LPS, and a third were a control. From those, samples were collected at 24 and 48 hours. If there is no affect of pesticide exposure on the fish, we would expect the treatment-control group to have the same level of gene expression as the control-control group. If pesticide exposure does affect immune response, the treatment-treatment (either viral or bacterial) group will have different gene expression from the control-treatment group. If there is a difference between the sampling periods, there will be a difference in gene expression between the 24 and 48 hour sample groups.
Literature Cited
Baldwin, D.H., J.A. Spromberg, T.K. Collier, and N.L. Sholz. 2009. A fish of many scales: extrapolating sublethal pesticide exposures to the productivity of wild salmon populations. Ecological Applications 19(8):2004-2015.
Eder, K.J., M.A. Clifford, R.P. Hedrick, H.R. Koher, and I. Werner. 2008. Expression of immune-regulatory genes in juvenile Chinook salmon following exposure to pesticides and infectious hematopoietic necrosis virus (IHNV). Fish and Shellfish Immunology 25:508-516.
Engelhaupt, E. 2008. Real-world pesticide mixtures harm salmon. Environmental Science and Technology, p. 4619.
Gilliom, R., J. Barbash, D. Kolpin, and S. Larson. 1999. Testing water quality for pesticide pollution: US Geological Survey investigations reveal widespread contamination of the nation’s water resources. Environmental Science and Technology News, 164-169A.
Nehlsen, W., J.E. Williams, and J.A. Lichatowich. 1991. Pacific salmon at the crossroads: stocks at risk from California, Oregon, Idaho, and Washington. Fisheries 16:4-21.
11/2/10 – Lab 5: Quantitative PCR and Epigenetics (continued)
Summary: Measured HSP27 gene expression in sockeye salmon using quantitative PCR (qPCR). Developed the methylated cytosine dot blot from last week to measure DNA methylation of acid treated sockeye salmon using chromogenic immunodetection.
Purpose: By running cDNA, RNA, and water for a negative control in the qPCR, we are measuring the amount of gene expressed and checking the quality of the sample. We also measured DNA methylation using immunodetection methods.
Materials/Methods:
Western Breeze Chromogenic Immunodetection
1. Place the membrane in 10mL of Blocking Solution (prepared last week), and incubate on the shaker set at 1 revolution/second for 30 minutes.
2. Decant the blocking solution.
3. Rinse the membrane with 20mL of water for 5 min; decant; repeat.
4. Prepare 10mL of Primary Antibody Solution (1:5000 dilution)
a. 10mL Blocking Solution
b. 2µL 5-MeC antibody
c. 10mL total volume
5. Incubate the membrane with 10mL of Primary Antibody Solution for 1 hour.
6. Decant Primary Antibody and wash the membrane for 5 min with 20mL of TBS-T. Decant and repeat three more times.
7. Incubate membrane in 10mL of secondary antibody solution for 30 min. Decant.
8. Wash the membrane for 2 min with 20mL of TBS-T. Decant and repeat two more times.
9. Rinse the membrane with 20mL of water for 2 min, then decant. Repeat twice.
10. Incubate the membrane in 5mL of Chromogenic Substrate until the color begins to develop. This process is supposed to take somewhere between 1 to 60 minutes. A faint color started to appear on the dots after about 10 min, but I am unsure what the actual completed developing time was.
11. Rinse the membrane with 20mL of water for 2 min; decant; repeat twice.
12. Dry the membrane on a clean piece of filter paper.
Quantitative PCR
1. Rehydrate primers to a 100mM concentration: add 230µL of nuclease free water to the forward primer, and 364µL of nuclease free water to the reverse primer. Label the tubes 100µM.
2. Create 100µL of solution by adding 10µL of primer to 90µL of nuclease free water and spin. Labeled tubes BB, HSP27-F, 10µM; BB, HSP27-R, 10µM.
3. Prepare a master mix for 7 reactions:
| Component |
Volume/Rxn |
Final Conc. |
Volume Added |
| Master Mix, 2X (Immomix) |
25µL |
1x |
175µL |
| Syto-13 dye (50µM) |
2µL |
2µM |
14µL |
| Upstream primer, 10µM |
2.5µL |
2.5µM |
17.5µL |
| Downstream primer, 10µM |
2.5µL |
2.5µM |
17.5µL |
| Ultra Pure Water |
16µL |
NA |
112µL |
5. Thaw cDNA samples.
6. Add cDNA, RNA, and water to the appropriate wells:
| Label |
Solution |
Well in qPCR |
| 1 |
cDNA |
B12 |
| 2 |
cDNA |
B11 |
| 3 |
RNA |
B10 |
| 4 |
RNA |
B9 |
| 5 |
Water |
B8 |
| 6 |
Water |
B7 |
8. Spin the strips to collect the volume in the bottom of the wells.
9. Load plate (as indicated in above table) and start the run (done by a lab research scientist).
Results:
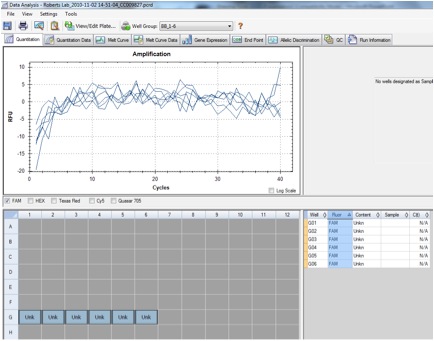
Figure 2. qPCR showing the amplification of the HSP27 gene in sockeye salmon.
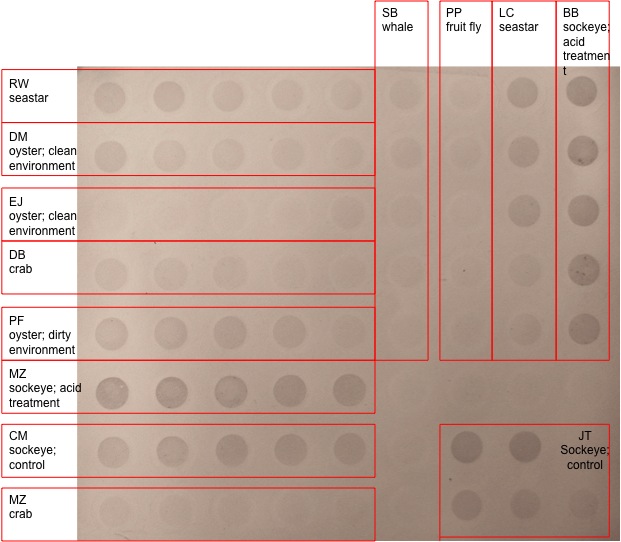
Conclusion: The dot blot visualizes the amount of methylated cytosines by comparing band densities in different lanes. These results are more qualitative, and are interpreted by comparing the dot density or darkness of band. My samples included acid-treated sockeye, and showed strong band representation with very little color attenuation between the top and bottom lanes following the increase in dilution. The other set of dark bands in the dot blot were also from acid-treated sockeye salmon, which were slightly lighter than my sample. Other sockeye salmon samples showed band coloration that wasn't as strong as the acid-treated samples, but were stronger than most other species sampled. Only one sea star sample and one dirty oyster sample were comparable in coloration to the sockeye control samples. The whale, fruit fly, (the second) sea star, crab, and (the second) oyster samples showed little to no methylation. It is interesting that the bands between the acid-treated sockeye samples were similar, yet slightly darker than the control sockeye samples indicating that the stressor may be responsible for the difference in methylation. The difference in band color between my sample and the other acid-treated sample may be negligible and explained by variations in sample treatment or individual differences.
The qPCR results differed greatly from the anticipated results. A graph with good gene expression would have three pairs of lines: if the gene had successfully amplified through the use of the primer, the two lines representing the cDNA samples would have an exponential line; the water samples would appear as straight lines at 0; the RNA samples would ideally have no representation, but may have some contamination indicated by a slight increase in amplification. My sample, however, had no amplification. This could be due to several potential reasons. There may have been no gene expression; my sample was the control sockeye sample with no stressor, which may result in no amplification of heat shock protein, and would serve as a good comparison with treated samples. There may have been a problem with the primers, if the solution didn't rehydrate correctly or if the coding region wasn't long enough. A third possibility is an error in protocol that would contaminate the samples or prevent them from amplifying. These results could be confirmed or corrected by optimizing the qPCR machine to the correct melting temperature or by using a new set of primers.
Reflections: The purpose of this lab was to measure the amount of cytosine methylation in an acid-treated sockeye sample and to measure the amount of gene expression of the HSP27 gene in a control sockeye sample. The effect of stressors on organisms can be measured by both of these methods. I wish there was more information on how to interpret the dot blot results. For example, I would like to know if there is more than just qualifying the results based on the darkness of color. I think Caroline mentioned using a computer program that quantifies the data in the dots, but I'm not positive. I was frustrated with this lab because the qPCR didn't work for my sample. I don't know if it is due to operator error (which I hope isn't the case!) or if there was a problem with my primers, which seems more likely, or if the machine just needs to be optimized. If there was time, I would be interested in finding out the reason behind my failed qPCR. It would also be interesting if we had the information to compare the qPCR results between different samples.
10/26/10 – Lab 4: Epigenetics
Summary: Ran agarose gel to visualize last weeks PCR results of the HSC71 gene in sockeye salmon using an ethidium bromide (EtBr) treatment. Began steps to measure cytosine methylation on acid-treated sockeye salmon. We used a 5-MeC antibody to detect methylated cytosines followed by chromogenic immunodetection.
Purpose: Last weeks PCR is visualized on agarose gel to check for contamination and presence of cDNA in the samples. Measure cytosine methylation using chromogenic immunodetection methods.
Materials/Methods:
Agarose Gel Electrophoresis
1. Place gel in the gel box and fill with 1x TAE buffer to fully cover the wells. Remove combs from wells.
2. Load 5µL 100 bp ladder in the first lane.
3. Load 25µL of the PCR sample into the gel (keep the remainder at -20ºC). Gel 1, bottom half, Rows 2-5.
4. Ran gel at 100V for 50 min (then 150V for 9 min, and 85V for 7 min).
5. Visualize the gel on the UV transilluminator. Initial visualization indicated we needed to run the gel for longer than 50 min, so we continued at 150V for 9 min. When the gel got too hot, we lowered the voltage to 85V for 7 min.
Methylated Cytosine Dot Blot Procedure
DNA Dilutions
1. Sockeye Acid 1
2. Label 5 snap cap 1.5 mL tubes: DNA BB .8, DNA BB .4, DNA BB .2, DNA BB .1, DNA BB .05.
3. Prepare five dilutions for a total volume of 200µL
| Dilution |
Target Concentration |
µL of H2O |
µL of 20X SSC |
µL of 50ng/µL of DNA Sample |
| 1 |
0.8 ng/µL |
124 |
60 |
16 |
| 2 |
0.4 ng/µL |
132 |
60 |
8 |
| 3 |
0.2 ng/µL |
136 |
60 |
4 |
| 4 |
0.1 ng/µL |
138 |
60 |
2 |
| 5 |
0.05 ng/µL |
139 |
60 |
1 |
Dot Blotting
1. Cut nylon membrane to fit 72 wells of manifold, and soak in 6X SSC for 10 min.
2. Cut filter paper to the same size and get wet in 6X SSC.
3. Assemble manifold with membrane on top of the filter paper.
4. Denature DNA in boiling water for 10 min. During the denature process, some caps popped off the tubes. For future procedures, use screw caps when denaturing in boiling water.
5. Immediately transfer DNA samples from boiling water to ice.
6. Apply 500µL of 6X SSC to each well and allow SSC to filter through with the vacuum. In wells 9A, 9B, 9C, 9D, and 9E.
7. Spin DNA for 5 sec.
8. Apply the entire 200µL of DNA to the wells and allow samples to filter through. Put sample 1 in 9A, 2 in 9B, 3 in 9C, 4 in 9D, and 5 in 9E. Some samples didn’t filter through. To alleviate the clog, pipetting up and down a couple times mixed the samples.
9. Soak filter paper cut to size in denaturation buffer.
10. Dismantle manifold and transfer membrane to filter paper soaked in denaturation buffer and let sit for 10 min.
11. Allow neutralizing for 5 min.
12. Place membranes on dry filter paper to dry.
13. Wrap dried blot in plastic wrap and place DNA side down on UV transluminator for 2 min at 120kJ to immobilize DNA.
WesternBreeze Chromogenic Immunodetection
1. Prepare 20mL of Blocking Solution with:
a. 14mL of ultra-filtered water
b. 4mL of Blocker/Diluent Part A
c. 2mL of Blocker/Diluent Part B
2. Due to time constraints, we stopped here and left the remaining procedure for next week.
Results:
Table 1. Information on lane use for Gels 1 and 2.
| Gel 1 |
Gel 2 |
|||
| Lane |
Top |
Bottom |
Top |
Bottom |
| 1 |
Ladder |
Ladder |
Ladder |
Ladder |
| 2 |
Chris |
Brianna |
Dave |
David |
| 3 |
Chris |
Brianna |
Dave |
David |
| 4 |
Chris |
Brianna |
Dave |
David |
| 5 |
Chris |
Brianna |
Dave |
David |
| 6 |
Peggy |
Ross |
Sammi |
Lauren |
| 7 |
Peggy |
Ross |
Sammi |
Lauren |
| 8 |
Peggy |
Ross |
Sammi |
Lauren |
| 9 |
Peggy |
Ross |
Sammi |
Lauren |
| 10 |
Paul |
Jason |
Erica |
Josh |
| 11 |
Paul |
Jason |
Erica |
Josh |
| 12 |
Paul |
Jason |
Erica |
Josh |
| 13 |
Paul |
Jason |
Erica |
Josh |
| 14 |
Model |
- |
- |
- |
| 15 |
Model |
- |
- |
- |
| 16 |
Model |
- |
- |
- |
| 17 |
Model |
- |
- |
- |
| 18 |
- |
- |
- |
- |
| 19 |
- |
- |
- |
- |
| 20 |
- |
- |
- |
- |
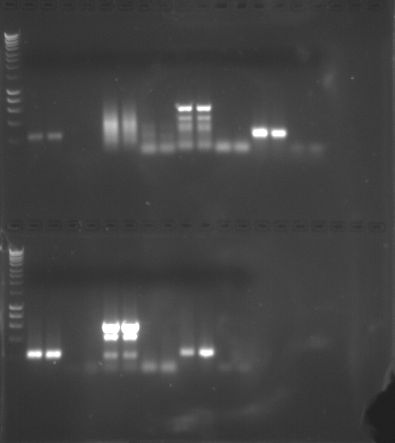
Figure 1. PCR Gel 1
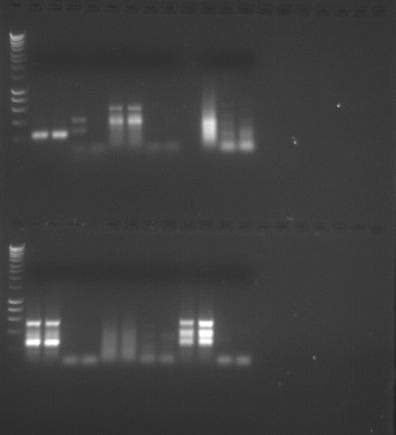
Figure 2. PCR Gel 2.
The cytosine methylation dot blot lab is not yet complete and therefore will not have results until next week.
Conclusion: We performed gel electrophoresis, which separates the amplified cDNA based on size, where the smaller strands move faster and further than the longer strands. The first two columns of the sample represent the amplified cDNA and the second column is the negative control. If there is a band in the first two columns, the PCR was considered a success. However, if there were also bands in the second two columns, the sample may be contaminated and the amplification may not be of the intended gene. My PCR amplification was successful based on the presence of two clear bands in the first two columns of my sample and the absence of bands in the second two columns. Based on these results, Quantitative PCR (QPCR) can be performed.
Reflection: The purpose of the lab was to visualize our PCR results on agarose gel to determine if PCR amplification was successful. We also began to measure cytosine methylation using dot blot and chromogenic immunodetection methods, but due to time constraints, have to finish up the process next week. This process is used to study heritable changes in phenotype or gene expression. We haven’t yet finished the lab procedure and will learn how to interpret the results next week.
10/19/10 – Lab 3: Reverse Transcription and End-Point PCR
Summary: reverse transcribed RNA into cDNA (complimentary DNA) using DNA polymerase; made an agarose gel; amplified HSC71 in sockeye salmon using end-point Polymerase Chain Reaction (PCR).
Purpose: RNA is reverse transcribed into cDNA to create a strand stable enough to withstand PCR. The cDNA is used in PCR to amplify the HSC71 gene. The agarose gel was prepared to later visualize the DNA in the next lab.
Materials/Methods:
Reverse Transcription
1. Mix stock RNA by inverting tube.
2. Label a 0.5 mL PCR tube cDNA BB
3. Combine the following into the 0.5 mL PCR tube:
a. 5µL of sockeye RNA
b. 1µL of oligo dT
c. 4µL of nuclease free H2O
4. Briefly centrifuge. Incubate the mixture for 5 min @ 70ºC on the thermocycler and immediately transfer to ice.
5. Add the following:
a. 5µL of M-MLV 5X Reaction Buffer
b. 5µL of dNTPs
c. 1µL of M-MLV RT
d. 4µL of nuclease free H2O
6. Incubate the mixture for 60 min @ 42ºC, then heat inactivate for 3 min @ 70ºC on the thermocycler.
7. Centrifuge, and store on ice.
Polymerase Chain Reaction
1. Label (4) 0.5mL PCR tubes: 1 BB, 2BB, 3BB, and 4BB on the caps and sides of the tubes.
2. Make Master Mix in a 1.5mL tube, label MM BB, and add:
a. 250µL of GoTaq Green Master Mix, 2X
b. 15µL of forward primer, 10µm
c. 15µL of reverse primer, 10µm
d. 108µL of nuclease free H2O
*Deviation from lab: I accidentally added too much reverse OR forward primer, so I had 30µL of one and only 15µL of the other. Because of this mistake, I later used Model’s Master Mix, who was also amplifying the HSC71 gene.
3. Pipette 48µL of Master Mix into each of the four labeled tubes.
4. Add 2µL of cDNA into tubes 1 and 2; add 2µL of nuclease-free H2O into tubes 3 and 4.
5. Centrifuge to pool liquid at the bottom of the tubes, and load samples into the thermocycler. The samples will go through the following thermal cycling profile and stored at 20ºC afterwards:
Step: Temperature: Time: Cycles
Denaturation: 95ºC: 5 min: 1
Denaturation: 95ºC: 30 sec
Annealing: 55ºC: 30 sec: 40
Extension: 72ºC: 90 sec
Final Extension: 72ºC: 3 min: 1
Hold: 4ºC: infinite: 1
Agarose Gel
1. Weigh 2g of agarose and mix with 150mL 1x TAE in a 1L flask.
2. Microwave solution for ~3 min. Watch sample while in the microwave to make sure the sample doesn’t boil over. If boiling starts to occur, take sample out and swirl while checking for clarity of solution.
3. Cool solution and add 12µL of ethidium bromide (EtBr). Warning: EtBr is a carcinogen, so use gloves and dispose of tips in appropriate recepticle.
4. Mix thoroughly by swirling, and pour into gel tray.
5. Add gel combs.
6. After gel is set, wrap in plastic wrap and place gel in the fridge for storage until next week.
Results/Conclusion: The cDNA was used in PCR and the full results will not be known until it can be visualized on the agarose gels made in lab. Results from this weeks lab will be determined in next weeks lab.
Reflection: The purpose of this lab was to use primers to amplify a gene of interest and create cDNA to run in PCR. The results will later be visualized on an agarose gel. These studies can be used to study gene expression and determine how gene expression may change with varied environmental conditions.
10/12/10 - Lab 2: RNA Extraction and Protein Analysis, Part 2
Summary: Separated proteins from each other them during Polyacrylamide Gel Electrophoresis and visualized using Coomassie Brillian Blue stain. RNA is extracted using TriReagent and is quantified using a spectrophotometer to measure absorbance the absorbance of the RNA sample at 260nm. -
Purpose: to complete the RNA extraction, quantify the amount of RNA extracted from the tissue sample, and separate proteins based on molecular weight in gel electrophoresis.
Materials/Methods:
SDS-Page
1. Boil water on a hot plate and thaw protein extract from last week. Mix well be inverting tube several times.
2. In a fresh 1.5mL screw-cap tube, add 15µL of your protein stock and 15µL of 2X Reducing sample buffer. Return protein stock to the box in the -20ºC.
3. Mix sample by flicking and briefly centrifuge (10s) to pool liquid in the bottom of the tube.
4. Boil sample for 5 min.
5. Observe assembly of gel box and gels. Rinse the gel wells as demonstrated.
6. When sample is finished boiling, centrifuge for 1min to pool liquid.
7. Slowly load the entire sample into the appropriate well using a gel-loading tip.
8. Pug lid on gel box and plug electrodes into appropriate receptacles on the power supply. Turn the power on and set voltage to 150V. Run for 45 min.
9. Add ~150mL (just enough to cover the gel) of Coomassie Stain to a designated container.
10. Turn off power supply and disconnect gel box from power supply.
11. Remove the lid from the gel box. Disengage the tension wedge. Remove gel from gel box. Carefully crack open cassette to expose get. Trim wells at the top of the gel.
12. Notch a designated corner of the gel to remember the correct orientation.
13. Place gel into container with Coomassie Stain and incubate in the shaker/rocker for 5 min.
14. Carefull pour stain back into the original container.
15. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
16. Add ~250ML 10% acetic acid to container with gel. Incubate on shaker/rock for 15min.
17. Change out buffer and repeat until bands become clearly visible. This may need to incubate overnight (if so, cover container with plastic wrap and leave on shaker/rocker).
18. Take a picture of the gel.
RNA Extraction, Part 2
1. Turn on heating block to 55ºC.
2. Incubate the homogenized tissue sample tube at room temperature (RT) for 5 min.
3. In the fume hood, add 200µL of chloroform to the sample and close the tube.
4. Vortex for 30s. Solution should become a milky emulsion.
5. Incubate at RT for 5 min. My sample incubated for approximately 20 min while I waited for a free microfuge.
6. Spin tube in refrigerated microfuge for 15 min @ max speed.
7. Gently remove tube from microfuge and be careful not to disturb the tube. Slowly and carefully transfer most of the aqueous phase (the top clear layer) to a fresh microfuge tube. DO NOT transfer any of the interphase (the white, cell debris between the aqueous and organic phase).
8. Add 500µL isopropanol to the new tube containing our RNA and close the tube.
9. Mix by inverting several times until the solution appears uniform.
10. Incubate at RT for 10 min.
11. Spin in refrigerated microfuge @ max speed for 8 min. When positioning tube in microfuge, place the tube hinge pointing up, away from the center of the microfuge.
12. A small, white pellet (RNA and salts) should be present. Small white pellet was visible.
13. Remove supernatant.
14. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. Pellet didn’t dislodge.
15. Spin in refrigerated microfuge at 7500g for 5 min.
16. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet.
17. Briefly spin tube (~15s) to pool residual EtOH.
18. Using a small pipette tip, remove remaining EtOH.
19. Leave tube open and allow pellet to dry at RT for no more than 5 min.
20. Resuspend pellet in 100µL of 0.1%DEPC-H20 by pipetting up and down until pellet is dissolved.
21. Incubate tube at 55ºC for 5 min to help solubilize RNA. Remove tube from heat, flick a few times to mix and place sample on ice. This will be the stock RNA sample.
RNA Quantification
1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm.
2. Click “Blank”, to zero the instrument.
3. Pipette 2µL of RNA sample onto the Nanodrop pedestal and lower the arm. Click “Measure” and record the A260 absorbance, RNA concentration (ng/µL), A260/280 ratio, and A260/230 ration.
4. Raise the arm and wipe off the sample with a KimWipe.
5. Clearly label RNA stock sample: RNA, sockeye, BB, 10/12
6. Store samples at -80ºC.
Results:
Table 1. List of sample types and lanes in gel electrophoresis.
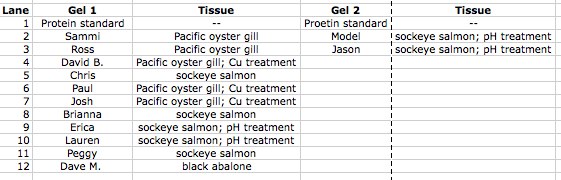
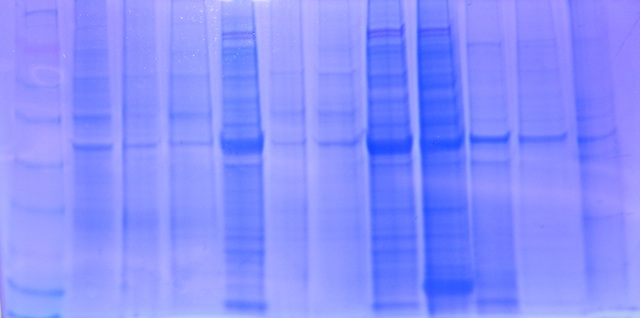
Figure 1. Gel 1 starting with Lane 1 on the left. My sample was in Lane 8

Figure 2. Gel 2 starting with Lane 1 on the left.
RNA Quantification:
A260: 278.8 ng/µL
A260/A280: 1.84
A260/A230: 1.85
Conclusion: The gel electrophoresis separated protein based on molecular weight. My sample for sockeye salmon with no treatment was in Lane 8. A similar sockeye sample was in Lane 5, and the two samples seemed very similar in protein band location and protein band widths. The sample in Lane 9 was a sockeye salmon with pH treatment. The sample appears to be extremely similar except for one main difference: the treatment sample has a thick band near the bottom that is not shown in the sockeye sample without treatment. Good observations! -
The RNA sample for sockeye had an absorbance of 278.8 ng/µL indicating RNA was present and successfully extracted. The ratio for A260/A280 was 1.84, between the acceptable range between 1.8-2.0 for a clean sample. A ratio of 2.0 is generally accepted as pure for RNA, therefore, since my sample is lower than the 2.0, it may indicate the presence of protein, phenol, or other contaminants, but is still within the approved range so it isn’t too contaminated. The ratio for A260/A230 was 1.85, between the acceptable range between 1.5-2.0. This shows the sample was not contaminated by co-purified contaminants. Based on these results, the sample is ok for further lab work, including QPCR and Reverse Transcription.
Reflection: The purpose of this lab was to familiarize us with the processes, complete the extraction of RNA for future procedures, and visualize the results of last weeks lab through gel electrophoresis. The gel provided a means to compare protein presence between samples with different treatments. I wish there was more information on the different ways a gel could be used in studies. I will find some examples. -
10/5/10 - Lab 1: RNA Extraction and Protein Analysis, Part 1
Summary: isolated RNA from juvenile sockeye tissue using TriReagent; protein was extracted from juvenile sockeye tissue using CelLytic MT; the concentration of protein was quantified using the Bradford Assay.
Purpose: to gain experience with RNA and protein extraction methods and quantify the amount of protein that was extracted from the tissue sample.
Materials/Methods:
RNA Extraction, Part 1
1. Received juvenile sockeye control Sample D (19mg). Store on ice when not in use.
2. Label tube: BB 10/5
3. Add 500µL of TriReagent to the 1.5mL snap cap tube with tissue sample.
4. Homogenize tissue with a disposable pestle. Sample was difficult to homogenize, so it was necessary to vortex a few times.
5. Add an additional 500µL of TriReagent to the tube.
6. Vortex for 15s.
7. Store sample at -80ºC.
Protein Extraction
1. Received juvenile sockeye control Sample D (23mg).
2. Label tube: BB 10/5
3. Add 500µL of CelLytic MT to the 1.5mL snap cap tube with tissue sample.
4. Homogenize tissue with disposable pestle. Proteins damage more easily, so did not vortex during homogenization.
5. Invert tube several times.
6. Spin in a refrigerated microfuge for 10min at max speed.
7. Label new 1.5mL snap cap tube: Protein, sockeye, BB, 10/5
8. Transfer supernatant to labeled tube and store on ice.
Protein Quantification
1. Label 2mL tube: Protein, BA, BB, 10/5
2. Dilute an aliquot of the sample by adding 15µL of protein and 15µL DI water into the 2mL tube. Mix by pipetting.
3. Label 2mL tube: Blank. Add 30µL DI water.
4. Add 1.5mL of Bradford Reagent to both tubes.
5. Mix by inverting several times and incubate at room temperature for 10min.
6. Mix Blank by pipetting and add 1000µL to a cuvette.
7. Zero the spectrophotometer using the blank sample. Wipe the cuvette with a KimWipe to remove any fingerprints.
8. Mix the ‘sample’ by pipetting and add 1000µL to a new cuvette.
9. Measure the absorbance at 595nm and record the value: 0.205
10. Remove the cuvette and mix by pipetting. Measure the absorbance: 0.205
11. Average the two values: 0.205
12. Back-calculate the protein concentration by using the standard curve.
13. Store protein sample at -20ºC
Results: RNA sample stored until extraction process can be completed. Protein was successfully extracted and stored. The concentration of protein was measured using the Bradford Assay, and back-calculated using the standard curve, with the equation
y = 1013.9x.
y = 1013.9 (0.205) = 207.8495µg/mL
Amount corrected to account for the dilution
2y = 415.699µg/mL
Conclusion/Reflection: Without studies for comparison, it is difficult to say if the results I received were expected; however, the presence of results while quantifying protein indicates there were protein and the extraction was therefore a success.
The purpose of this lab was to familiarize us with the extraction techniques for proteins and RNA. We will use these techniques later in our research project as we examine the effect of external environmental factors on an organism’s physiology. The protein will be separated using gel electrophoresis and later analyzed using the Western Blot. The RNA we extracted will be used in PCR and reverse transcription.
To familiarize myself with NCBI for later use in primer design, I used NCBI to choose the following genes for study:
Heat shock protein (Accession#: FJ226381) Good job. This is an mRNA sequence so it will be good for designing primers to measure gene expression using qPCR. The sequence is short, but you should be able to design primers off of it. -
Type-1 growth hormone gene (Accession#: U14551) While an interesting gene this is the entire gene sequence (not just mRNA or cDNA) which contains both the introns and exons (the expressed regions transcribed into RNA). We will only be able amplify exonic (since it is RNA) regions using qPCR so primers designed off of this sequence might not work because we do no where the exons are. For this class, I would stick to one of the other genes because there is a better chance that the primers will work. -
Gonadotropin-releasing hormone (GnRH), (Accession#: D31868) Good. -