http://cmar451.wikispaces.com/Research+Project+Draft
I wasnt sure how to post the wikipage. Hopefully this counts!
SEA ANENOME OUTLINE
11/10/09 EXPERIMENT, TISSUE EXTRACTION and RNA ISOLATION
8 abalone
4 control
4 subjected to stress
Control Volumes
- 4.3442 cm3
- 6.4352 cm3
- 0.8046 cm3
- 49.2693 cm3
Stress Induced Volumes
- 64.3520 cm3
- 28.9575 cm3
- 9.6518 cm3
- 6.4352 cm3
COLLECTION AND EXPERIMENT
Eight sea anemones were taken from the Fisheries aquarium. Samples were removed from rocks and placed into 2 separate containers distinguishing 4 as controls, and 4 as stressed samples.
Stressed sample organisms were subjected to mechanical stress by being placed in a plastic container with 700 mL of water on a rocker with a tilt level of 15 and speed of 30 for 30 minutes. Tissue samples were then taken from the pedal disk and tentacles. Samples ranged from .63 grams to .116 grams.
TISSUE EXTRACTION & RNA ISOLATION
500uL of TriReagent was added to 1.5mL snap cap tube (repeat 16 times). 50-100mg of tissue sample was extracted from either tentacle or pedal disk tissue from both control and stress induced groups using a clean razor. Tissue sample was placed in snap cap tube and homogenized using a plastic pistol. 500uL more of TriReagent was added to each tube. All snap cap tubes were vortexed for approximately 16 seconds and subsequently placed in –80C freezer.
11/17/09 RNA ISOLATION (cont) & RNA QUANTIFICATION
RNA ISOLATION (cont)
Tubes were incubated at room temperature for 5 minutes. Once thawed, 200uL of chloroform was added to each tube in the fume hood. Tubes were vortexed for 30 seconds until solution turned opaque and milky followed by additional incubation at room temperature for 5 minutes. Tubes were then spun in a refrigerated microfuge for 15 minutes at the maximum speed. Being very careful not to disturb the tube, the clear aqueous phase from the top (RNA) was removed from the tubes and placed into new snap cap tubes. Rest of sample was disposed of as tissue waste. 500uL of isopropanol was added, and then each tube was inverted until solution was uniform in appearance. Tubes were then left at room temperature for 10 minutes before once again being placed in a refrigerated microfuge for eight minutes at maximum speed. A small white pellet appeared at the bottom of the tube. Supernatant was carefully pipetted away from the white pellet. 1mL of 75% EtOH was added to the pellets and then placed in a refrigerated microfuge for 15 seconds at 7500g to displace the pellet. Supernatant was removed once again before tube was spun for approximately 15 seconds to pool residual EtOH (which was removed). Snap cap tube was left open at room temperature to allow the pellet to dry for 5 minutes. Pellets were resuspended in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet dissolved. Tube was incubated at 55C for 5 minutes to allow RNA to solubilize. After removing from heat, tubes were flicked and consequently placed on ice. These tubes were used as RNA stock sample.
11/19/09 DNAase and RNA QUANTIFICATION
To make sure no genomic carryover exists in the RNA stock, samples were DNAased to break up any genomic DNA. I added 2.5 uL of DNAase buffer, 1uL of Turbo DNaAasewas added to 20.% uLof RNA. the 24UL tubes were placed at 37 degrees C for 30 minutes. An addition 1 uL of Turbo DNAase was added and then placed at 37 degrees again for another 30 minutes. Once both cycles were completed, 2.5 uL of Inactivation Reagent was added to the tubes. They were allowed to wit at room temperature for 2 minutes and then spun at 10,000 rcF for 1.5 minutes. The supernatent was added to new tubes before being quantified in the spectrometer.
Using RNA stock sample, 2uL were placed onto previously blanked Nanodrop pedistal. Absorbencies for A260, A260/280, A260/320, and RNA concentration (ng/uL) were recorded. Samples were then stored once again at –80C.
11/24/09 RNA stock normalization-
Based on RNA quantification, the samples were normalized to 150 ng/uL which was the baseline derived from Control tentacle tissue samples 1 & 2 which were 159.11 and 142.42 ng/uL respectively. Control tissue samples from the pedal tissue #4 and Stressed tentacle tissue #1 were not normalized since their concentrations were well below 150 ng/uL. Stressed pedal tissue #3 was also undiluted because its concentration of 162.66 ng/uL was too close to the normal pretein concentration to dilute.
12/1/09
No new updates, as I am still waiting for primers
12/4/09 Reverse Transcription and cDNA
25 ug of my stock RNA samples were placed in PCR tubes, with an additional 5uL of water. After spinning breifly, they were placed in the thermocycler for 5 minutes at 75C. before immediately being placed on ice. A Master Mix of
4uL of MMV RT buffer, 8uL of dNTPS, 1uL of MMV RT transcriptase, 1uL of oligo dT primer, and 1uL of RNase free water were made for a total of 14 samples. the MM water addd to the RNA, vortexed, spot spin and placed in the thermocycler for 1 hour at 37C. and then heat activated for 3 minutes at 95C. Samples were then kept cold in the thermocycler until I was able to run PCR.
PCR MM consister of 17 rxns for 5 primers at 25uL size reactions a piece. Based on a paper by Rodriquez Lettney et al. 2006, I designed 5 primers for proteins believed to be produced at higher rates under stress (von Willebrand Factor, Sphigosine 1-phostphate phosphotase 2, Beta-actin, Copper/zinc superoxide dimutase, and Putative senescence-associated protein). In each tube wen 12.5uL of 2x Immunnomix, 1.0uL of sytodye, 0.5 uLprimer forward, 0.5 uL primer reverse, 9.5 of PCR water, and either 1.0uL of cDNA or water (sample or negative control). Tubes were then placed into the thermocyler for replication. (Denature(~94C), Anneal (50-60C), and Extension (~72C)). Results for PSAP, SOD, and SIPP2 were useable but Beta-actin and vWF appeared to be contaminated, due to replication in the negative controls. However, upon analyzing the data, there wasn't any statistically valuable trends based upon the sample size. And there did not appear to be any increase in protein production in the stressed samples compared to the control samples. In fact concentrations, appeared to decrease under stress implying that energy was not being applied to my specific proteins, but some other cellular process. In replicate or future studies, increased sample size is a must for statistically valuable data. Similarly, more proteins should be analyzed, to figure out exactly what cellular process is actually occuring. Other erros could be improved by increasing the amount of mechanical stress actually inflicted upon the organisms, and possibly for a longer period.
.
October 6, 2009
Protein Extraction Procedure
1. 0.5 mL of CellLytic MT solution was added to 1.5 ML snap cap tube
2. ~50 mg sample of Hard clam foot was cut and homogonized with pestle (due to the nature of clam foot, small amount of sample remained unhomogonized).
3. Tube containing tissue sample was spun in refrigerated microfuge for 10 minutes at maximum speed.
4. Supernatent was extracted and placed on ice.
5. In a fresh 2mL tube labeled 'sample', diluted alequots 15 uL of protein sample and 15 uL of DI water was added and mixed.
6. In a second 2mL tube labeled 'blank', 30 uL of DI was added.
7. 1.5 mL of Bradford reagent was added to both 'sample' and 'blank' tubes.
8. Both tubes were then transfered to 1 mL cuvette tubes and place in the spectrometer, and measured for absorbance at 595 nm.
9. Protein sample was stored in -20 degree freezer.
10. Back calculate with Bradford protein assay kit curve
Results
Two absorbances were recorded for 'sample' average x = 1.0835
y=1011.9(x)*2
y= protein (um/mL)
x= absorbtion
*mulitply by two to account for dilution factor
1011.9(1.0835)*2=2192.787 ug/mL
RNA Isolation Protocol
1. 500 uL of TriReagent was added to 1.5 mL snap cap tube and then stored on ice.
2. ~50 mg sample of Hard clam foot was added to tube containing TriReagent.
3.Sample was homogonized using a pestle (due to the nature of clam foot, small amount of sample remained unhomogonized)
4. Addition 500 microliters were added to the 1.5 mL snap cape tube.
5. Tube was vortexted for 15 seconds and then stored in -80 degree C freezer.
October 12, 2009
When deciding to examine gene expression, I should to create primers used in physiological response of detoxification. Gene selected for PCR is glutathione S-transferase Pi-1 mRNA
Primer foward: TTCCGCTGAGGGGTCGTGGC
Primer reverse: GGGGCTTGGCCAAATGGCATC
October 13 2009
RNA Isolation Protocol II (continued)
6. Removed sample from freezer and incubated at room temperature for 5 minutes
7. Added 200 uL of choloform to sample and closed tube and then vortexed for 30 sec, if done correctly the sample should look milky
-Observation: sample looked opaque
8. Incubated tube at room temperature for 5 minutes and then spun in refridgerated microfuge at for 15 minutes at maximum speed.
9. Being careful not to disturb the tube, the most aqueous phase at the top was transferred to a fresh microfuge tube
-Observation: there was a thin layer of aqueous, it may be possible that some of the white interphase was transfered into new tube, however careful I tried to be
10.Disposed of tube containing organic and interphase
11. 500 u: of isopropenol was added to the new tube containing RNA and was inverted until solution looked uniform
-Observation: a small white pellet appeared before incubation and microfuge, even after inverting for several minutes
12. Incubated at room temperature for 10 minutes and then spun in refridgerated microfuge at maximum speed for 8 minutes
13. Small white pellet appeared (RNA and salts) but supernatant was removed
14. 1 mL of of 75% EtOH was added to pellet and the vortexed briefly before spinning tube in refridgerated microfuge for 5 minutes at 7500g
15. Remaining supernatent was removed and tube was spun for approximately 15 seconds and again additional EtOH was removed using P20 pipette
16. Tube was left open in room temperature for 5 minutes to allow pellot to dry
17. Pellet was resuspended in 100uL of 0.1%DEPC-H20 by pipetting up and down until pellet was dissolved
-Observation: pellet remained stubborn when trying to dissolve, very small flakes remained
18. To help solubilize RNA, tube was incubated at 55C for 5 minutes, once removed tube was flicked to resuspend sample
19. Stock sample was placed on ice and will be used to quantitate RNA yield using spectophotometer (Nanodrop)
Results:
RNA was successfully isolated and will be be quantified using Nanodrop spectrophotometer.
Protein Gel Protocol
1. Protein extraction sample was thawed and inverted several times to mix
2. 15 uL of protein sample and 15 uL of 2X Reducing Sample Buffer were added to a fresh 1.5 mL screw cap tube
3. Flicked sample to mix and centrifused for 10 seconds to pool liquid in bottom of tube
5. Sample boiled for 5 minutes and immediately centrifuged for 1 minute to pool liquid
5. Used rinsed 4-20% Tris-HEPES gels according to procedure (done by Mac)
6. Loaded entire sample into appropriate well (30 uL), along side molecular ladder SeeBlue Plus 2 Prestained Standard 1x (Invitrogen)
7. Gel was run for 30 minutes at 150 V, and once gel was removed placed on shaker/rocker with Coomassie stain for 5 minutes
8. Liquid stain was dumped back into container while being careful not to distrub gel
9. Gel was rinsed with 10% acetic acid before adding 250 mL of 10% acetic acid to gel in shaker/rocker for 15 minutes, this was repeated until bands in gel became visible (procedure finished by Mac)
Results
Amount of protein loaded into gel box: 2192.787 ug/mL * 15 uL* 1mL/1000uL= 32.892 ug of protein from Hard Clam foot tissue

My protein was loaded into the 4th row from the left. While it the bands aren't completely distinct, there does appear to have distinct bands near the Myosin protein band as well as Glutamic Dehydrogenase, and Alcohol Dehydrogenase. Other distinct bands appear thoughout the gel, but I am unsure as to what the protein expression represents.
10/20
LAb 3: RNA Quantification, Reverse Transcription, and Agarose Gels
-
RNA Quantification
Purpose: to quantify RNA sample
Samples prepared during the previous weeks were thawed, and 2 uL were pipetted onto the Nanodrop pedistal (after it was blanked). Pedistal was wiped down with a kim wipe. The Nanodrop uses Beer-Lambert Law to calculate RNA concerntrations ([RNA]=40ug/mL x Dilution Factor). Clean RNA samples should have absorbance A260/A280 range from 1.8-2.0 and A260/A320 should range between 1.5-2.0.
Results:
A260 Absorbance=3.007
A260/A280= 2.70
A260/A320= 0.44
RNA concentration = 120.3 ug/uL
Neither of my samples fall into the clean RNA absorbance ranges, which means I must assume there was contaminiation during extraction or quantification protocols.
Reverse Transcription Protocol
1. Invert RNA samples several times to mix, and then transfer 5 uL or RNA and 5 uL of water to a fresh PCR tube. Mixture was then incubated for 5 minutes in thermocycler at 75C. The samples was then immediately placed on ice for around 5 minutes.
2. 4 uL of AMV RT buffer 5x, 8uL dNTPs, 1uL AMV RTranscripase, 1uL Oligo dT Primer, 1 uL RNase free water was then added to RNA/water sample which totaled in 20 uL in volume. Sample was vertexed, spot spun, and incubated at room temperature for 10 minutes.
-Observation- these final steps were performed by a lab partner after I had to leave lab early.
3. Mixture was placed in thermocycler for 1 hour at 37C, and then subsequently heat inactivated for 3 minutes at 97C
5. Sample mixture was spon spun and then stored at -20C.
Primer Preperation for glutathion s-transferase
Forward Primer: 5' TTCCGCTGAGGGGTCGTGGC (36.7nMoles)
Reverse Primer 5' GGGGCTTGGCCAAATGGCATC (38.4 nMoles)
1. Primers were reconstitued for a working 10uM stock soultin by adding 367 uL of water to forward primers and 384 uL of water to reverse primers.
2. 10 uL of each primer and 90 uL of water was added to 2 seperate snap cap tubes for a total volume of 100 uL in each tube.
PCR Protocol
Reason: this will help quantify gene expression for glutathion s-transferase
1. A master mix was composed 25 uL GoTag Green Master Mix (2x), 10 uM forward primer (2.5uL), 10 uM of reverse primer (2.5uL), 2 uL cDNA (for samples) or PCR water (used for negative controls), and 18 uL of PCR water. Amounts shown are for one reaction, though we created a master mix for 5 reactions to ensure enough was made.
-Observation: Primer amounts were determined using (10mM)(Vol1)=(.5mM)(50uL)
2. 2 tubes used cDNA (for samples) and 2 tubes used water (for negative controls. All 4 tubes were placed into thermocycler for PCR.
Agarose Gel Preparation
1. 2g of agarose was weighed and mixed with 150 mL 1x TAE buffer in 1 L flask. Mixture was microwaved for 2 minutes on high power.
2. 30 mL of buffer was added to flask (to compensate for evaporation) and was microwaved for an additional 30 seconds.
3. Solution was cooled, and 12 uL of ethidium bromide was added to flask.
4. Flask was swirled to thoroughly mix solution, and then contents were pourd into a gel tray with gel combs.
5. Gel was allowed to set, then wrapped in plastic wrap and stored in the fridge.
-Observation: this protocol was performed by other students in lab as I had to leave early.
Results and future investigations:
Products from PCR will be run on gel to determine if primers actually worked for our samples. This will help us determine if the protein coded in our primers is expressed in our organisms tissue sample.
October 27, 2009
Lab 4 PCR products and Western Transfer - Immunoblots
Runnning the PCR products in a gell with help determine whether or not the designed primers worked for out samples, which would confirm that the coded protein in our primers is expressed in our tissue sample.
PCR METHOD
1. Gel was placed in gel box along enough 1x TAE buffer to fully cover the wells.
2. Remove combs and load 7 uL of 100bp ladder into the first lane.
3. 25 uL of my PCR samples were loaded onto top row 2-5 (2 & 3 were control, 4 & 5 were sample)
4. Gel was run at 100 V for one hour and then visualized on a UV transilluminator.
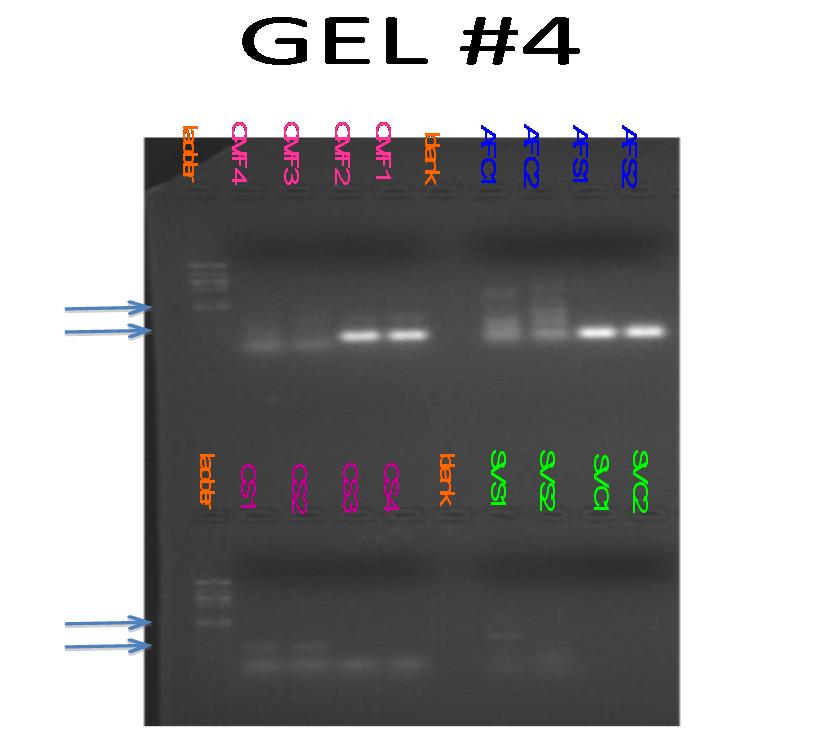
-Observations and results:
Rows 2 and 3 show bands at appoximately 100 bp. While this are the control lanes, banding could be due to primer dimer. lanes 4 and 5 show banding in between 100 bp and 150bp. The approximate amplicon size is 130, so this is exactly the results we were looking for.
METHODS FOR PROTEIN TRANSFER TO MEMBRANE:
1. Transfer buffer was cooled to 4 degrees C
2. Soaked the filter paper, membrane, and gel in Transfer Bufer for 15 miuntes.
3. The blotting sandwich was assenbled in a semi-dry blotting apparatus as follows:
-positive anode
-filter paper (2 sheets for each gel)
-nitrocelluolse membrane
-gel
-filter paper (2 sheets)
-negative cathode
4 for 20 minutes at 20V, blot was allowed to transfer
5. removed gel, and rinsed with transfer buffer
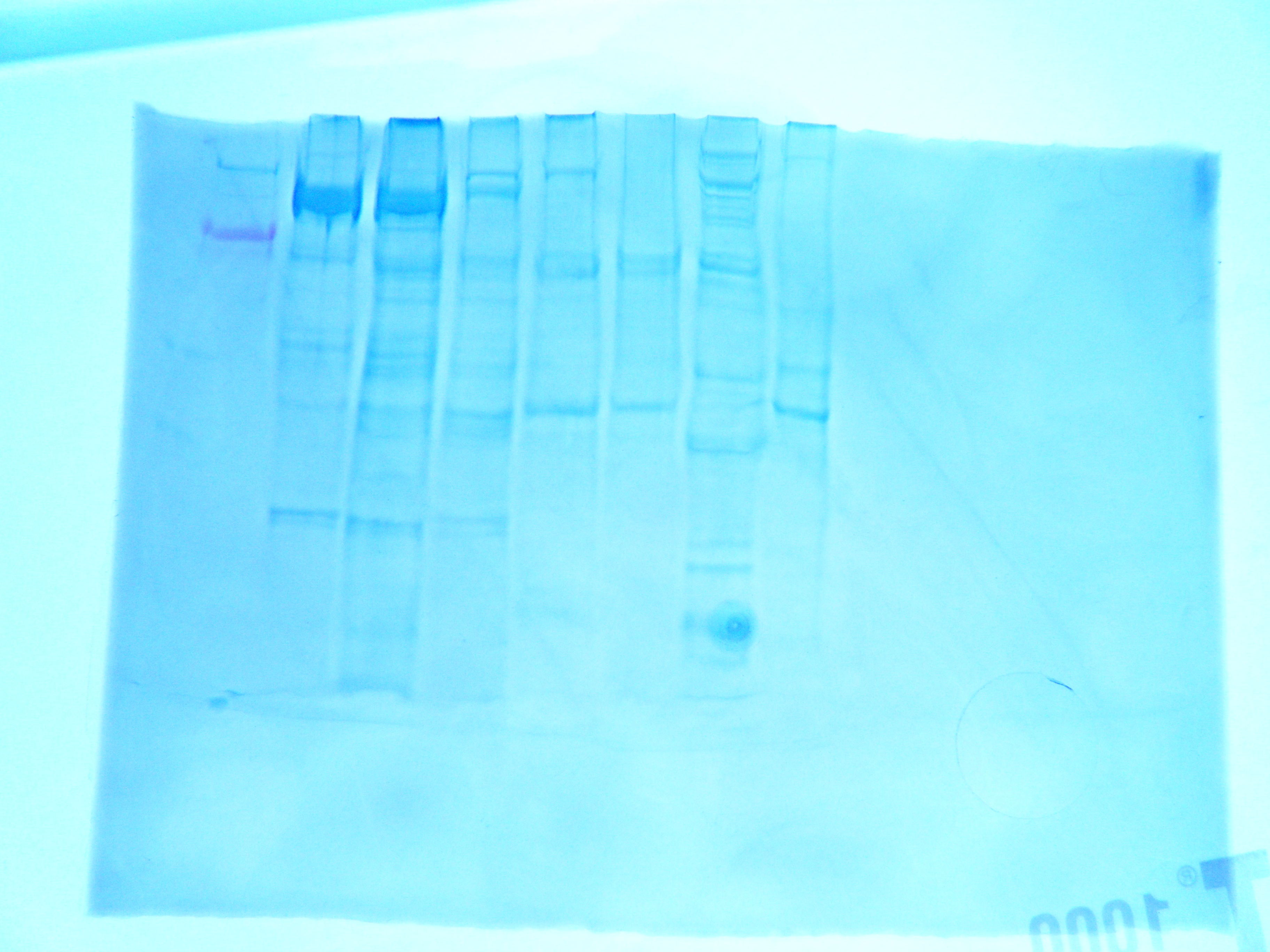 |
| FIgure 1. Gel for western blot. Hard clam sample was in lane 7 |
FIGURE 1: Western gel, hard clam sample is in lane 7.
RESULTS: in lane 7 the largest bands are prettty small. Howver banding means that PCR amplified cDNA sample. Futher studies could test quality of cDNA to check for DN A contamination.
.
.
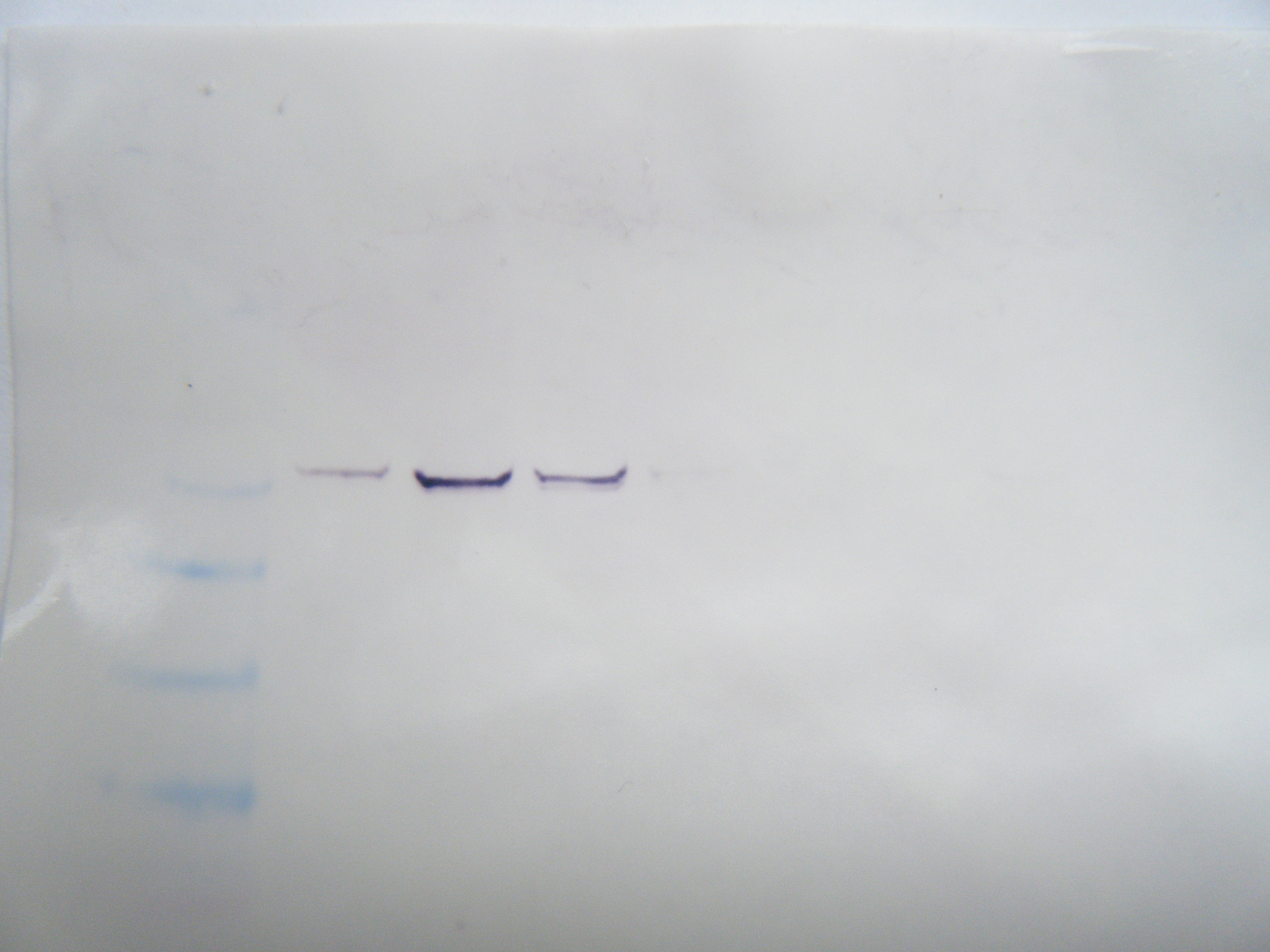
FIGURE2: Western Blot Result: Hard clam sample was in row 7. No observable protein presence indicatiors.
METHODS FOR WESTERN BLOTTING:
1 prepared 20mL of Blocking Solution with
-ultra filtered water 14 mL
-Blocker/Dilutent (Part A) 4 mL
-Blocker/Dilutent (Part B) 4 mL
2. Membrane was placed in 10 mL of appropriate Blocking Solution in a covered, plastic dish and incubated for approximately 30 minutes in a rotary shaker set at 1 rev/sec
3. Blocking solution was decarted
4. Membrane was rinse with 20mL of water for 5 minutes, then decarted (repeat twice)
5. 10 mL of primary antibody solution (1:3000) was made
-10 mL of Blocking solution
-3.3 uL HSP 70 antibody
6. Membrane was incubated with 10mL of primary antibody solution overnight. the next morning, primary antibody was decanted.
7. MEmbrane was washed for 5 minutes with 20mL of prepared Antibody Wash, then decant. Repeat 3 times.
8. Incubated membrae in 10 mL of secondary antibody solution for 30 minues, then decant.
9. Membrane was washed with 20mL of Antibody Wash, then decanted. Repeat 3 times.
10. Membrane was rinsed fir two minutes with 20 mL of water the decanted. Repeat 2 times.
11. Membrane was incubated in 5 mL of Chromogenic Substrate until purple bands developed on the membranes. Allow one hour to complete.
12. Membrane was dried on a clean peice of filter paper in the open air. under a infared lamp or by a stream of slightly warm air.
Western Blot ResultsL the antibodies were unsuccessful in binding to proteins for my samples. This could be that the primers were not successful in bidning to the RNA, that the proteins were not expressed in the tissue, or that the proteins did not successfully transfer to the membrane
November 3, 2009
Lab 5: Quantitative PCR
Purpose:
To amplify a desired region cDNA using a polymerase, species specific primers, and dNTPs.
Methods:
1. Master Mix was prepared with the following recipe for 7 reactions for a total of 48 uL per reaction (6 total reactions desired with an extra reaction for pipetting errors)
a.Master Mix, 2X (Immomix): 25 uL
b. Syto-13 dye (50uM): 3.5 uL
c. Upstream primer (from previous stock): 1.5 uL
d. Downstream primer (from previous stock): 1.5 uL
e. Ultra Pure Water: 16.5 uL
2. 48 uL of Master Mix were added to 6 wells of a PCR plate.
3. first two wells containted 2uL of cDNA
4. 2 uL of ultra pure water were added to second two wells as negative controls.
5. 1:4 dilution was made for RNA with 5 uL of RNA: 15 uL of water.
6. 2 uL of RNA dilution were added to last 2 wells to test for genomic DNA carry-over.
7. The plate was loaded onto a real-time PCR thermocycler.
RESULTS:
According to my PCR amplification, there appears to be genomic carryover in my RNA as well as slight contamination in my negative controls. Wells 1 &2 were negative controls, 3 & 4 were cDNA, and 5 & 6 were RNA. Samples of RNA were amplified indicating genomic carryover. Melting points of cDNA were slightly lower that the melting points of the Negative control. I am unsure of what this contamination could consist of.