For results and analysis see the google doc
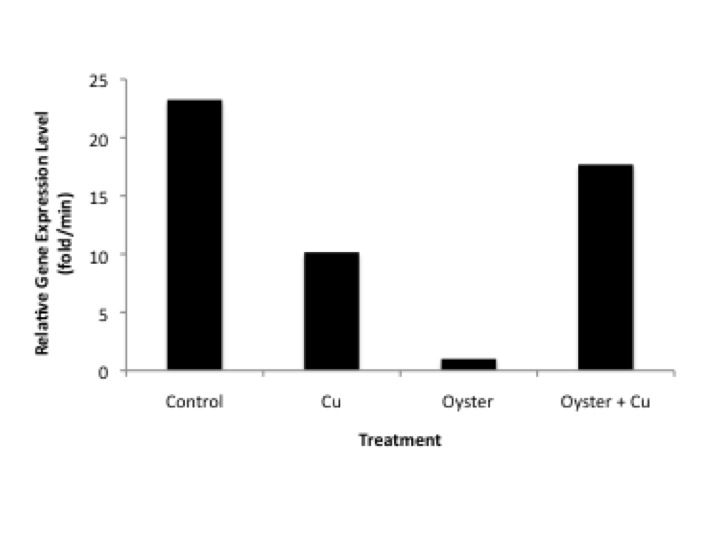
Further analysis (excerpt from paper discussion)RseA is a transcription factor that negatively regulates the expression of sigma factor E, part of the rpoE operon (Missiakas et al., 2003). Sigma factors are typically involved in heat stress response pathways and the management of improperly folded proteins. Sigma factor E (sE) in particular is involved in the maintaining the integrity of the cell envelope and expression is increased in response to protein degradation in the preiplasm (Ades, 2004). Expression of algUmucABCD (AlgU), the functional equivalent of sigma factor E in E.coli (Yu et. al., 1995), is implicated in the regulation of stress response proteins as well as alginate production. Alginate is an exopolysaccharide (EPS) that has been shown to be the primary EPS in the biofilms of Cystic fibrosis pathogenesis (Govan and Deretic, 1996). Biofilms are important mediators between a bacteria and their external environment, providing protection from compounds such as antibiotics and host immune response factors. Knockout of the RpoE operon increased suseptability of Burkholderia pseudomallei to environmental stressors such as reactive oxygen species and osmotic stress (Korbsrisate et. al., 2005). It has been shown that RpoE is essential for maintenance of extracellular proteins in E.coli upon treatment with Cu(II) (Egler, et. al., 2005). Our findings are similar to those of Egler, et. al. 2005, where they saw a decrease in the levels of anti-sigma factor Rse-A in response to Cu(II) treatment of E.coli. The decrease in RseA after exposure to Cu(II) may indicate that Cu has a negative effect on the integrity of the bacterial envelope of V. tubiashii. A decrease in RseA would indicate that higher levels of sE(Sigma E?) are required to maintain the structural integrity of proteins in the periplasm. Interestingly, the addition of a pathogenic host in this experiment had the opposite effects on RseA regulation. The presence of oysters alone almost eliminates the expression of RseA. This may suggest that the stress from compounds secreted by the host’s immune system have a more severe impact on the structural integrity of the pathogen’s cellular envelope compared to CuSO4, resulting in decreased expression of RseA so as to maximixe SigmaE expression. However, the higher expression levels of RseA mRNA in the combined treatment of Cu and host tissue compared to either Cu(II) or host tissue alone is troublesome.
December 3, 2010
qPCR repeat from December 1, 2010
Notes: Rerunning samples to correct for discrepancies in reps from previous run. Also running dilution curve to calculate mRNA levels instead of using to universal curve.
Columns1-4 = 16s
Columns5-8 = RseA
Columns9-12 = AlkSerPro
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
|
| A |
0 |
0 |
Vt |
Vt |
0 |
0 |
Vt |
Vt |
0 |
0 |
Vt |
Vt |
| B |
1:5 |
1:5 |
Vt+Cu |
Vt+Cu |
1:5 |
1:5 |
Vt+Cu |
Vt+Cu |
1:5 |
1:5 |
Vt+Cu |
Vt+Cu |
| C |
1:10 |
1:10 |
Vt+oyster |
Vt+oyster |
1:10 |
1:10 |
Vt+oyster |
Vt+oyster |
1:10 |
1:10 |
Vt+oyster |
Vt+oyster |
| D |
1:50 |
1:50 |
Vt+oyster+Cu |
Vt+oyster+Cu |
1:50 |
1:50 |
Vt+oyster+Cu |
Vt+oyster+Cu |
1:50 |
1:50 |
Vt+oyster+Cu |
Vt+oyster+Cu |
| E |
1:100 |
1:100 |
NTC |
NTC |
1:100 |
1:100 |
NTC |
NTC |
1:100 |
1:100 |
NTC |
NTC |
| F |
1:500 |
1:500 |
NTC |
NTC |
1:500 |
1:500 |
NTC |
NTC |
1:500 |
1:500 |
NTC |
NTC |
| G |
||||||||||||
| H |
||||||||||||
| 1x |
26x |
|||||||||||
| Immunomix |
12.5 |
325 |
||||||||||
| Syto 13 |
1 |
26 |
||||||||||
| F primer |
1.25 |
32.5 |
||||||||||
| R Primer |
1.25 |
32.5 |
||||||||||
| Water |
8 |
208 |
December 1, 2010
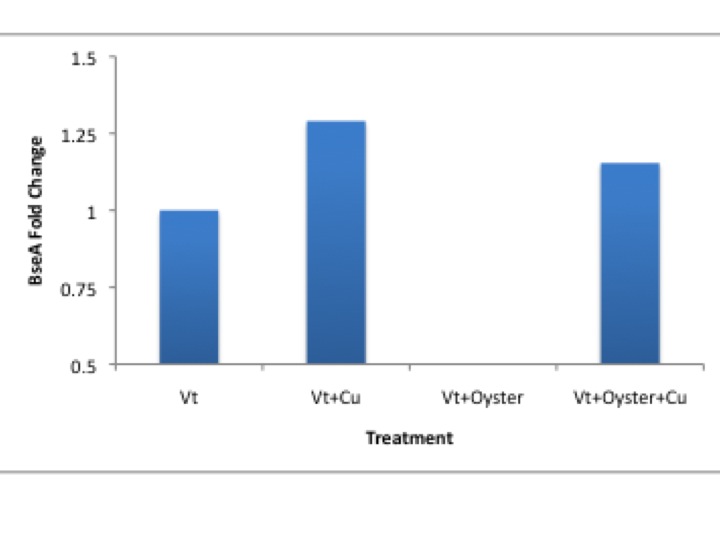
Results from qPCR on November 30, 2010
I would rerun RseA as big diff in rep for "Vt", also how about 16data reps? and melting curve data? -
| Sample |
Gene |
Ct rep1 |
Ct rep 2 |
Avg Ct |
16s Norm |
Fold Change |
| Vt |
16s |
23.12 |
23.16 |
23.14 |
||
| Vt+Cu |
16s |
25.56 |
25.73 |
25.65 |
||
| Vt+Oyster |
16s |
31.14 |
31.15 |
31.14 |
||
| Vt+Oyster+Cu |
16s |
26.19 |
26.09 |
26.14 |
||
| Blank |
16s |
38.15 |
- |
|||
| Blank |
16s |
- |
- |
|||
| Vt |
BseA |
22.94 |
31.75 |
27.35 |
1.18 |
1.00 |
| Vt+Cu |
BseA |
39.30 |
38.95 |
39.13 |
1.53 |
1.29 |
| Vt+Oyster |
BseA |
- |
- |
|||
| Vt+Oyster+Cu |
BseA |
35.83 |
35.49 |
35.66 |
1.36 |
1.15 |
| Blank |
BseA |
- |
- |
|||
| Blank |
BseA |
- |
- |
|||
| Vt |
SerProt |
- |
- |
|||
| Vt+Cu |
SerProt |
- |
- |
|||
| Vt+Oyster |
SerProt |
- |
- |
|||
| Vt+Oyster+Cu |
SerProt |
38.30 |
38.66 |
38.48 |
1.08 |
|
| Blank |
SerProt |
- |
- |
|||
| Blank |
SerProt |
- |
- |
|||
| Vt |
Chitinase |
- |
- |
|||
| Vt+Cu |
Chitinase |
- |
- |
|||
| Vt+Oyster |
Chitinase |
- |
- |
|||
| Vt+Oyster+Cu |
Chitinase |
- |
- |
|||
| Blank |
Chitinase |
- |
- |
|||
| Blank |
Chitinase |
- |
- |
November 30, 2010
Vt qPCR with cDNA made on November 29, 2010
Primers:
Chitinase
16s (version 1)
SerineProtease
RseA
| Component |
Volume 1X |
Volume 8X |
Final Con. |
| Master Mix, 2X (Immunomix) |
25ul |
200ul |
1x |
| Syto-13 dye (50uM) |
2ul |
16ul |
2uM |
| F Primer (10uM) |
2.5ul |
20ul |
2.5uM |
| R Primer (10uM) |
2.5ul |
20ul |
2.5uM |
| Sterile H2O |
16ul |
128ul |
N/A |
Diluted cDNA 1:5 and added 2ul to each reaction.
Plate Map:
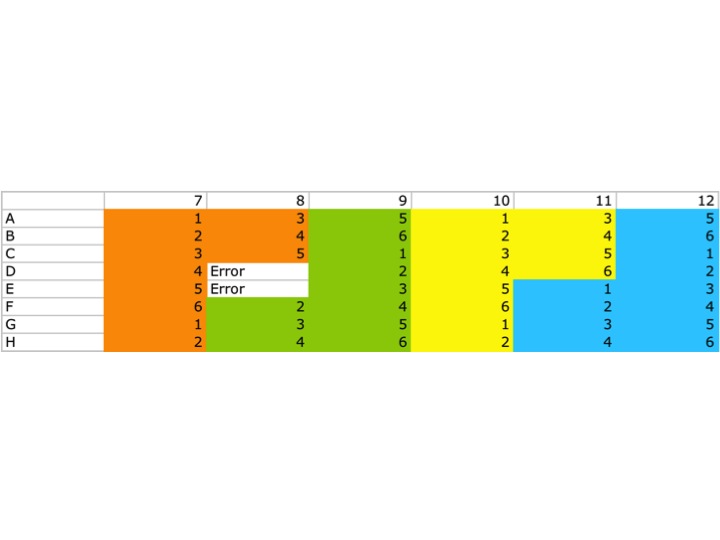
Orange: 16s
Green: RseA
Yellow: SerineProtease
Blue: Chitinase
November 29, 2010
DNase and RT of Vt RNA from November 24, 2010
DNase protocol:
Combine the following...
45ul RNA (no more than 10ug)
5ul 10x DNAse Turbo buffer
1ul DNas Turbo enzyme
-Incubate at 37 degrees C for 30min
add 5.5ul of inactivation reagent. mix by flicking tube with finger and incubate at RT for 2min
spin 10,000xg for 1.5min
transfer supt. to new tube and save.
NanoDrop results after DNase treatment
| Sample ID |
User ID |
Date |
Time |
ng/ul |
A260 |
A280 |
260/280 |
260/230 |
Constant |
Cursor Pos. |
Cursor abs. |
340 raw |
| Vt |
Default |
11/29/10 |
12:14 PM |
136.39 |
3.41 |
1.917 |
1.78 |
0.61 |
40 |
230 |
5.566 |
0.011 |
| Vt + Cu |
Default |
11/29/10 |
12:15 PM |
106.15 |
2.654 |
1.738 |
1.53 |
0.48 |
40 |
230 |
5.524 |
0.186 |
| Vt + Oyster |
Default |
11/29/10 |
12:16 PM |
103.03 |
2.576 |
1.602 |
1.61 |
0.4 |
40 |
230 |
6.48 |
0.116 |
| Vt + Cu + Oyster |
Default |
11/29/10 |
12:17 PM |
106.76 |
2.669 |
1.464 |
1.82 |
0.88 |
40 |
230 |
3.036 |
0.079 |
RT Protocol
Combine 1ug RNA with 1ul OligoDT
-Incubate at 70 degrees C for 5min. Ice immediately
Add: 5ul 5x reaction buffer, 5ul dNTPs, 1ul M-MLV RT, and 4ul water
Incubate 42 degrees for 60min followed by 70 degrees for 3min.
store at -20 degrees C.
Sample #s
1. Vt only
2. Vt + Cu
3. Vt + Oyster
4. Vt + Oyster + Cu
5. Water/negative control
November 24, 2010
Vt RNA extraction from water filters collected on November, 19, 2010
Followed Standard TriReagent RNA extraction
NOTES:
1. Extracted RNA from entire filter. Filter breaks apart (much like tissue) and forms homogenous mixture of filter pieces of the same size.
The filter dramatically increases the volume of the TriReagent that is added for homogenization so I only homogenized in 500ul so that I would have enough space in the 1.5ml tube to add the remaining reagents. Volumes for the remainder of the protocol were cut in half to maintain appropriate proportions to the amount of TriReagent used.
2. Samples were resuspened in 30ul of water. For the Vt only and Vt + Cu I only had 1 filter to extract. I extracted 2 filters for the Vt+Oyster+Cu and 2 filters for Vt+ Oyster. The idea was to try and extract as close to 500ml of filtered material as possible. (see filter sampling from November 19, 2010 for more info). For samples where more than one filter was extracted I pooled the extractions in a single tube for a total of 30ul RNA.
NOTE: RNA pellet for Vt + Cu sample was HUGE. Resuspended in 150ul water.
NanoDrop Results:
| Sample ID |
User ID |
Date |
Time |
ng/ul |
A260 |
A280 |
260/280 |
260/230 |
Constant |
Cursor Pos. |
Cursor abs. |
340 raw |
| Vt only |
Default |
11/24/10 |
11:48 AM |
143.97 |
3.599 |
2.28 |
1.58 |
0.5 |
40 |
230 |
7.132 |
0.225 |
| Vt + Oyster |
Default |
11/24/10 |
11:49 AM |
143.65 |
3.591 |
2.292 |
1.57 |
0.44 |
40 |
230 |
8.126 |
0.139 |
| Vt + Oyster + Cu |
Default |
11/24/10 |
11:50 AM |
137.98 |
3.45 |
2.056 |
1.68 |
1.08 |
40 |
230 |
3.208 |
0.143 |
| Vt + Cu |
Default |
11/24/10 |
11:53 AM |
154.88 |
3.872 |
2.57 |
1.51 |
0.52 |
40 |
230 |
7.452 |
1.278 |
Looks like I have RNA!!!! Although its not very clean/pure. I only have 4 independent samples so I'll go ahead and DNase these.
November 19, 2010
Tissue Sampling
Today the group is collecting oyster tissues from the Cu/Vt challenge that has been going on all week. Tissues to be collected are the gill (for its concentration of hemocytes), mantle (mostly for DNA/methylation analysis), hemolymph (alternative source for hemocytes), and size dimensions (Lenth, width, height) of the oysters.
Tasks
Oyster Shucker - split between Jason and Ross
Gill tissue - Sammi
Hemolymph - David
Mantle -
Size measurements - Dave and Jason
Water filtering - Dave
My primary collection task was to filter the water containing the different treatments of V.tubiashii.
Treatments:
Note: all Vt treatments are in a volume of 5L at a concentration of 7.5x10^5cfu/ml
Vt + seawater
Vt + seawater + Cu (130ppm)
Vt + seawater + oysters (Big Beef Creak only)
Vt +seawater + Cu (130ppm) + oysters (Big Beef Creak only)
Filtering Seawater:
Supplies:
Vacuum flask
Vacuum tubing
Milllipore filtering aparatus
2um filter membrane
100um filter screen
transfer beaker (250ml)
Methods:
1. Use transfer beaker to collect a 250ml sample of water from the 5L bucket
2. Hold 100um screen directly over filter apparatus reservoir.
3. Pour 250ml of sample water directly onto 100um screen so that it will be collected by the reservoir.
4. Turn on vacuum pump to filter water.
Note: Samples containing oysters had a lot of debris which caused filters to clog. All 5L were not filtered. The following is a breakdown of filter samples collected:
Samples
Vt + seawater + oysters (Big Beef Creak only) = 4 filters, 1L total volume, 250ml each.
Vt + seawater + Cu + oysters (Big Beef Creak only) = 2 filters, 400ml total volume, 200ml each
Vt + seawater = 1 filter, 500ml total
Vt + seawater + Cu = 1 filter, 500ml total
November 18, 2010
V.tubiashii challenge
Vt culture from 11/17/10 was spun down at 4,800rpm for 20min in a swing bucket rotor.
Remove supt. and resuspend Vt in 1L of filtered seawater.
Spin again 4,800rpm 20min.
Remove supt. and resuspended in 110ml filtered seawater (11 treatments, 10ml/treatment)
Spec at OD550.
Spec Reading = 0.750
Calculating CFU
Estimated CFU by assuing 1U at OD550 = 5x10^8 cfu/ml
0.750 x (5x10^8) = 3.75x10^8cfu/ml
3.75x10^8cfu/ml x 10ml/sample = 3.75x10^9cfu
3.75x10^9cfu = total amount of Vt added to each bucket
3.75x10^9cfu/5000ml = 7.5x10^5
7.5x10^5cfu/ml = concentration of Vt in each bucket
November 17, 2010
Tending to the Vt cultures...
After o/n incubation of 100mL cultures of Vt and Vp at 37degrees C, it looks like the Vt has not grown at all.
Started three new 5ml cultures of Vt
One culture was inoculated from a Vt plate into the LB (1%NaCl) that was prepared yesterday
Another from the same plate but into Marine Broth
and the third was from a -80 glycerol stock into the LB made yesterday.
NOTE: After innoculation, I added another 10g (1%) NaCl to the LB, split it into two 500ml aliquots, and reautoclaved.
At the end of the day today, I will take all of my Vt cultures and combine them into one of the 500ml flasks of LB, and add the Vp culture that is growing so well into the other 500mL aliquot of LB.
November 16, 2010
So far so good...
Today marks the beginning of the CuSO4 challenge.
Method:
Who? C.gigas collected on 11/11/10 and acclimated to the system beginning 11/12/10.
Water volumes were adjusted to 5L for all treatments.
6.5mg of CuSO4 was added to the appropriate treatments (1.3mg/L)
Oysters will be challenged for either 72hours in CuSO4 only, or 48hours followed by an additional 24hours with V.tubiashii(to be added 11/18/10)
V.tubiashii cultures
Diluted starter cultures prepared by sam on 11/10/10 into 100ml LB(2%NaCl)
Prepared 1L LB(1%NaCl) to expand cultures with tomorrow.
NOTE: Labeling scheme on LB was confusing. "LB +1% NaCL" could mean 1% total, which would be the normal recipe, or 2% total. Turns out it means 2% total NaCl
LB Recipe
10g NaCl
10g Yeast Extract
10g Tryptone
800ml Water
pH 7.0
adjust volume to 1L with Water
Autoclave!
November 12, 2010
Oyster samples collected on November 11, 2010 put into system.
There was a minor disaster this morning when I went to put the oysters into the basement system. Apparently the recirculating rates for the two sumps was not even which resulted in all of the water from one sump being pumped into the other. The problem is two fold:
1. One of the sumps was dry and not being held at 12 degrees.
2. The other sump was too full which lifted and tipped the buckets over.
All tipped buckets were drained and refilled with pure sea water. Half of the water from the full sump was pumped back to the dry pump. The chiller was disconnected from one of the sumps so it is now only circulating water for one system. I hooked up another chiller to the other system. This chiller is slighty different in that water is not flowing through it, instead there is a cooled wired that chills the water directly in the sump. This may or may not induce a tank effect. The pump was left in the water of the coil chiller to help mix the sump water.
Result: everything seems to be working for the moment and both sumps are holding at ~12-13 degrees C.
FINALLY the oysters can be put into buckets. Four oyster were put in each bucket. Blue/green label are from Big Beef Creek and Red labels are from the park. Treatments are separated so that all buckets that will eventually contain V.tubiashii will be kept together. Extra oysters were put in separate buckets and placed in the system with the coiled chiller.
No food has been added to the buckets.
Sammi will check on the systems before she leaves tonight.
November 11, 2010
Field Sampling!!
Low tide for the two sites were at 3pm. Low tides were a +7 which didnt allow us to collect oyster that were very far out on the tidal flats. Oysters at Big Beef Creak were collected closer to 4pm. ~40 oysters were collected from each site. Oysters were kept in coolers in the back of Steven's truck o/n and transported to the lab on friday morning.
November 10, 2010
Wet lab set-up for Oyster treatments:
With the help of Sammi and Ross we managed to hash out a system in the quarantine room in basement of FSH. Currently we have two large white totes (like the kind they use on fishing boats) that contain 8 buckets each. This gives us a total of 16 buckets which is how many we will need to carry out our 4 treatments in duplicate for each site.
At the moment we have seawater in all of the buckets with airstones only. There is no recirculation/filtration happening on the buckets. The waterbath totes have fresh water that is circulating through a chiller. Initial temperature measurements have one waterbath at 17 degrees C and the other at 19 degrees C. I will check the temps before I leave tonight to see if they are dropping towards our target temp of 12 degrees C.
(hopefully I can post a picture once we put animals in on friday)
Lose ends:
1. Will have to decide how we want to separate treatments. My vote is to keep all Vt treated animals in one tote and all control and copper only treated animals in the other.
2. Lids need to be made with labels for each bucket.
Research Proposal:
Introduction
Shellfish are essential components to the Pacific northwest landscape and economy. Their sedentary lifestyle, and life history strategies expose them to some of the harshest marine environments. The process of filter feeding in addition to having open circulatory systems results in their tissues being constantly exposed to and influenced by their surrounding environment. Consequently, shellfish are ideal model systems to study and monitor the impacts of environmental contaminants (Rittschof and McClellan-Green, 2005). The goal of this study is to determine if long-term exposure to pollutants has an effect on the pacific oyster’s (Crassostrea gigas) susceptibility to Vibrio tubiashii. V. tubiashii is a bacterial pathogen in bivalve mollusks and is responsible for massive die-offs in hatcheries around the world. Understanding and improving disease resistance of Pacific oysters is a topic that has grabbed the attention of federal agencies (see pacific shellfish institute website http://www.pacshell.org/projects/summerMortality.htm) as well as local fisheries.
Research Objective
Previous experiments have shown that a one-week exposure to heavy metals prior to infection can have detrimental effects on the oysters immune system (Morley 2010). More specifically, hemocyte function has been shown to be effected by contaminants such as PCBs (Pipe et al., 1995; Canesi et al., 2003) and heavy metals (Auffret et al., 2002; Gagnaire et al., 2004). However, the molecular cause for this is still unknown. In addition, no studies have been conducted on populations that are under continuous selection in these high pollutant environments. It is possible that pollutant exposure spanning several generations would select for individuals more tolerant to these conditions, therefore resulting in a population that is equally adept to dealing with V.tubiashii infection as oysters from a clean site. Gene expression in V. tubiashii will also be monitored to elucidate whether any changes in susceptibility of the oysters from different water quality treatments is due to a change in the gene expression of the host or the pathogen. This study is designed to tease apart the underlying molecular mechanisms of the oyster’s immune system that are being affected by industrial and agricultural pollutants, and to ask whether or not these changes are persistent after contaminant exposure.
Methods
Oyster collection
Oysters will be collected form a site in Puget Sound that has been previously identified as containing high levels of industrial contaminants and a site isolated from industrial and agricultural contamination. Water will also be collected from each site for contaminant analysis. Enough water from the contaminated site will be brought back to the lab to use during challenge experiments. Filtered seawater from the aquarium will be used on the oysters from the “clean site”
V. tubiashi challenge
Oysters from the contaminated site will be divided into two groups. One group will be challenged with V. tubiashii while the other ten will serve as controls. Oysters from the clean site will be divided four groups. One group will be challenged with V.tubiashii. the other will serve as controls. A third and fourth group of oysters from the “clean” site will be exposed to heavy metals for 7 days to test for differences in short term versus long term (contaminated site) exposure to pollutants. These oysters will then be separated and one will be challenged with V. tubiashii. while the other will serve as controls.
Six separate trials will also be conducted to monitor V. tubiashii gene expression. V. tubiashii will be exposed to the following conditions: clean water, clean water plus oyster tissue, contaminated water, contaminated water plus oyster tissue, heavy metal water, and heavy metal water plus oyster tissue.
Tissue sampling
Hemolymph and gill tissue will be taken from each oyster for subsequent analysis. V. tubiashii will be isolated by centrifugation.
Gene expression analysis
RNA will be isolate from gill tissue and transcribed in to cDNA for qPCR analysis. We will be analyzing genes from a variety of stress and immune response pathways including, but not limited to, proteins involved in stress response such as ACTH, proteins expressed in response to oxidative stress, metallothionine and other metal binding proteins, and proteins involved in immune response and antigen recognition such as cell surface receptors and genes involved in phagocytosis. Of particiular interest will be expression levels of metalloprotease mRNA in V. tubiashii, which has been linked to its pathogenicity (Hasegawa, et al. 2008).
Timeline
Week one of the experiment will consist of field sampling in which the oysters are collected from the designated sites. Once brought back to the lab, contaminant exposure will be conducted immediately on a subset of animals for one week. Week two will consist of the V. tubiashii challenge and tissue collection. Weeks 3-4 will be devoted to sample work up in the lab. Week five will be data analysis and writing.
References
Auffret M, Mujdzic N, Corporeau C, Moraga D. 2002. Xenobiotic-induced immunomodulation in the European flat oyster, Ostrea edulis. Mar Environ Res. 54: 585-589.
Canesi L, Ciacci C, Betti M, et al. 2003 Effects of PCB congeners on the immune function of Mytilus hemocytes; alterations of tyrosine kinase-mediated cell signaling. Aquat Toxicology. 63: 293-306.
Gagnaire B, Thomas-Guyon H, Renault T. In vitro effects of cadmium and mercury on Pacific oyster, Crassostrea gigas (Thunbert), hemocytes. Fish Shellfish Immunol. 16: 501-512.
Hasegawa, H, Lind, E.J., Boin M.A., Hase C.C. 2008. The extracellular metalloprotease of Vibrio tubiashii is a major virulence factor for the Pacif Oyster (Crassostrea gigas) larvae. Appl Environ Mocroboil. 74(13): 4101-4110.
Morley, N.J. 2010. Interactive effects of infectious diseases and pollution in aquatic mollusks. Aquatic Toxicology. 96(1): 27-36.
Pipe RK, Coles JA. 1995. Environmental contaminants influencing immune function in marine bivalve mollusks. Fish Shellfish Immuno. 5: 581-595.
October 27, 2010
Lab 5:
Intro:
Today is a very exciting day in lab because we are going to be finishing the dot blot protocol that we started last week to look at genome wide DNA methylation as well as testing the primers we designed on October 12, 2010 by performing a qPCR assay on our cDNA. The portion of the dot blot protocol we will complete is the chromogenic immunodetection protocol in which we will use two sets of antibodies to detect the genomic methylation levels of our DNA samples.
Chromogenic Immunodetection:
1. Unwrap membrane from the plastic wrap that it was stored in for a week.
2. Add 20ml of blocking solution to the membrane in a covered plastic dish and incubate 30min at room temp on a rotary shaker (~1rev/min)
NOTE: blocking solution was the previous week by adding 14ml sterile water, 4ml blocker/diluent A, and 2ml blocker/diluent B.
3. Pour off blocking solution and rinse the membrane 2x in 20ml sterile water for 5min.
4. Incubate membrane in 10ml primary antibody solution (antibody dilution 1:5,000) for 1hr at room temp. on rotary shaker.
NOTE: Primary antibody solution = 10ml blocking solution, 2ul 5-Methyl Cytidine primary antibody
5. Primary antibody solution was removed, saved, and stored at 4 degrees C for possible reuse.
6. Membrane was washed in 20ml TBS-T for 5mins.
7. Added 10ml secondary antibody solution and incubated for 30min
8. Secondary antibody was also removed and saved for possible reuse.
9. Membrane was washed 3x for 3min in TBS-T
10. Rinsed membrane with 20ml water for 2min.
11. Incubated membrane in 5ml Chromogenic Substrate.
NOTE: I waited around after class for ~15min until color began to develop!
12. TA Carolyn dried the membrane and took a picture for us.
Results:
Note: My samples are DM oyster; clean environment
(See discussion for analysis)
Primer resuspension:
-Primers were delivered in a dehydrated form, so before we do anything we need to resuspend them at the appropriate concentration.
-Standard stock concentration of primers is 100uM
-To make 100uM stock, identify the amount of nM of primer in the tube, move decimal once place to the right, and the equivalent number of ul of sterile water to the primers.
SPI Forward primer: 44.9nM was resuspended in 449uL sterile water for a 100uM stock
SPI Reverse primer: 29.0nM was resuspended in 290uL sterile water for a 100uM stock
Quantitative PCR:
NOTE: cDNA was generated yesterday from my own personal samples of hard clam RNA.
1. Make working concentration of pimers (10uM) by diluting 100uM stock 1:10 for a final 10uM stock (10ul primer in 90ul water)
2. Make master mix:
NOTE: We are going to be running a total of 6 qPCR assays but will make enough master mix for 7 to compensate for any volume loss due to pipetting.
| Component |
Volume 1X |
Volume 7X |
Final Con. |
| Master Mix, 2X (Immunomix) |
25ul |
175ul |
1x |
| Syto-13 dye (50uM) |
2ul |
14ul |
2uM |
| F Primer (10uM) |
2.5ul |
17.5ul |
2.5uM |
| R Primer (10uM) |
2.5ul |
17.5ul |
2.5uM |
| Sterile H2O |
16ul |
112ul |
N/A |
4. Add 2ul RNA to two tubes, 2ul cDNA to two tubes, and 2ul water to two tubes.
NOTE: the RNA will control for DNA contamination from cossover of our RNA extractions in our reverse transcription reactions. The water will serve as a negative control to test for contamination in our reagents.
5. Cap tubes and handed over to TA to load/run on qPCR machine.
NOTE: Plate location of qPCR samples are B7-12
Sample 1: RNA
Sample 2: RNA
Sample 3: cDNA
Sample 4: cDNA
Sample 5: water
Sample 6: water
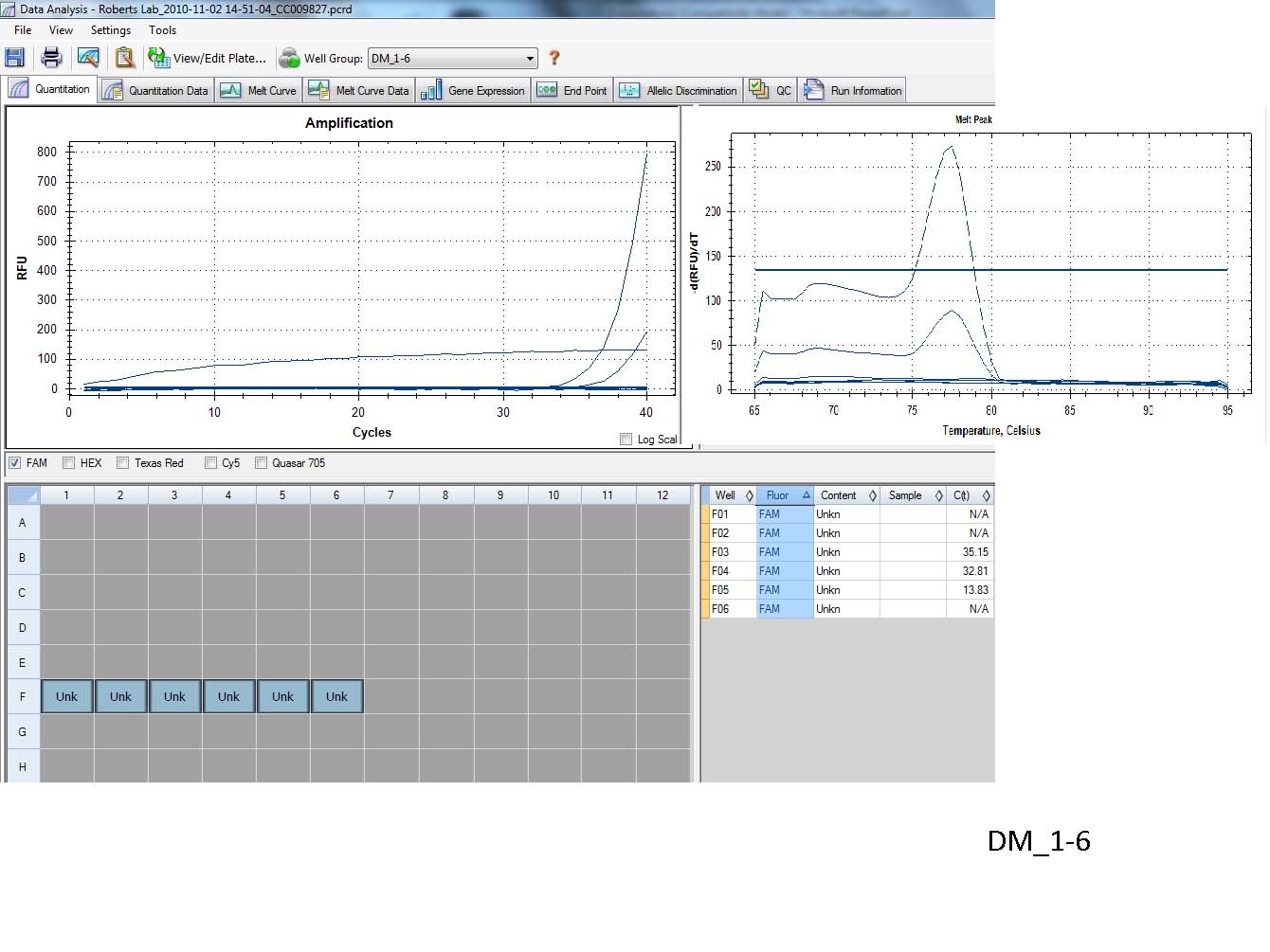
Results:
Discussion:
Dot Blot:
1. It worked...hooray!
2. My oyster samples from a "clean" site look quite different from Peggy's that were from a "dirty" site. w/o any kind of quantitative measurement of the the dot intensities it is difficult to say exactly how different they are, but just by looking at the dots I would say that perhaps the highest concentration in both samples are somewhat similar, but where the differences start to pop out are after you start comparing the dilutions. Peggy's samples maintain a higher level of staining through the dilutions than mine do which suggests that her "dirty" oysters have a higher amount of genomic DNA methylation. Also in support of this are EJ's clean oyster samples that show very low levels of methylation when compared to both mine and Peggy's. So consistently, oysters from "clean" sites have lower methylation while "dirty" sites have higher.
qPCR
1. Success!? The goal of this experiment was to test the specificity and function for a new set of primers that target the SPI gene in hard clams. From the small amount of amplification that I had it looks like the primers are specific which is indicated by the presence of a single peak in the melting curve.
2. At the moment I am not too concerned about the low level of amplification because I would expect SPI to be induced as part of an immune response and I have yet to confirm whether or not the MAX 4 sample used in this experiment is under any kind of immune stress.
What about the dissimilarity of the replicates? -
Reflection:
The goal of todays lab was to carry a number of ongoing experiments to completion and exercise our skills in data analysis. The dot blot was particularly exciting to me since I have never performed on of these before. Just seeing what the data looks like (ie. purple dots on a paper) was a valuable experience that will help with future discussion on epigenetic topics and experimental design. However, I am taking my analysis of the data with a grain of salt, in that I cannot be 100% confident that the differences I am seeing between clean and dirty oysters are "real." There are too many hands working in the lab at one time which can introduce user induced variation. The qPCR was also exciting in that the primers I tested are actually for a project happening outside of class. This information is extremely valuable in deciding how to proceed with this project. The next step in testing these primers will be to test them on samples that are most likely expressing immune response genes. Unfortunately, we do not have the data at this time to confirm whether or not the clam samples are responding to an infection, so a best guess is all we can do at the moment. Hopefully I will see higher, more robust expression in the next experiment so I have a better idea of how the primers behave.
October 26, 2010
Lab 4:
Intro:
Today in lab we will be analyzing our results from the PCR reactions that were run on October 19, 2010 by using gel electrophoresis to separate our DNA fragments by size. What we are all hoping for is that our primers are specific to the genes we are amplifying which means that we will see a single band of the predicted amplicon size as predicted by the number of nucleotides that separate the primers. We will also begin a Dot blot/chromogentic immunodetection assay that will allow us to visualize the genome wide DNA methylation intensity of DNA samples.
PCR Gel electrophoresis:
Notes: The agarose gel we will be using to analyze our PCR samples was prepared on October 19, 2010.
1. 1X TAE running buffer was poured into the gel box until gels were covered.
2. Combs were removed from gel.
3. 5ul of DNA ladder was loaded in the first lane of each well. (HyperLadder1 from Bioline).
4. Loaded 25ul of PCR sample into each well
NOTE: My samples are in lanes 2-5 along the top of Gel#2. Lane 2 & 3 are my PCR reactions and lanes 4 & 5 are my negative controls.
5. Gel was run at 100V for 55min, 150V for 10min, and 85V for 20min. The variation in voltages was because our samples had not run far enough to clearly separate from the primer dimers or whatever else was present so we increased the voltage so the gel would run faster and hopefully complete by the end of class. Once the gel had run far enough we turned the voltage down to continue separation until end of class but no too high as to lose track of it and have our samples run off the end.
6. Gels were visualized on a UV transilluminator.

Cytosine Methylation Dot Blot:
Note: I selected the "clean oyster" DNA sample on the board and received a DNA sample at a concentration of 50ng/ul.
1. Made the following dilutions
| Dilution |
Target Concentration |
ul Water |
ul 20X SSC |
ul of 50ng/ul DNA sample |
| 1 |
0.8ng/ul |
124 |
60 |
16 |
| 2 |
0.4ng/ul |
132 |
60 |
8 |
| 3 |
0.2ng/ul |
136 |
60 |
4 |
| 4 |
0.1ng/ul |
138 |
60 |
2 |
| 5 |
0.05ng/ul |
139 |
60 |
1 |
2. Nylon membrane was cut to a size that would cover 72wells of a 96well plate.
3. Nylon membrane was soaked in 6X SSC for 10min.
4. Filter paper was cut to the size of the nylon membrane and soaked in 6x SSC.
5. Vaccum/Dot blow manifold assembly: first the filter paper was place on the manifold followed by the nylon membrane on top. The 96well plate looking portion of the manifold was placed on top of the nylon membrane and clamped down.
6. DNA dilutions were boiled for 10min to denature and form single stranded DNA. In the mean time 500ul of 6X SSC was pipetted into each well and filtered through by turning on the vaccum manifold. NOTE: Remember to cover any unused wells in the manifold in order to pull a proper vacuum!
7. After the DNA was boiled, it was placed immediately on Ice to "freeze" the DNA in a denatured state.
8. The entire volume of our DNA dilutions (200ul) was added to each well.
NOTE: My samples were loaded in wells B1-5.
9. DNA samples were filtered through membrane by turning on the vacuum manifold.
10. Another piece of filter paper was cut to size and soaked in denaturation buffer.
11. Once filtration was complete, the manifold was disassembled and the nylon membrane was incubated on the filter paper soaked in denaturation buffer for 10min.
12. The membrane was then soaked in neutralization buffer for 5min.
13. The membrane was then transferred to dry filter paper and let to dry in the hood.
14. Once dry, the membrane was wrapped in plastic wrap and placed DNA side down on the UV transiluminator for 2mins @ 120Kj to effectively adhere the DNA to the membrane.
At this point we ran out of time in lab and will leave the WesternBreeze chromogentic immunodetection protocol for next week. The blot should be stable while it's dry until we can pick up where we left off next week.
Results/Discussion:
PCR results look great!! The band obtained is of the predicted size falling between 200-400 base pairs on the DNA ladder indicating an amplicon of around 300 base pairs which is what the primers were designed to do. This PCR in particular looks very clean and specific for the following reasons:
1. Only a single band is present in the first two lanes.
2. The negative control lanes are completely negative.
3. No primer dimer "haze"
-
Reflection:
The purpose of todays lab was to complete our PCR protocol from last week by running our PCR DNA on an agarose gel and visualizing the samples using a UV transiluminator. We were also exposed to the dot blot technique which is an immunodetection based assay that can be used to visualize genome wide DNA methylation trends/concentration between separate samples. I am particularly interested in seeing the fluorescence emitted by the two people in lab that have the "dirty oyster" DNA to see if there is an affect of clean vs. dirty. I do not know the details of the samples, but presumably the "clean" samples came from an area with little water contamination and the dirty came from a more polluted region. I am excited to see if there is any difference and if so what it is?!
October 19, 2010
Lab 3:
Intro
In today's lab we will be using the RNA that the lab purified on October 5, 2010 in reverse transcription reactions that will utilyze the activity of a reverse transcriptase enzyme to generate complimentary DNA or cDNA that will encode the expressed sequenced of a gene. What I mean by that is any post translational modifications (ie. exon splicing) will be represented by the cDNA. Once the cDNA is synthesized, we will essentially be testing the quality of our cDNA using primers provided by the TA to ensure our templates are of sufficient quality for when our designed primers arrive.
Notes:
1. The TA has provided primers for O. nerka and C. gigas. Since the RNA that I isolated is from M. mercenaria, I will be using O.nerka RNA isolated by Chris Gru (Monson?) -
Methods:
Reverse Transcription
1. 5uL of RNA was combined with 1uL oligo dT and 4uL nuclease free water in a 0.5ml PCR tube.
2. Sample was spun to collect contents at the bottom of the tube and then incubated at 70 degrees Celsius for 5min.
3. Sample was spun down again and kept on ice.
3. 5uL of M-MLV 5x reaction buffer, 5uL dNTPs, 1uL M-MLV RT (the reverse transcriptase), and 4uL of nuclease free water was added to the sample.
4. Sample was spun again and then placed in the thermocycler and we used program 44170 (42 degreesC for 60min followed by 70 degreesC for 3min).
5. Samples were spun down and stored on ice.
Polymerase Chain Reaction:
1. Made the following PCR master mix in a 1.5ml centrifuge tube:
250ul GoTaq green 2x mastermix
15ul forward primer HSC71
15ul reverse primer HSC71
108ul nuclease free water
2. Added 48ul of my mastermix to 4 pcr strip tubes labeled 1-4 w/ my initials on the tab.
3. Added 2ul template:
Tube 1&2 received cDNA
Tubes 3&4 received nuclease free water to serve as a negative control (amplification would indicate contamination).
4. Spun tubes to collect sample at bottom and put in the thermocycler. Ran the following program:
1. Denaturation 95 C for 5min
2. Denaturation 95 C for 30sec
3. Annealing 55 C for 30sec
4. Extension/Elongation 72 C for 90sec
5. Cycle steps 2-4 40x
6. Final extension 72 C for 3min
7. Hold 4 C forever!
NOTES: Initially I was using forward and reverse primers for GnRH (gonadotropin releasing hormone) until I pipetted what I thought was GoTaq mastermix which was actually somebodies mastermix from class. I was unable to remake my mastermix because we ran out of GoTaq green so I used the remaining mastermix from Chris which contained forward and reverse primers for HSC71.
Making Agarose Gel:
This was more of a group activity.
My responsibility was to secure the gel plate in the gelbox so that it would not leak when the agarose was added and positioning of the gel combs.
Once that was accomplished, the following procedure was carried out by the rest of the class...
1. 2g of agarose powder was combined with 150ml of 1xTAE in a 1L flask and was microwaved for 3min with constant swirling to ensure homogenous mixing of agarose. Solution turned from opaque white to clear when agarose was fully dissolved.
2. Solution was left to cool for several minutes before addition of 12ul EtBr.
3. Solution was poured in the expertly prepared gel boxes and left to cool/polymerize into a gel. -
4. Once polymerized, gel was wrapped in plastic and kept in the fridge until next week when we will use it to test the success of our PCR.
Results/Conclusions:
Stay tuned...
Today was full of procedural techniques to amplify cDNA from a target gene, the results of which we will not know until we visualize the PCR product on the agarose gel next week.
Reflection:
Todays lab was all about familiarizing ourselves with more standard laboratory practices and reagents such as the use of thermocyclers, gel preparation, and enzymatic reactions involved in DNA replication. Next week we will know the outcome of these processes upon examination of our agarose gels. We will then be able to proceed with our experiment to design and test primers that we designed ourselves on the cDNA we generated today. If all goes well, these primers will ultimately be used to quantify the cDNA that was reverse transcribed from our RNA in an attempt to quantify the level of transcription for our chosen genes.
October 12, 2010
Lab 2:
Intro: The purpose of todays lab is to finish the RNA extractions using Tri-Reagent that we started last week. RNA will be quantified using a nanodrop. We will also further analyze our protein extractions from last week by running them on an SDS page gel.
Methods
RNA extractions using TriReagent:
1. I will be isolating RNA from eight gill tissue samples from hard clam Mercenaria mercenaria.
2. Tissues were homogenized in 200uL of Tri-Reagent using a sterile blue pestle. 800uL of Tri-reagent was added after homogenization to make a total of 1ml. Note: squirted the 800uL of Tri-Reagent around the tip of the pestle into the tube to wash off any residual tissue pieces that were attached to the pestle.
3. Samples were kept on ice until homogenization was complete.
4. Once all samples were homogenized, they were incubated at room temperature for 5min.
5. Added 200ul chloroform (in the hood) to each tube and vortexed for 15s.
6. Samples were incubated at room temp. for 2min and then centrifuged at 12,000xg for 15min
7. Centrifugation resulted in the separation of the sample into two layers. The upper clear layer contains the RNA while the lower contains DNA and cell debris.
8. The upper layer was transferred to a new tube where 500ul of isopropanol was added to precipitate the RNA.
9. Samples were incubated at room temp for 5min and then centrifuged at 12,000xg for 8min.
10. Centrifugation resulted in the pelleting of the RNA. Isopropanol was removed by pipetting and the RNA was washed by the addition of 1ml 75% EtOH.
11. Samples were centrifuged again for 5min at 12,000xg to recollect any RNA that was dislodged from the bottom. EtOH was removed by pipetting and samples were left to air dry.
12. Once samples were dry, they were resuspended in 50ul of sterile 0.1% DEPC treated water.
13. Samples were kept on ice for 30min to re-suspend.
14. 2ul from each sample was used to determine their concentrations using a Nanodrop spectrophotometer.
15. RNA was stored at -80 degrees celsius.
SDS page gel electrophoresis:
1. Combined 15ul of my protein samples (prepared on October 5, 2010) with 15ul 2x SDS reducing sample buffer in a screw cap tube.
2. Remaining protein was frozen at -20 degrees.
3. Sample containing 2x sample buffer was boiled for 5min.
4. Sample was spun for 1min @14k to collect sample at the bottom of the tube.
5. Used a gel loading tip to load all 30ul onto an SDS gradient gel (8-20%).
6. Gel was run at 150 volts for 1hr.
Note SAMPLE LOCATION = Gel 1 Lane 12
7. Gel was removed from cassette and stained in coomassie stain for 5min.
8. Coomassie stain was removed by pouring back into container.
9. Destain was added and continuously replaced ever 10-20min for ~1hr and then left o/n to finish destaining.
Results/Conclusions:
1. Most samples were in the acceptable 260/280 absorbance ratio range of 1.8-2. Samples MAX6-8 and CA-10 have lower 260/280 ratios than I would like. This result could be due to DNA contamination among other things. For now I am not too worried and will wait and see what the absorbance ratios are after I DNase treat the samples.
2. It looks like everybody's protein extractions worked well and share a similar protein pattern. It is interesting that the pH treated sockeye sample that was run in lane 9 of gel 1 has a major band at ~20kDa that does not appear in other sockeye lanes. A few possibilies for this are that a new protein is showing high levels of induction from pH treatment, the major bands in the other lanes that are slightly below 20kDa has undergone a conformational change in response to pH treatment (may or may not be upregulated as well, it looks like significantly more protein was loaded in that lane), or the band shift is an artifact of loading too much protein in that lane. Good. -
Reflection
The purpose of this lab was to learn the techniques used to isolate pure RNA samples that can be used in an array of studies from qPCR gene expression, to next generation sequencing. The SDS protein gel electrophoresis portion of the lab was performed to familiarize us with the practices involved in various protein based assay such as western blots and immunoprecipitations. In this case, we were visualizing the results from the protein extractions performed last week.
Primer Design
The sequence for the serine protease inhibitor was finally tracked down....although it is not yet posted in genebank. I used Primer3 to help design the following primers:
Start Len Tm GC% any 3' Seq 6 LEFT PRIMER 135 20 60.51 55.00 4.00 0.00 CGCTGGTGTAGCACTGTTTG
RIGHT PRIMER 289 20 59.72 50.00 4.00 2.00 CGCATTTCCTGGTACAGTCA
These primers were chosen over others for the following reasons:
1. Tm close together.
2. They lack a long string of GGGGGs and CCCCCs that were present in other primers.
3.. They are not 100% homologous to any sequences when using BLAST
4. They dont end on G or C which can affect how they primer lays down on the DNA
5. They also dont end on a repeat which can cause the polymerase to skip and make errors.
October 5, 2010
Intro:
Today in lab we will be performing RNA extractions using TriReagent and Protein extractions using a cellLytic solution and quantified using a Bradford assay on a tissue of our choice. My tissues will be different for each extraction since I already have hard clam (Mercenaria mercenaria) for my own research that I need to extract RNA from. For the Protein extractions I will use black abalone heart tissue provided by the TA. At the end we will select three candidate genes and obtain their mRNA/cDNA sequence for subsequent PCR assay development.
Materials and Methods:
RNA extractions:
Today I only proceeded as far as cutting the gill tissue (stored in RNAlater) of my clam samples in half, one as a back up and one for extractions. Both pieces were frozen at -80 degrees C. They will be homogenized and extracted using Tri-Reagent during next weeks lab.
Protein extractions:
1.Obtained black abalone heart tissue from TA
2. Total weight of tissue = 10mg
3. Added 500ul CellLytic MT solution to the tube containing the tissue.
4. Using a sterile blue pestle, I homogenized the tissue until no visible chunks remained.
5. I inverted the tube several times to mix and then spun for 10min @ 14K.
6. Supernatant was transferred to a new tube labeled “Protein/DM/10-5-10”
Protein Quantification Using Bradford Reagent:
1. Diluted “Protein” sample generated above 1:2 in diH2O so that the spec readings would fall in the range of the standard curve.
2. 1.5ml of Bradford reagent was added to the tube containing 30ul of diluted protein, and another tube labeled “blank” containing 30ul of diH2O.
3. Samples were incubated at room temp. for 10min.
4. The spectrophotometer was set to ab595 and 1ml of the “blank” samples was used as the “zero” reading.
5. Measured absorbance of 1ml from the diluted “protein” sample. Removed from spec., mixed with pipette, and took a second reading of the same sample to calculate an average.
Note: The remaining protein not used for protein quantification was stored at -80 degrees C.
Calculating concentration from standard curve:
- To calculate the concentration of my sample, I substituted my absorbance value in place of “x” in the equation for the best-fit line of the standard curve. I then multiplied by two to account for the initial dilution of the sample from step1 of the protein quantification method listed above. I also calculated the total amount of protein that was extracted from the tissue by multiplying by the amount of CellLytic solution the tissue was homogenized in.
- Best fit line equation: y=1013.9x
- Calculation: y=1013.9(0.134), y=135.86ug/ml
- Final protein concentration = 135.86 x 2 = 271.72ug/ml
- Total protein = 271.72 x 0.5ml = 135.86ug
Results:
135.86ug of protein was extracted from a 10mg piece of black abalone heart tissue.
Discussion/Future Experiments/Reflection
Overall the protein extraction was a success. Time will tell for what happens with the RNA extractions next week. I think the overall goal of the lab was accomplished in that we were all (re)exposed to the laboratory setting and were familiarized with how to use pipettes, centrifuges, and general lab safety techniques. These basics will be invaluable in both future classroom exercises and as we proceed with our own research. It’s always nice to see concepts that we talk about in lectures unfold before us at the lab bench!
In preparation for future labs I used the NCBI EST database to obtain mRNA/cDNA sequence for three possible genes of interest to use for primer development. Genbank accession numbers are as follows: Alternative oxidase (Genbank#FJ607016), Rel (Genbank#DQ673622), P53-like protein (Genbank#EU621671). Any of these sequences can be used to design primers for qPCR gene expression assays -