TUES 5 OCT 2010
LAB 1RNA extraction and protein analysis, part 1
Tissue sample: tail clips from juvenile Sockeye salmon A control
RNA isolation protocol
Sample weight 14mg (huge amount of tissue - whole tail severed prior to caudal peduncle)
add 500ul TriReagent, homogenize with disposable pestel
add additional 500ul TriReagent -> vortex -> -20C --> -80C
Results: RNA isolation to continue next week
Conclusions: Next week
Reflection: Would rather use Qiagen RNeasy mini kit ha! $$$ -
continued:
See lab 2 (TUES 12 OCT 2010)
Protein Extraction protocol
Sample weight 19mg (huge amount of tissue - whole tail severed prior to caudal peduncle)
add 500ul CellLytic MT, homogenize with disposable pestel
spin at top speed for 10min at 4C
transfer supernatant to clean tube, dispose of pellet
supernatant --> ice, remove 15ul for quantification, rest --> -20C
Results: see protein quantification
Protein Quantification (Simple) Bradford Assay
1:2 dilution of sample in CellLytic MT supernatant in DI water
15ul sample + 15 ul DI water
additional 30ul DI water in separate tube
add 1.5ml Bradford reagent (coomassie blue in EtOH (with acid)) [sigma? Yes -
invert several times, incubate at RT for 10min
spec with [spectrophotometer name/brand] at A595nm
fit to standard curve generated by [????] TA-
Results - protein quantification
A595nm-1 0.204
A595nm-2 0.202
average = 0.203
standard curve: y=mx+b
y=1013.9x
R^2=0.97738
y=1013.9(0.203)
y=205.82ug/ml
*2 dilution factor = 411.6ug/ml or 411.6ng/ul
Conclusions: can use protein sample for enzyme assays, westerns, protein-protein/DNA/RNA interactions... etc. To be determined by future labs.
Notes: In the past I have used Qiagen RNeasy mini kit for RNA extraction. No phenol or chloroform.expensive but quick and easy.
In a true Bradford assay a standard curve should be created each time to account for the changes in pH of the water, temp and air pressure day-to-day. True -
Genes of interest
Interesting genes, however you have provided protein sequences which cannot directly be used to design primers for qPCR gene expression assays. We will only be able amplify exonic (DNA transcribed into RNA) so you need to find nucleotide cDNA, mRNA, or exonic sequences for your genes of interest -
I was blasting the protein sequences between Danio rerio and O.nerka since protein tends to be conserved even when nucleotide sequences are not. There is not a lot of sequence information on O.nerka, but blasting back to zebrafish gives me an idea if the sequences actually code for a homologous gene.
NADH dehydrogenase (subunit 1)- accession number: YP_908764
COX1-subunit 1 - YP_908766.1
UPR genes: XBP1, S1P/S2P, ATF4
ATF4 is preferentially translated by eIF2a after phosphorylation by PERK. XBP1 is spliced upon dimerization and autophosphorylation of IRE1a, the splice form is a potent transcription factor that translocates to the nucleus and activates the translation of other UPR genes. And S1P and S2P are upregulated to cleave ATF6 which then upregulates target UPR genes. By looking at the expression of the genes ATF4, XBP1s and S1P, you can understand if protein folding in the ER is not functioning properly. The UPR is a mechanism that the cell uses to reduce the load of unfolded or misfolded proteins. Chronic upregulation of the UPR is a sign of ER Stress and can lead to apoptosis.
XBP1 - accession number: can't find it for sockeye but for Atlantic salmon: BT045205, Danio rerio: NM_131874, O. mykiss: EX764677 --> use for primer design
ATF4 - accession number: can't find for sockeye salmon, Danio rerio: BC067714
Site-1 Protease (S1P) - accession number: can't find for sockeye salmon, Danio rerio (mbtps1):NM_199213.1
Danio rerio sequences for these genes do not align significantly with anything in sockeye salmon.
Primer design, O.mykiss xbp1
LEFT PRIMER 870 20 60.00 50.00 3.00 2.00 tgtcaccacgaccatcttgt RIGHT PRIMER 1024 20 60.04 50.00 4.00 0.00 tagcgagcggtactcgtttt product size: 154or
LEFT PRIMER 574 20 59.99 55.00 4.00 1.00 gcgagaacttggtcttccag RIGHT PRIMER 767 20 60.19 45.00 4.00 0.00 tcgaaccaagccaattcttc PRODUCT SIZE: 194I will have to do more work to fish a good primer set for the spliced version of xbp1 (xbp1s) as the ratio of the two is the indicator of upregulated UPR.
TUES 12 OCT 2010
LAB 2SDS-Page
15ul protein from quantified stock - 411.6ng/ul = 6.174ug protein + 15 ul 2X reducing sample buffer
centrifuge 10s
boil sample for 5mins
(set up gel box while boiling)
short spin to pool liquid
load entire sample (30ul) to well#5 of premade 8-20% gradient gel
run at 150v ~45mins
open and mark gel for directionality
stain with coomassie on rocker for 5 mins
2x 15 min rinse with 10% acetic acid
incubate in 10% acetic acid until bands visible
Results
ladder ..............................................CHRIS
conclusions
I have significant bands, although I don't have the ladder information, nor do I know how to edit and upload images with this wiki. I got lucky with this one after 10 or so tries.
RNA isolation continued (from LAB 1 TUES 5 OCT)
thaw homogenized sample (trireagent)
add 200ul chloroform -> vortex
incubate at RT for 5 mins
spin at top speed for 15 mins
transfer top layer to new tube, dispose of organic phase and interphase
add 500ul isopropanol to precipitate RNA -> invert several times
incubate at RT for 10 mins
spin at top speed for 8 mins
remove supernatant
add 1ml 75% EtOH, invert to dislodge pellet
spin at 7500g for 5 mins
remove EtOH -> brief spin -> remove remaining EtOH
dry on benchtop (covered with kimwipe) or could have used hood
resuspend in 100ul 0.1%DEPC water -> pipette to dissolve pellet
incubate at 55C for 5 mins
quantify using nanodrop @ A230
Results
sample: 117.5ng/ul
conslusions
Less total RNA than I expected give tissue volume, however, homogenization was difficult as pestel did not quite fit tube being used.
Was your protein sample similar to any others? How was it different? -
Reflections? -
TUES 19 OCT 2010
LAB 3PCR
Reverse transcription
Thaw RNA
in PCR tube mix:
5ul template RNA
1ul oligo dT
4ul RNase free water
incubate at 70C for 5 mins
spin down
add:
5ul M-MLV 5x reaction buffer
5ul dNTPs
1ul M-MLV RT reverse transcriptase
4ul RNase free water
incubate at 42C 60mins
heat inactivate at 70C for 3 mins
put at -20C
PCR
Master mix:
250ul GoTaq
15ul primer F (HSC71)
15ul primer R (HSC71)
108ul DNase/RNase free water
add 48ul master mix to each PCR tube (4)
add 2ul cDNA or water to tubes
1. cDNA
2. cDNA
3. neg. control H2O
4. neg. control H2O
(total 50ul rxn x 4)
gave remaining master mix to Dave M.
PCR at 55C
1. 95C 5'
2. 95C 30"
3. 55C 30"
4. 72C 1.5'
5. repeat at step 2, 40x
6. 72C 10'
7. 4C forever. (could be 14C as PCR product is very stable)
Agarose Gel
1.3%
2g Agarose
150ml 1x TAE
bring to boil
add 12ul EtBr
results: we'll see next time. I have run PCR reactions maybe 800 times over the last 4 years. All conclusions are dependent on the gel image and based on image I can optimize reaction.
Previously I used Invitrogen or Denville taq which come with Buffer w/o MgCl2. Varying the amount of MgCl2 can greatly affect PCR reactions. MgCl2 is necessary for the taq enzyme to activate but too much can cause excess primer dimerization. This can be aided by the addition of DMSO. dNTP's are also subject to degradation with repeated freeze/thaws and in the past I was very careful about the use dNTP aliquots. taq is also easily degraded at RT and should be kept on ice at least, although pre-made master mix taq's tend to be more stable at RT. If there are any problems with this PCR reaction, my first troubleshooting step would be to vary the primer amounts, the dNTP amounts, MgCl2 concentration, and the template DNA (I would add a standard concentration rather than a standard volume).
will load gel for Lab 4 - OCT 26
TUES 26 OCT 2010
Epigenetics and gel electrophoresisGel Loading
1.3% gel from 19 October
Load ladder (hyperladder 1) 5ul into well 1 of top row of gel 1
Load 25ul PCR product into wells 2-5
wells 2 - 3 : sample with HSC71 primers
wells 4 - 5 : negative control, no cDNA.
Ran at 100V for about an hour, increased to ~150V for 20 mins and back down to 85V.
Imaged in Young Lab
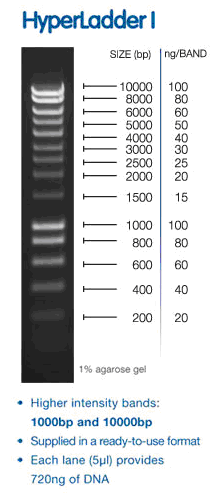
bands in sample lanes ~ 220bp although I need ladder information. We used hyperladder I (I think) (and i figured out how to steal pictures from other websites).
I don't have primer sequences yet or the expected product size.
Methylated Cytosine Dot Blot
Immunostaining methylated cytosinein DNA sample
DNA: Sockeye Salmon control I
Dilute DNA to:
1. 0.8ng/ul
2. 0.4ng/ul
3. 0.2ng/ul
4. 0.1ng/ul
5. 0.05ng/ul in 200ul 6x SSC (saline sodium citrate)
boil DNA dilutions for 10 min to denature DNA strands, ice immediately following (prevents DNA from re-annealing)
soak nylon membrane (cat#?) in 6x SSC
soak filter paper in 6x SSC
soak another piece of filter paper in denaturation buffer
soak another piece of filter paper in neutralization buffer
set up manifold with nylon membrane on top of 6x SSC soaked filter paper
apply 500ul 6x SSC to wells 1-5 of row F
vacuum pull through membrane
load entire DNA samples (200ul) to each corresponding well, 1-5.
apply vacuum and pull through DNA sample
remove membrane and transfer to denaturation buffer soaked filter paper - 10 min
remove membrane and transfer to neutralization buffer soaked filter paper - 5 min
dry in hood on dry filter paper
once dry, wrap blot in plastic wrap and UV transluminate for 2 min at 120kJ (DNA side down)
this binds the DNA to the membrane
Results: stopped here.....the rest of the protocol would take 2.5 hours, no more class time.
Next lab, 2 NOV 2010 - block and antibody stain.
Conclusion: No problems so far, just slow going with so many students working the same blot.
TUES 2 NOV 2010
qPCR and Epigenetics (continued)Epigenetics continued
continued with dried membrane from 26 Oct.
blocking solution (western breeze kit):
14ml H20
4ml Blocker/dilutent A
2ml Blocker/dilutent B
total 20ml
Primary antibody solution
10ml Blocking solution
2ul 5-MeC antibody
total 10ml
place membrane in blocking solution
incubate for 30min on rocker at RT
Rinse with H2O 2x 5min
Incubate with Primary antibody solution for 1 hour at RT
Rinse 3x 5min with TBST
Incubate with 10 ml Secondary antibody solution for 30min *supplied western breeze kit
Rinse 3x 2min with TBST
Rinse 3x 2 min with H2O
Incubate in Chromogenic Substrate until dots visible
Dry membrane and image
Loaded 1 2 3 4 5
0.8ng/ul 0.4ng/ul 0.2ng/ul 0.1ng/ul 0.05ng/ul
no difference in intensity between samples, likely saturated at the lowest concentration.
no discernible difference (by eye) between intensity of my (sockeye controls) and either acid treatment. would need to measure with image j or like program to quantify blots and statistically compare them.
While this will give us a general view of methylation of the genomic DNA, it gives no indication of the specific methylation pattern. Another way to get this answer (a little quicker) is a restriction digest with HpaII and MspI. This allows for a control (5'-AZA) if we had live samples. Another, more specific, methylation assay is bisulfite sequencing, if we had specific target genes.
cDNA samples from previous experiment (Sockeye control A)
50ul rxn
1x Master Mix 7x
25ul 2x Immomix 175ul
2ul Syto-13 dye (50uM) 14ul
2.5ul 10uM primer F 17.5ul
2.5ul 10uM primer R 17.5ul
16ul nanopure H2O 112ul
2ul Sample 2ul each tube
Add 48ul mastermix to each (6) tube
add 2ul cDNA to tubes 1-2
add 2ul RNA to tubes 3-4 (genomic. control)
add 2ul H2O to tubes 5-6 (neg. control)
loaded 6-1 in lanes 7-12 row G
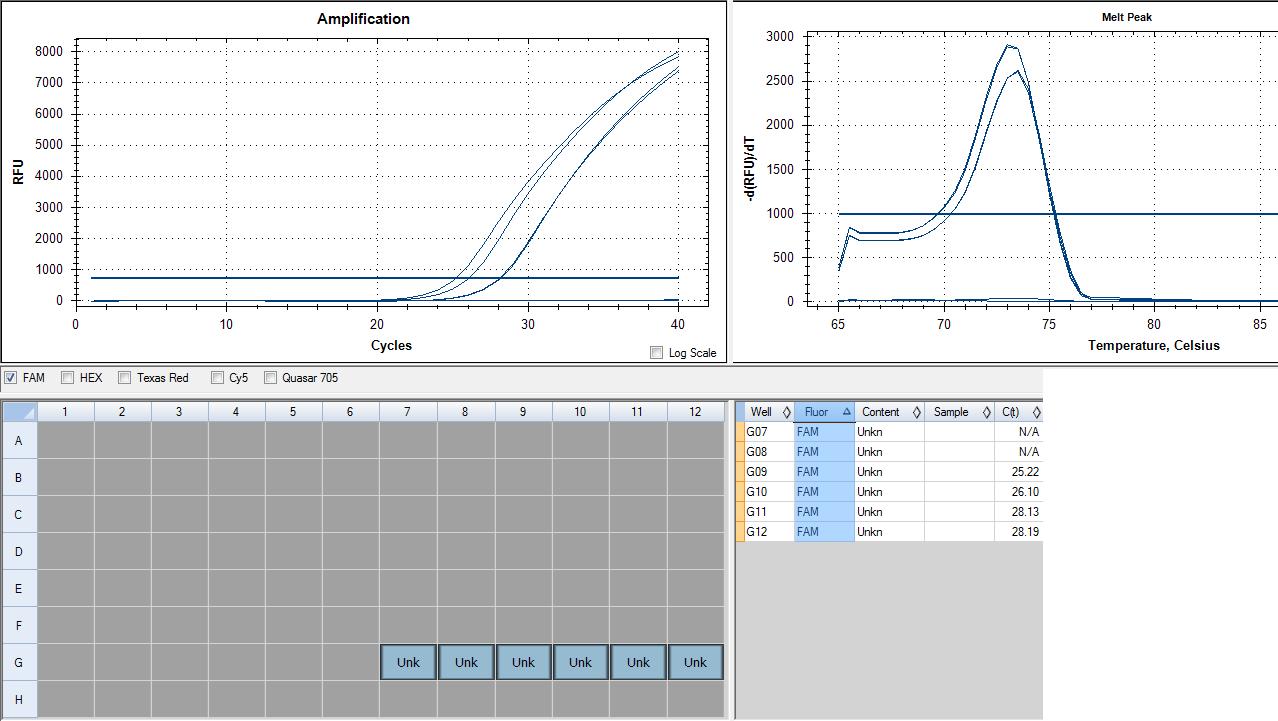
Genomic contamination in RNA samples, but interesting because it peaks at an earlier cycle number. This could mean a higher concentration of genomic DNA contamination in my RNA than there is cDNA in my cDNA sample. MY cDNA sample peaks are together, indicating good samples, however 28 cycles indicates fairly low expression. xbp1 is a transcription factor involved in the unfolded protein response and the immune response, so it shouldn't be highly expressed in a non-stressed sample such as mine (sockeye control A). In the future, DNase treatment will be used to ensure no genomic contamination.
TUES 9 NOV 2010
Salmon Pesticide Project
Picked several primer sets to look at genes related to the immune response (IL6 - in response to LPS injection) and to the stress response (CRH, SOD and xbp1) and a liver control (liver fatty acid binding protein).
CM mykiss_IL6_F TTTCAGAAGCCCGTGGAAGAGA
CM mykiss_IL6_R TCTTTGACCAGCCCTATCAGCA
CM mykiss_SOD_F TGAAGACGGGCAACGCTGGC
CM mykiss_SOD_R ACCGATGTCCGTGAGGTGCCT
CM mykiss_Lfabp_F caagtgtatgcgcaggagaa
CM mykiss_Lfabp_R aggagttggtgacggacttg
CM mykiss_xbp1F GGTCAGGTGTTGGAGTCCAG
CM mykiss_xbp1R GTCCTCAGAGTCTGTAGAGTCAGTCTG
CM mykiss_CRH_F TCGCAGCGTCTTCTGCAAGGTA
CM mykiss_CRH_R CAGCAGATGGAACGTCAGGTCCA
RNA samples from dosed and LPS injected fish are available for quantification/RT and qPCR
TUES 16 NOV 2010
Salmon pesticide project
RNA samples were quantified by nano-drop (posted on salmon project message board) monday 15-NOV.
Test PCR for genomic DNA contamination
RNA samples were aliquoted in new tubes of 200ng/ul (10ul).
0.5ul of 200ng/ul RNA samples were added to 50ul qPCR rxn
1x Master Mix 80x
25ul 2x Immomix 2000ul
2ul Syto-13 dye (50uM) 160ul
2.5ul 10uM primer F 200ul
2.5ul 10uM primer R 200ul
17.6ul nanopure H2O 1408ul
0.5ul of RNA samples were used as a simulated dilution from RT rxn. Dilutions were not made directly of RNA to save tubes and template.
12 samples are in need of DNase.
18 NOV 2010
DNase reaction.... each sample: 11, 25, 28, 32, 41, 42, 45, 58, 59, 61, 76, 773ul TURBO buffer
1ul TURBO DNase
30ul RNA
incubate 30min at 37C
add 1ul TURBO DNase
incubate additional 30min.
add 3ul DNase inactivation reagent
incubate at RT for 5 min, mixing occasionally
spin down
freeze at -80 for quantification
concentrations are less than expected but can be used. 1ug of each sample (where available) will be used for RT. Where 1ug is not available, dilution of cDNA will account for concentration difference.
19 NOV 2010
Samples from DNase treatment were quantified by nano drop... see salmon group spreadsheet for detailsSamples were normalized to 1ug RNA per RT reaction and used in following protocol:
- Mix your NORMALIZED RNA sample by inverting tube several times.
- In a 0.5 ml labeled PCR tube combine the following:
- 1ug of RNA
- 0.5 μl of oligo dT
- H2O to 15ul
- Incubate the mixture for 5 min at 70C on the thermocycler then immediately transfer to ice. Briefly centrifuge you tube and the add the following:
- 5 μl of M-MLV 5X Reaction Buffer
- 5 ul of dNTPs
- 1 μl of M-MLV RT
- Incubate the mixture for 60 min at 42C and then heat inactivate at 70C for 3 min on the thermocycler.
- Spin down the sample in a desk top centrifuge.
- Store on ice or at -20C
Samples are now ready for qPCR starting next week
cDNA will need to be diluted by adding 225ul to most samples and specified amount (spreadsheet) to specified samples. Total dilution factor for normal samples was 1ng RNA used in reaction to 4ul total cDNA after reaction. Total RNA used was 1000ng, total cDNA was diluted to 250ul.
Next step: finish diluting samples to be used in gene expression assays; qPCR. Check primers with several samples of cDNA in qPCR rxn.
30 NOV 2010
Remaining samples were reverse transcribed following RT protocol described above.
All samples that were previously RT'd were normalized for the amount of RNA used in the standard RT reaction. The normal amount used according to the protocol was 1ug RNA/25ul reaction. Several samples deviated from this based on RNA concentration and volume used and are noted in the spreadsheet. These were normalized in diluting cDNA to the same initial total RNA used for total cDNA volume. normal samples were diluted by a factor of 1:10, to compensate for lower total RNA amounts used in some samples, dilution factors varied from the norm.
All primer pairs were reconstituted at 100uM and diluted to working stocks of 10uM. Each primer was run in a standard qPCR reaction, as described above, using mixed cDNA samples.
results of primer test are pending as are dilutions of the newly discovered samples that were RT'd.
1 DEC 2010
Set up plate: duplicates of samples 49-80, liver fatty acid binding protein primers.
50ul rxn x 68
25ul 2x Immomix 1700ul
2ul Syto-13 dye (50uM) 136ul
2.5ul 10uM primer F 170ul
2.5ul 10uM primer R 170ul
16ul nanopure H2O 1088ul
2ul cDNA 2ul each ran in duplicate in neighboring columns (ie sample 49 - A1, B1; sample 50 - A2, B2; etc.)
PCR at 55C
LFABP is a liver specific protein and is an indicator or liver function at the translational level. Depressed expression would indicate reduced liver function and reduced overall health. This may have implications across all other genes of interest in this study. Since we have liver RNA from exposed samples, depressed expression of LFabp could indicate events that may mask or inflate results from expression profiles of other genes. Poor liver health could mask all other gene expression results.
Plate Layout:
A1 - 49
A2 - 49
B1 - 50
B2 - 50
etc.
A3 - 57
A4 - 57
B3 - 58
B4 - 58
etc.
8 DEC 2010
ran ef1a (elongation factor 1 alpha) as a normalizer for the rest of the groups qPCR.
25 ul rxn x 32 sample x 2 + 6 blanks
12.5ul immomix
1ul styo
1.25ul 10um primer F
1.25ul 10um primer R
7ul H2O
2ul cDNA
PCR at 55C
Plate Layout:
A1 - 49
B1 - 49
A2 - 50
B2 - 50
etc.
C1 - 57
D1 - 57
C2 - 58
D2 - 58
etc.
ef1a should be a gene that is expressed at constant levels through our treatment protocol and as such, easily used as a normalizer. All expression values will be divided by ef1a expression values and subsequently compared to mock control group. Fold change in expression compared to mock control will be reported.