November 16 2010
Last week in lab we calculated the amount of copper needed for 1.3 ppm unfortunately due to a slight error in the calculations we accidently ended up adding 130 ppm of copper while the oysters didn't die immediately which is what I thought might happen. Friday we commenced tissue sampling my station was sampling of the hemolymph from the heart. The sampling went very well. With the exception of 2 oysters that were already dead before we started the experiment and were mistaken for alive. I expect that the rest of our process will start picking up now that we have our samples to analyze. For this process I will be doing mainly qpcr which i believe we will be starting Tuesday. I also took pictures which i will be posting soon.
November 9 2010
Today we discussed the setting up of our class projects. The project i am involved with is the oyster stress project. We set up a date for collecting sample organisms. This we did Thursday and Friday we put the oysters in to the experimental set up. So far our timetable will be good. On tuesday we start the experiment and on friday we will have our tissue sample that we can analyze.
Proposal
IntroductionSalmon are an economic staple for many countries and an environmental necessity for many ecosystems. So understanding how a changing environment in combination with disease and pollution is affecting salmon eggs from hatching till fry stage, their most vulnerable and important stage of development, is of critical environmental and economic importance. This is also important in a hatchery were the goal is to insure that developing eggs are healthy and less prone to disease and crippling development mutations. While there is evidence that temperature difference has major impacts on development (Wedekind, Kung 2010) and that exposure to heavy metals and toxins affect egg fertility and later development (Stekoll, et al 2009). This lead me to the idea of testing combinatios of different stressors on egg development. Since in the natural environment especially one that has been particularly disturbed these eggs would come in contact with a cocktail of different chemicals and diseases. So I am proposing in my experiment to test the weather these combinations of problems are a sum of each other or wether certain ones can combine and become much more deadly than either would have been on its own.
Materials and Methods
With the salmon run coming back to the hatchery fertilized salmon eggs would be rather easy to procure the question is in the number needed. Roughly between 100-200 eggs per set and at least 2 sets for each condition would be appropriate each set would be in its own incubation tray. The conditions would be: control exposure to heavy metals or other contaminants(PCBs) exposure to warmer temperatures(20-22 C) exposure to pathogens exposure to heavy metals or other contaminants, warmer temperatures, and pathogens exposure to heavy metals and other contaminants, and pathogens exposure to warmer temperatures and pathogens and finally exposure to warmer temperatures and to heavy metals or other contaminants The fish embryos changes noted and photographed while dead embryos would be removed and die-offs would be recorded also tissue samples would be taken to analyze for gene expression through taking the RNA reverse transcribing to cDNA and then performing quantitative PCR on it.
TImeline
The first week would be devoted to experiment set up and egg collection. After that it would be a matter of periodic checking for 3 weeks or so with weekly tissue samples and analysis. The final week would be reserved for data analysis and writing of the paper.
References
Stekoll, et al. (2009). Response of the early developmental stages of hatchery reared salmonids to major ions in a simulated mine effluent. Aquaculture, 172-181.
Wedekind, C., Kueng, C. (2010). Shift of Spawning Seaon and Effects of Climate Warming on Developmental Stages of Grayling (salmonidae). Conservation Biology, 1418-1423.
November 2 2010
Introduction
Today was an exciting day of lab because we finally got to use the primers we had designed in testing for gene expression. We also finished up our dot blot analysis.
Method
Chromogenic Immunodetection/Dot Blot Analysis
We started by taking the dried membrane and adding 20 ml of blocking solution and set for 30 min Pour out blocking solution and then rinse twice with 20 ml of pure water for 5 min put the 10 ml of primary antibody solution on the membrane for 1 hour pour off the antibody solution and save wash with 20 ml of TBS-T then add 10 ml of secondary antibody solution for 30 min wash three times with TBS-T for 3 minutes rinse for 2 min with 20 ml of water Finally incubate in 5ml of Chromogenic substrate it then developed and a picture was taken the picture below is the resulting development
Below is the results from the dot blot analysis
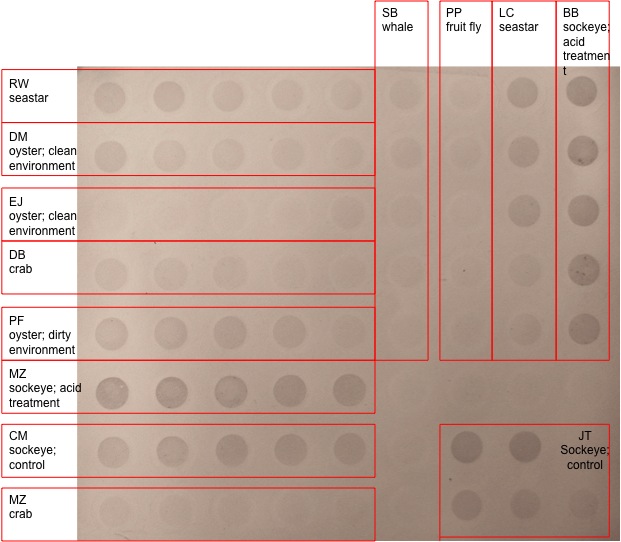
MeDotBlot_FISH441_F10_LABLED.jpg
Quantitative PCR
First the primers were resuspended this was done by moving the decimal over in the nM and adding that many ul of pure water then we mixed the master mix enough for 7 so there was a margin of error These are the quantities for 1 mix and then the amount we used along with the final concentrations of each
| Component |
Volume 1X |
Volume 7X |
Final Con. |
| Master Mix, 2X (Immunomix) |
25ul |
175ul |
1x |
| Syto-13 dye (50uM) |
2ul |
14ul |
2uM |
| F Primer (10uM) |
2.5ul |
17.5ul |
2.5uM |
| R Primer (10uM) |
2.5ul |
17.5ul |
2.5uM |
| Sterile H2O |
16ul |
112ul |
N/A |
4. Then we added 48 ul of the master mix to each of 6 PCR tubes 5. I added 2 ul of RNA to tubes 1 and 2; 2 ul of cDNA to 3 and 4; and 5 and 6 i added 2 ul of water 6. then i spun the tubes down and handed them over to be run in the PCR Below is a picture of my PCR results
Results My results were mixed my dot blot analysis came out good you can see a good gradient from the different concentrations. My PCR results were a bit of a problem while my pcr went great a mistake in one of the earlier labs made it hard to tell if the copper oxidase gene was actually being expressed. My error was most likely when pipetting out the RNA when we separated it. This led to DNA contamination and more of a showing that the gene is in the genome and not weather its being expressed.
Discussion While it was really cool to see that my primers worked it was unfortunate that i couldnt tell about gene expression because of the contamination. Though mabe i will get another chance to try out my primer if we test copper on oysters for our class project. As for the dot blot methylation tests they were interesting especially when contrasting the salmon with acid treatment with then untreated one
I know you had problems with your lab notebook, but could you add the dot blot picture? -
October 26 2010
Introduction
Today in lab we ran 2 tests we did a gel electrophoresis test on our PCR products from last time and we started a dot blot/chromogenetic immunodetection assay which detects methylation on DNA this we will finish next lab.
Materials and Methods
Gel Electrophoresis
- Add 1X TAE buffer until gel is completely covered
- Gels were then loaded with our PCR samples 25 ul and the sample ladder 5 ul
- The gel was then run at 100V for 55min then 150V for 10 min and finally 85V for 20 min
- Gels were then set in UV light to show where the DNA was and a pic was taken and posted on the message board
Here is a link to the picture of the ladder and my gel runs the first 2 are the tests the last 2 the blanks
http://2.bp.blogspot.com/_MdDM57wDmt8/TM5ZFUc0TyI/AAAAAAAAABI/lDVpyuoqCnc/s1600/Gel2_PCR_FISH441F10-1.jpg
Dot Blot/Chromogenetic Immunodetection Assay
- First we selected tissue samples mine was of a crab
- Next we made different dilutions mine went like this
| Dilution |
TARGET
|
ul of H20 |
ul of 20X SSC |
ul of 50ng/ul
|
- || 1 || 0.8 ng/ul || 124 || 60 || 16 ||
- || 2 || 0.4 ng/ul || 132 || 60 || 8 ||
- || 3 || 0.2 ng/ul || 136 || 60 || 4 ||
- || 4 || 0.1 ng/ul || 138 || 60 || 2 ||
- || 5 || 0.05 ng/ul || 139 || 60 || 1 ||
- A nylon membrane that was cut to size was soaked in 6X SSC for 10 min
- Filter paper cut to the same size was soaked in 6X SSC as well
- the vacuum dot blot assembly was put together so that the filter paper was under the nylon which was under the wells
- first run each well was filled with 6X SSC
- we then boiled our DNA for 10 min to denature then put on ice
- then the entire amount of each sample 200 ul
- My samples are in D1-5
- the vacuum was turned on to filter the samples through
- after this was done the nylon membrane was placed on a piece of filter paper with denaturation buffer for 10 min
- then for 5 min the nylon was soaked in neutralization buffer
- the nylon was then dried in the hood this is where we left off and the rest will be completed next lab
Results
I was happy with my results on the Gel run since it showed my PCR had gone well with clear differences between my blanks and my tests. As for the dot blot everything seems to have gone well so far but we will see for sure next time.
Discussion
My results from the gel test were great news since it shows I could correctly run a PCR this is heartening since we will be getting to use our own primers soon and i wouldnt want to screw that up. The dot blot mechanism was interesting mostly cause I've never seen or heard of that particular method before I am interested to see its completion.
October 19 2010
Introduction
This week we reverse transcribed our RNA to cDNA so we could practice PCR techniques in preparation for next lab where we will be using our own primers that we designed. We also made agarose gel to analyze the DNA after we have run the PCR. Good -
Materials and Methods
- PCR and Reverse Transcription
- Thaw RNA sample and add 5 ul to a tube with 1 ul of oligo, and 4 ul of nuclease free water
- Put RNA sample back in the freezer
- Put mix in the thermocycler for 5 min at 70 C
- Centrifuge and then add 5 ul of M-MLV 5X, 1 ul of M-MLV RT, 5 ul of dNTPs, and 4 ul of nuclease free water
- Put back in the thermocycler at 42 C for 60 min with 3 min at the end at 70 C to inactivate it
- Spin down and put on ice
- In the Master Mix tube add 250 ul of GoTaq Green Master Mix 2X, 15 ul MnSD (?) forward primer and 15ul of the p450 reverse primer, and finally add 108 ul of nuclease
- get 4 PCR tubes label 2 test and 2 blank
- put 48 ul of master mix in each tube
- in the blanks put 2 ul of nuclease free water
- in the tests samples put 2 ul of cDNA
- then put in the thermocycler with this program in place first 95 C for 5 min, then 55 C for 30 seconds, 72 C for 90 seconds and repeat those steps about 40 times after that run 72 C for 3 min
- after this put on ice for testing next time
- Since we ran out of GoTag GoTaq-
storercg I shared with someone else.
Agarose
- Mix 2 g of agarose gel with 150 ml of 1X TAE
- Heat until agarose is completly disolved
- when solution cools enough to touch add 12 ul of ethidium bromide
- pour gel in a mold
- after it has set store for next time
Results
The agarose gel set nicely and we will use it next time to see how our PCR and reverse transcriptions went by running a gel electrophoresis.
Discussion
So far I felt this is gone well and that I will be ready for when our primers come in. Though we will see for sure next week how well my PCR turned out. I have enjoyed the chance to design our own primers since i have never done that myself and cant wait to test mine.
October 12 2010
Introduction
This week we performed 2 procedures. We continued the RNA extraction from last lab and we performed further analysis of the protein we extracted last lab by running a SDS-PAGE. My sample was the Pacific Oyster C. gigas Treated with Cu treatment C.
Materials and Methods
SDS-PAGE
- Thaw protein solution in snap cap that we prepared last time
- Add 15µL of protein solution to 15µL of 2X Reducing Sample Buffer
- Put protein solution back in freezer
- Briefly centrifuge
- Boil sample for 5 minutes
- Put together gel box
- Centrifuge for 1 minute
- Load entire sample into assigned well
- Turn power supply on and run gel for roughly 45 min
- Turn off power supply and remove gel
- Use Coomassie stain on gel
- Put on shaker to set stain
- Rinse gel with acetic acid to remove excess stain.
- Continually replace acetic acid till gel can be clearly read
- Once bands are visible record results
RNA extraction part 2
- Take RNA gathered last lab and Thaw
- Add 200µL of chloroform under the fume hood
- Vortex sample for 30 seconds
- Keep sample at room temperature for 5 minutes.
- For 15min
- Gently remove tube from the microfuge without disturbing it
- Transfer the super
- Add 500µL isopropanol to the RNA
- Stir and then store at room temperature for 10 min
- Spin for 8 min at max speed
- Without disturbing the RNA pellet remove the supernatant
- Add 1 mL of EtOH to pellet.
- Vortex to dislodge the pellet from side
- For 5 min spin at 7500g
- Remove as much EtOH as possible and then let dry
- Add 100µL of 0.1% DEPC-H2O and the re suspend the pellet
- Put in a 55 degree water bath for 5 min
- Check the amount of RNA with the nanospectrometer
Results
SDS-PAGE
The gel went really well with everyones results coming out really well clear. My sample seems well since it matches some of the other oyster samples. The link below has a picture of my results. The 1st lane is the size standard and the the 4th is my result.
Protein+Gel+1+FISH441+F10.jpg
RNA extraction
My RNA extraction also seemingly went well my results were very high compared to some of the others I saw with 1692.9 Ng/µL I believe these are good results though these high numbers could be potentially from contamination. True -
Reflection? -
Oct 5 2010
Introduction
In Lab we performed two main tests procedures -
Materials and Methods
RNA Extraction
- Got my sample Pacific Oyster Cu treatment C
- prepared a labeled tube with initials and date DDB 10/5/10
- I then proceded to add 500 µl of trireagent to the tissue sample
- Then I homogenized the sample with a provided pestle
- I vortexed for 15 seconds and turned it in to be stored at -80 degrees C
Protein Extraction and Analysis
- Got sample Pacific Oyster Cu treatment C and noted recorded weight 13 mg
- Added 500 µl of CellLytic solution
- once again used a blue pestle to homogenize the solution
- inverted the tube a couple of times and the spun it in a refrigerated centrifuge for 10 min
- I then transfered the supernatant to a new tube that I labelled Protein DDB 10/5/10
- then we ran Bradford Analysis
- 2 tubes were used in the first the protein sample was diluted in half by adding 15 µl of protein to 15 µl of water this was labelled Test. In the second 30 µl of water was addedthis was labelled blank>
- then 1.5 ml of Bradford reagent was added to each tube and samples were left at room temperature for 10 min
- The sample was then analyzed twice in the spectrophotometer that was set to 595 nm after zeroing it with the blank
- an average of the 2 reading was taken and this was used to calculate the final amount of protein (0.117)
- Leftover protein was turned in to store at -80 degrees C
Calculations
My calculations were accomplished by plugging our Bradford assay results into the equation we got from the standard curve provided.
The equation Y=mx+b with b=0 it could be changed to Y=mx with m provided as 1013.9 x is where our analysis info goes
so Y=1013.9(0.117)= 118.626
since the the original sample of protein was diluted by half we double the number from this equation
118.626 * 2= 237.252 µg/mL
Results
The RNA extraction will be stored for use next lab.
From 13 mg of tissue I was able to extract 237.25 µg/mL
Discussion
While the first part of the lab was part of a series we will be continuing the following weeks so the effectiveness of my RNA extraction wont be seen till later. On the protein extraction I felt I did well since my numbers somewhat matched those of people working around me. The main point of this lab was getting us used to common molecular biology techniques. In addition to this it also helped orient us with the lab and procedure. In preparation for next lab I will be finding 3 gene sequences to bring to class.
Do you have any genes of interest? I know you provided this info for the weekly quiz, but could you could please post them here? -
281 Candidate genes for larval growth heterosis (Crassostrea gigas) Crassostrea gigascDNA, mRNA sequence
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;">**IDENTIFIERS** **dbEST Id:** **49616663** EST name: 281 GenBank Acc: EX151622 GenBank gi: 291621473</span>
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;">GATCAACTGCGGAGTTTGCCAACGTCTCTATGCTATGTGCATCAACTGCTGCATGACGCC
ATGCTTCGAATCTTGTGGTGGAATCTTCCATCATTTTAAGAAATGAACATAGCAACTGAT
TAAACTTTTCTAGTCTTCGTCTGACTTGTTTTTCAAACTTTATATGTGATTCATTTTTCA
CATTGCATCTGTGATAAATAATATTTATATTTCATGTCCAAAGTTTCGGATTTTTCGCCG
ATGTGTACATGTTTTTATGTAAGAGAGAAATAAAGTTCCTTTTTATTTAAAGAC</span>
281 Candidate genes for larval growth heterosis (Crassostrea gigas) Crassostrea gigas cDNA, mRNA sequence
Primer
| 1 |
ATGC |
| 51 |
|
| 101 |
|
| 151 |
|
| 201 |
| Left Primer 1: |
|||||
| Sequence: |
|||||
| Start: 5 |
Length: 21 bp |
Tm: 59.9 °C |
GC: 42.9 % |
ANY: 6.0 |
SELF: 3.0 |
| Right Primer 1: |
|||||
| Sequence: |
|||||
| Start: 192 |
Length: 20 bp |
Tm: 60.0 °C |
GC: 45.0 % |
ANY: 6.0 |
SELF: 1.0 |
| Product Size: 188 bp |
Pair Any: 4.0 |
Pair End: 0.0 |
|||
ACATGTACACATCGGCGAAA
Crassostrea gigas multicopper oxidase mRNA, complete cds
| Pair 1: |
|||||
| Left Primer 1: |
|||||
| Sequence: |
|||||
| Start: 385 |
Length: 20 bp |
Tm: 60.0 °C |
GC: 40.0 % |
ANY: 5.0 |
SELF: 2.0 |
| Right Primer 1: |
|||||
| Sequence: |
|||||
| Start: 586 |
Length: 20 bp |
Tm: 60.0 °C |
GC: 45.0 % |
ANY: 6.0 |
SELF: 0.0 |
| Product Size: 202 bp |
Pair Any: 4.0 |
Pair End: 2.0 |
|||
Forward:tttgtgacgcaatgtccaat
Reverse:cccaattgtgattccattcc
Crassostrea gigas HSP70 mRNA for 70kDa heat shock protein, complete cds
Crassostrea gigas HSP70 mRNA for 70kDa heat shock protein, complete cds
2,278 bp linear mRNA
AB122063.1 GI:46359613
Primer
| Left Primer 4: |
|||||
| Sequence: |
|||||
| Start: 564 |
Length: 20 bp |
Tm: 60.1 °C |
GC: 50.0 % |
ANY: 3.0 |
SELF: 0.0 |
| Right Primer 4: |
|||||
| Sequence: |
|||||
| Start: 767 |
Length: 20 bp |
Tm: 60.0 °C |
GC: 50.0 % |
ANY: 4.0 |
SELF: 2.0 |
| Product Size: 204 bp |
Pair Any: 5.0 |
Pair End: 2.0 |
|||
tcaccgttcctgcctatttc
tggtaaggatggacacgtca
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;">caagtttgaa acttcgaaac tttgagtcaa ctagactacg attcagtaac agtacataca
61 ataatttgtt ttatctacaa ttgaaaaatt tattcaatcg ctactttcac tttagtgttt
121 acaaacaagc atggcaagta aagctccagc catcggcatc gacctgggaa cgacgttttc
181 ctgtgtcggt gtatttcaac atggaaaagt ggaaatcatc gccaacgacc aaggaaacag
241 aacgacgccc agctacgtcg ccttcacaga cacagaaaga ctgataggag atgcggctaa
301 aaaccaggta gccatgaacg ccaacaatac aatctttgac gccaagaggc tgataggccg
361 caagttcaac gacgacagtg tacagtccga catgaaacat tggccgttca cggtgatcaa
421 tgatggagga aagcccaagc tagaagtgga gttcaagaac gagaaaaaga gatttacccc
481 cgaagaaatc agctcaatgg tgctgaccaa aatgaaggag acagcagaag cctacttggg
541 acaaactgtc cgagacgcag tcgtcaccgt tcctgcctat ttcaacaacg cccagagaga
601 ggccactaaa gacgccggag tgatagccgg tctcaatgtt ctcaggatag taaacgaacc
661 cacagctgct gctctggctt atggtctgga taagaacatt tccggtgaga aaaacgttct
721 catctttgac ctcggaggtg gtacatttga cgtgtccatc cttaccattg atgagggttc
781 catatttgag gttcgctcca ctgcagggga tactcacctg ggaggtgaag atttcgacaa
841 cagaatggtc aatcactttg tgcaggaatt caagcgcaag tacaacaaag acatttcaaa
901 aaataaccgt tcactgagac gactcaggac ggcttgcgag cgtgccaaga gaacactgtc
961 cagcagttct gaggccaaca ttgagatcga ctcgctgttt gagggaatgg acttctacag
1021 caaaatcaca cgggctagat ttgaagaaat gtgtgcagat ttgttccgtg gaaccttaga
1081 acctgtagag aaagccctga gggacgccaa aatggacaaa tctaaaattc acgaggtggt
1141 tttggttgga gggtcgacaa gaattccaaa gatccagaaa atgctgcaag acttcatggg
1201 cggaaaagaa ctgaacaagt cgatcaaccc agacgaagct gtggcctatg gtgccgctgt
1261 ccaagcggct attttgaagg gagataagag tgatgccatc aaggacgttc tcctggtcga
1321 tgtcactcca ctgtctctag gcattgaaac tgctggaggg gtcatgacaa agatcgttga
1381 acgaaatgcc aagattccta ccaaagcatc acagaccttc accacatact ccgataacca
1441 gcccggtgta tccattcagg tgtttgaagg tgaacgagcc atgacaaaag acaataacaa
1501 actaggtact tttgaactga atggaattcc tcccgcccct cgaggtgtcc cacagattga
1561 cgtggagttt gacattgacg ccaacggtat cctgaacgta tcagccaagg acaagagcac
1621 gggaaagtcc aataaaatca ccatcaccaa cgacaaaggt cgtctgagta aggccgacat
1681 tgagagaatg gtgaacgagg ccgagacata caaagaggaa gacgataaac agcgtcagag
1741 gatagcggcc cgcaatcagt tagaatccta tgtcttcacc gtcaaacagg cagcagagga
1801 cacaggagac aaacttcagt ctgaagacaa agagacaata tccagggtgt gcagtgaaac
1861 agtgtcctgg ctcgacaaca atgccctggc agaggttgac gaatatgaat tcaaactgaa
1921 ggaggttcag aaggtgtgtt cacccatcat ggccaaacta caccagaacg ggtccacggg
1981 aaatccagga ccagctagtt ctagtcaggg tccgactgta gaggagatgg attaagtgtg
2041 aagaattact taatgataat aaaacctcaa attgtattat gaacttagat tggtactttc
2101 tatgaaagat gtattaaatg ttgaatgtag tattaaaatt tgtagttgtt aatttatttg
2161 agagcattat tttttttttt tacagattac gatatttttt tttacttata caattgttgt
2221 tattcaatgt tgataaaata aaatcatttg aaaattatct aaaaaaaaaa aaaaaaaa</span>