-Summary: Isolation of V. tubiashii RNA from cell pellet.
Procedure:
Add 0.5 mL TriReagent to pellet and mix with pestle.
Add additonal 0.5 mL TriReagent to the same tube.
Vortex the tube for 15 sec.
Give to Mac for -80 storage.
Results:
N/A
Conclusions/Next Steps:
Precipitate of RNA will be washed and eluted
Quantification
10-6-09
Summary: Protein extraction of V. tubiashii
Procedure:
Add 0.5 Ml of CelLytic MT to Vt pellet in labeled tube
Homogenize with pestle
Invert several times to mix tube
Centrifuge for 10 min at max speed at refrigerated temp
Put supernatant into new labeled tube
Dilute 15 ul of supernatant with 15 ul DI water
Freeze rest of supernanant at -80
Add the 30 ul of 1:2 diluent to 1.5 ml Bradford Assay
Incubate at RT for 10 min
Make Blank for cuvette: add 1.5 ml Bradford Assay to 30ul of DI water and label as "blank". Incubate for 10 min
Calibrate spectrophotometer with 1 ml Blank made in pervious step (mix well)
Pipette 1 ml of sample into cuvette (pipette up and down to mix)
Read absorbance of sample
Perform duplicate readings and average the values
Results:
Absorbance results: 0.301 and 0.303 = 611.19 ug/mL total protein in sample
Conclusions/Next steps:
Readings were within value ranges of standard curve.
Looks like I have sufficient total protein in supernatant to move forward with SDS-PAGE gel.
10-11-09
Summary - Picking Vt primers to use for RT-PCR
Procedure:
Lit search for interesting genes. I was looking for genes responsible for adherence to host tissue and possibly some gene that is expressed in response to different environmental conditions, especially pH.
Found two genes of interest:
1. TcpA -
toxin-coregulated pilus in several Vibrio species; located on major subunit
2. RpoS -
may be expressed in response to starvation, osmotic shock, oxidative stress and temperature in related species. May respond to pH shifts as well
To develop primers:
Used NCBI website to search for these genes in Vt
No luck
Searched for these genes in other Vibrio species on NCBI
BLASTed the nucleotide sequence I found against nucleotide DB and ESTs looking for any Vt sequence similarities
No luck, but found related Vibrio sp. appeared to have similar sequences
Used ClustalW website to align the Vibrio species that had these genes sequenced
Then using the alignment data I looked for regions of sequence that had similar regions between strains, then used those regions (or approx regions) to develop primers
Used related species to develop primers for both genes using Primer 3 website:
- For TcpA, I used V. mimucus to develop primers
- And for RpoS, I used V. cholerae to develop primers
Specified: Tm of 58-62C, product size of 100-150 bp, Max self - 4, Max self complementary - 1
Results:
Primers:
RpoS:
forward: CAATGCCGATCCTGAGTTT
reverse: TCTTCCAAGGTCGATGGTTC
Tm: 60C
Product size: 144 bp
TcpA:
Forward: AGTTGCACTGACACAGACATACC
Reverse: ACCTAAACTAACCAAGCCTGAAGT
Tm: 59C
Product size: 100 bp
Next Steps:
Put on class spreadsheet
Order primers
Reconstitute and use on Vt for PCR to test primer sets
10-13-09
Summary:
RNA Extractions (cont'd from 9-6-09) of Vt pellet
Procedure:
Thaw tube for ~5 min
add 200uL of chloroform to tube and vortex for ~30 sc
incubate at RT for 5min
Centrifuge for 15 min at full speed
Collect aqueous phase of the tube, do not disturb any other layers, and put aqueous phase into new tube
add 500uL of isopropanol to tube containing RNA, invert to mix
Incubate at RT for 10 min
Spin for 8 min at high speed
Remove supernatant, leaving the pellet undisturbed
Add 1 ml 75% EtOH and briefly vortex
Centrifuge 5 min at 7500 g
Remove supernatant, leaving the pellet undisturbed
Spin again briefly and remove any left over EtOH
Leave tube open to dry pellet at RT for no more than 5 min
Resuspend pellet in 100uL of 0.1% DEPC-H2O and pipette up and down to resuspend the pellet
Incubate in 55 degree C water bath for 5 min
Flick a few times and quant sample using spectrophotometer
TO QUANT:
use nanodrop reader in Roberts Lab
Place 2 uL of DEPC-H2O onto the pedestal to blank the spectrophotometer
Pipette 2 uL of RNA sample onto pedestal
Press "Measure" and record findings
Results:
Note: I never really saw a pellet in the extraction process
130.5 ng/uL in RNA sample
A260: 3.262
A280: 1.804
A260/A280: 1.81
A260/A320: 1.26
Next Steps/Conclusions:
Reverse transcription for RT-PCR!
Looks like even though I couldn't see a pellet in the extraction process, there was a good amt of RNA in my sample. My ratio of A260/A280 could've been closer to 2, meaning it might not be a perfect RNA sample, but we'll see what happens in the PCR process.
10-13-09
Summary:
SDS-Polyacrylamide Gel Electorphoresis from protein extractions done on 9-6-09
Procedure:
Start a boiling water bath
Thaw protein sample
Add 15uL of protein sample and 15 uL of 2X Reducing Sample Buffer into new tube and flick to mix
Boil the sample for 5 min in water bath and briefly spin down immediately afterwards
Set up gel (4-20% Tris-Hepgs Gel) with Running Buffer and clean out wells by pipetting some of the buffer in and out
Load 10uL of See Blue Ladder +2 (invitrogen) into Lane 1 for reference molecular weight
Load entire sample (30uL) in lane (I used Lane 5 in Group 1)
Put lid onto gel box and plug electrodes into power supply
Run for 45 min at 150V
Once gel is done, turn off power supply and take gel out of plastic casing
Remember to note orientation
Place gel into tray and pour enough Coomassie Stain to cover gel
Place on rocker and let sit for 5 min
Discard stain and rinse with 10% acetic acid
Place gel into tray and pour enough 10% acetic acid to cover gel in order to destain
Place on rocker and let sit for 15 min
Every 15 min or so, dump acetic acid and add new acetic acid to speed up the destaining process
Once done destaining, dump acetic acid and place gel onto light box to view results
Take picture of gel for notebook/future reference
Results:
I placed 9.165 ug of protein into lane 5 gel
Gel was run for ~30 min, not the specified 45 min, because the dye had hit the bottom of the gel at this time point
Picture of gel...


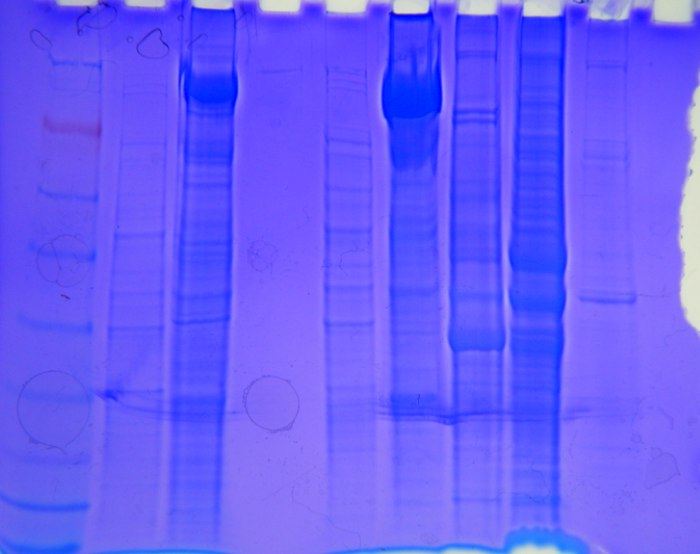
Next Step/Conclusions:
Gel looked good. There are multiple bands on my lane and the bands seem to correlate to Lane 1 which was Emma's Vt sample. I still need to evaluate my lanes to estimate the molecular wgts in relation to the ladder.
Comparing my lane to the ladder, bands on my lane seem to match up with: 250, 2 bands in between, 148, 1 band between, 98 (looks like Emma's lane doesn't show this band), 1 band between, 50, 36, and possibly 22 kDa, then the lane gets fuzzy at the end.
The brightest bands were 148, ~ 75, ~57, and 50kDa.
Next step - Western blot to detect specific proteins
10-20-09
Reverse Transcription and PCR
Summary: Reconstitute Primers, Reverse transcribe RNA to cDNA and prepare PCR gel for next week's lab.
Procedure:
To reconstitute primer sets:
Find nM on tube and multiply that number by 10
Add that number of uL to the powder vial for a primer concentration of 100uM per uL
Then make a 1:10 dilution of the 100uM mix to make a 10uM dilution
We will use the 10uM dilution of the primers for our PCR rxns
Make cDNA:
Turn thermocycler on and get ice bucket ready
Mix stock RNA by inverting tube
Put 5 ul (652.5 ng total for this sample) of RNA sample into fresh PCR tube
Incubate tube at 75C for 5 min in thermocycler
Immediately after the incubation, place tube on ice for at least 5 min
Make Master Mix:
Recipe per Reaction:
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10mM total)
1 ul AMV RTranscriptase
1 ul Random Primer (prokaryotes do not have Poly A tail) 500ug/ml
1 ul RNase free water
Total = 15 ul
Add MM components above to the PCR tube with the diluted RNA sample (total vol should be 20ul)
Vortex briefly and spin down quickly
Incubate at RT for 10 min
Place in thermocycler and incubate at 37C for 1 hr
Heat inactivate at 95C for 3 min
Spin down briefly
Store at -20C or on ice until ready to PCR
Make PCR Reaction:
Samples will be prepared in duplicate with 2 negative controls for each primer set
Recipe for a 50ul reaction.
25 ul of 2x GoTaq Green Master Mix
2.5 ul F Primer (10uM)
2.5 ul R Primer (10uM)
2 ul DNA Template (<250ng)
18.0 ul pH2O
Multiply by 5 to make Master Mix for EACH PRIMER SET:
125 ul of 2x GoTaq Green Master Mix
12.5 ul F Primer (10uM)
12.5 ul R Primer (10uM)
20 ul DNA Template (<250ng)
90 ul pH2O
Total vol. 260
Thermocycler settings are:
95C for 10 min
40 cycles of:
95C for 30 sc
55C for 30 sec
72C for 90 sc
then 72C for 3 min
4C forever
Place PCR tube in thermocycler with the settings above
Prepare 1.75% agarose PCR gel:
Place 2 g of agarose powder with 150mL of 1xTAE
Microwave for ~ 2 min or until rapid boil
Cool the solution and add 12uL of ethidium bromide (make sure to wear all PPE)
Swirl solution and pour into tray with gel combs of choice
Gel sets in ~20-30 min
Wrap in plastic and place gel into fridge for the next lab
Results:
Gel turned out well, nevermind the brief boil over in the microwave.
RT to cDNA - still need to see PCR results in order to visualize success of the rxn
Next Steps/Conclusions:
Visualize on cDNA on gel next week.
10-27-09
PCR and Western Blot
Summary: Visualize primer bands on PCR and start Western Blott
PRC Procedure:
Make sure tubes were run according to the theromocycler settings above
Take out gel out of the fridge that was made last week
Place gel onto gel tray and cover with TAE buffer (make sure gel is completely covered)
Remove combs if present
Load 7 uL of 100bp ladder into row one on the left
Add 25uL of PCR product into the well (in duplicate)
Add 25uL of blank into the next well (in duplicate)
(One ladder + Four wells of product and blanks)
I have 2 primer sets so I have 8 wells total plus one ladder
Once gel is loaded completely, put cover on box and plug electrodes into power supply
Run gel for 100v for 1 hour
After time is up, visualize bands under UV light and take picture
Examine band size in relation to ladder and record
PCR Result:
My lanes are 2-9 on the bottom half.
Lanes 2-5 are RpoS and 5-9 are TcpA.
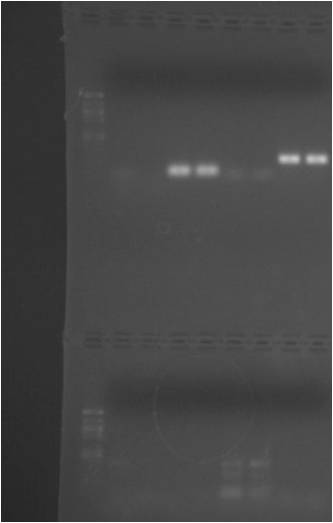
Boo! RpoS - Bad bands on lane 2, ie wrong size ~500bp (should be 144 bp), stutter on TcpA although brighter band at the bottom that is approx the right size.
Next Steps/Conclusions:
RpoS: looks like there's genomic contamination and the correct band did not show up. I think I may need to go back to the drawing board on this one. I will try to develop new primer sets for this one and make the product a bit bigger and re-run gel. I may be able to do run a conventional PCR on Vt template at home base to make sure the primers are working. This is the gene I am most interested in for the future, so it's important to get this accurate.
TcpA: I think I may be able to get this to work. Not sure if there's any genomic contamination there. I can try to re-run the thermocycler rxns to anneal at 60C instead of what was used here (55C). That may eliminate the stutter that appeared on the gel. If that fails, try new primer sets and re-do procedure over again.
Also, the problem could have been trouble amplifying the product with tubiashii since both primer sets were developed off of different species.
Western Blott Procedure:
To transfer the protein gel (my lane was loaded with 9ug (15ul) of protein in lane 2, group 2) to the membrane:
Transfer buffer should be at 4C
Soak filter paper, membrane and gel in the transfer buffer for 15 min
Assemble in the semi-dry blotting machine like this:
- Anode (+)
- Filter Paper
- Nicroccellulose Membrane
- Gel
- Filter Paper
- Cathode (-)
Remove the gel and rinse with the transfer buffer
Remove any extra gel with a cotton swab
Western Blott:
Reminders:
Avoid touching the membrane too much
Keep membrane moist when not working with it to avoid drying
Add solutions to the tray (slowly), starting at the edge of the membrane to avoid bubbles under the membrane
Prepare Blocking Solution (20ml total volume):
- 14ml filtered water
- 4 ml Part A Blocker/Diluent
- 2 ml Part B Blocker/Diluent
Incubate for 30 min on rotary shaker at 1 rev/sc
Decant solution from the dish
Rinse the membrane with 20mL of water for 5 min
Decant
Repeat water step once
Prepare 10ml of 1:3000 Primary Antibody Solution:
- 10 ml Blocking Solution
- 3.3ul of HSP 70 antibody
THE NEXT DAY:
Wash the membrane for 5 min with 20ml Antibody Wash
Decant
Repeat 3 times
Incubate the membrane with 10 ml Secondary Antibody Solution for 30 min
Decant
Wash membrane for 5 min with 20 ml Antibody Wash
Decant
Repeat 3 times
Rinse membrane with 20 ml water for 2 min
Decant
Repeat 2 times
Incubate membrane in 5 ml Chromogenic Substrate until you see purple bands on the membrane
you may need to wait 1-60 min
Look in amazement as your band appears!
Rinse membrane with 20 ml water for 2 min
Repeat 2 times
Air dry membrane on filter paper or under lamp.
Conclusions/Next Steps:
Waiting for the antibody solution of hsf70 to incubate with the gel overnight. I'll swing by the FTR in the morning to help with the next steps and see the results of the "Western Breeze."
10-28-09
Results:
No purple band on the membrane in my lane (2, next to ladder). Not surprising. I didn't think that hsp70 is present in prokaryotes. But interesting to see the process nonetheless. Other lanes on the membrane did show up, all similar size in molecular wgt.
10-27-09
Searched for new primer sets:
Rpos: Forward: TGCGTCTTAACGAGCGTATT; Reverse: TCGACGTAAACCCTCCACTT
Product size 299
Tm 59
GC 45-50
TcpA: Forward: AGTTGCACTGACACAGACATACC; Reverse: TGGGAACATATCACCGACAC
Product size 265
Tm 59
GC 48-50
I will order and re-run my PCR to see if this will work any better. Used same criteria in Primer 3 as I did above but extended the primer product size to 250-350 bp. TcpA was a pretty small sequence, only 600 bp and half that (at the end) didn't align well with the other Vibrio species I looked at. We'll see what happens.
11-3-09
Summary: qPCR
Procedure: qPCR using SYBR green chemistry
Develop Master Mix recipe
Prepare mix
Dilute RNA 1:4 (add 2 ul of RNA into 6 ul water)
Devy out mix and put the cap on for running qPCR
qPCR Cycling Parameters:
Rxn volume: 50ul
95C for 10 min
95C for 15 sec
55C for 15 sec
Read Plate
Repeat 39 times from #2
95C for 1 min
55 for 1 sec
(Manual ramp rate of 0.5C per sec)
Melting curve 55 to 95C, read every 0.5C, hold for 30 sec
Incubate at 21C for 10 min
Master Mix Recipe:
For 50 ul Reaction:
25 uL Immomix 2X
5 ul Syto-13 dye (50uM)
2.5 ul F Primer (10 uM)
2.5 ul R Primer (10 uM)
13 ul pH2O
2 ul cDNA
For 7 rxns (for each primer set):
175 ul Immomix 2x
35 ul Syto-13 dye (50uM)
17.5 ul F Primer (10 uM)
17.5 ul R Primer (10 uM)
91 ul pH2O
Load the plate
Verify the parameters (above)
Run
Results:
Graphs shown here:
qPCR Result
So neither of my qPCRs worked and it looks like there's genomic carryover in the RNA. The primers did not amplify the cDNA. My blanks were in fact negative controls. The only thing fluorescing is my RNA samples. It's interesting how the carryover just showed up on the first round of samples (1-6) and not the second (7-12).
Next Steps/Conclusions:
Just for the hell of it, I ran my primers I tried to amplify here on cPCR to see if they would work on Vt period. I used three strains: ATCC 19109, ATCC 19106 and RE100. The tcpA gene amplified well on conventional with the expected band size. No luck on the rpoS gene.
Where to go from here? Well, I could try multiple primers for rpoS and try it again. Talking to CF, we might want to shift direction and work on a couple genes from the SK. I will talk to SR on Mon to see if he has any Vt sequences for any of the GOIs and go from there.
As for genomic carryover, I will DNase my samples prior to reverse transcribing to be on the safe side.
PROJECT TIME LINE:
11/10
Run experiment and take samples (no needed supplies here, all provided by Friedman Lab)
Test primer sets and modify if necessary
11/17
Begin sample processing:
RNA extractions, quanting and cDNA making
qPCR for genomic DNA
11/24
Begin development of standard curve for the two genes
Run qPCR for genomic copies
Data analysis
12/1
Continue sample processing
12/8
Analysis and write-up
Needed supplies: (just reagents I think)
AMV RTranscriptionase
DNase
SYBR
Immomix
Go Taq green master
11-10-09
Set up for experiment:
Experimental Design:
Set up air tight container (in dup) with four 250ml beakers filled with T1N2 (1% tryptone and 2% NaCl) media.
2 control beakers
2 Experimental beakers
Rig up with bubbling Co2 and air in one set and just bubbling air in the other set.
Will inoculate one control beaker and one experimental beaker with 10^3 of Vt after 24 hours of equilibrating time.
After setting up the experiment I realized I had forgotten to set the pH of the media.
After 24hrs the pH of the experimentals was 6.2 and the control was 7. These values are too low to represent "real world" senario.
11-12-09
Try2:
Simplified the experiment a bit by using larger, 1L erlenmeyer flasks and canned the container system.
We used filtered sterile seawater with 1% tryptone for media and set the pH to ~7.8 by adding 200ml of supersaturated sodium bicarbonate solution per liter of media.
I aliquoted 400 ml into 6 flasks. Three experimental with bubbling CO2 and air, and three control with air only. I will leave one control and one experimental flask with uninoculated with bacteria.
Same routine as above...start bubbling and wait 24 hours to equilibrate.
Sampling protocol:

11-13-09
Went downstairs to inoculate each beaker with 10^3 of Vt and 4/6 flasks were contaminated with growth!
Could be the air lines (no filter), but my guess was that the media was contaminated since we only 0.2um filtered the media.
No time for any trial and error, now time for Plan C.
Talked to SR this morning and came up with a quick and dirty sampling plan:
2 tubs of seawater
One set at 7.3pH and one control at 8 pH
Grow up 4 liters of bacteria for 24 hrs - 2 in marine broth (7.6 pH) and 2 in 0.2um filtered seawater (8 pH)
Divide the bacteria the next day: Dump 500ml marine broth bacteria and 500 ml of seawater grown bacteria into each bucket.
Grow for 24-48 hrs and take some samples.Spin down 1 liter of sea water from each bucket to isolate bacterial pellets.
Ended up with:
Time 0: 2 samples (marine broth only)
24 hrs: 1 sample control, 1 sample experimental
48 hrs: 2 samples control, 2 samples experimental
Stored in -80 until the 19th and stuck in -20 in the lab until processing.
11-23-09
Extracting RNA:
Since I had only one set of 24 hrs samples and my pellets were decent size, I decided to split them up.
I realize that this is psuedoreplication of my samples, but I wanted to make sure I had duplicate samples throughout the process of extraction through PCR to see if they were similar. It was a test of my own technique I guess.
Procedure:
Add 0.5 mL TriReagent to pellet and mix with pestle.
Add additonal 0.5 mL TriReagent to the same tube.
Vortex the tube for 15 sec.
add 200uL of chloroform to tube and vortex for ~30 sc
incubate at RT for 5min
Centrifuge for 15 min at full speed
Collect aqueous phase of the tube, do not disturb any other layers, and put aqueous phase into new tube
add 500uL of isopropanol to tube containing RNA, invert to mix
Incubate at RT for 10 min
Spin for 8 min at high speed
Remove supernatant, leaving the pellet undisturbed
Add 1 ml 75% EtOH and briefly vortex
Centrifuge 5 min at 7500 g
Remove supernatant, leaving the pellet undisturbed
Spin again briefly and remove any left over EtOH
Leave tube open to dry pellet at RT for no more than 5 min
Resuspend pellet in 100uL of 0.1% DEPC-H2O and pipette up and down to resuspend the pellet
Incubate in 55 degree C water bath for 5 min
Flick a few times and quant sample using spectrophotometer
Place in -80
TO QUANT:
use nanodrop reader in Roberts Lab
Place 2 uL of DEPC-H2O onto the pedestal to blank the spectrophotometer
Pipette 2 uL of RNA sample onto pedestal
Press "Measure" and record findings
11-24-09:
Preliminary Quant (NG/UL) Results before DNasing:
T0-1: 285.7
T0-2: 428.2
E1-24: 327.6
E2-24: 157.5
C1-24: 136.9
C2-24: 159.7
E1-48: 324.0
E2-48: 427.7
C1-48: 141.7
C2-48: 128.1
All showed good peaks at 260, with exception of C2-48. That curve was a bit shaky, but turned out fine on the subsequent quant.
DNase Procedure:
Add to PCR tube:
2.5 ul DNase Buffer
1 ul turbo DNase
20.5 ul RNA sample
TOTAL - 24 ul
Incubate samples for 30min at 37C
After 30 min, add 1 ul of turbo DNase
Incubate samples for 30min at 37C
After 30 min add 2.5 ul Inactivation Reagent
Leave at RT for 2 min, mixing occasionally
Spin down at 10000 ref for 1.5 min
Transfer supernatant to new tube.
Quant and normalize to the lowest RNA value.
Results:
Quanting and Normalizing results:
| Samples |
ng/ul |
260/280 |
260/230 |
Normalizing Calculation |
uL Water Added |
Total Volume (ul) |
| T0-1 |
280.5 |
2 |
1.53 |
9.3 |
25.7 |
35 |
| T0-2 |
349.7 |
1.99 |
1.53 |
7.5 |
27.5 |
35 |
| E1-24 |
94.7 |
1.84 |
1.01 |
27.6 |
7.4 |
35 |
| E2-24 |
104.1 |
1.9 |
1.05 |
25.1 |
9.9 |
35 |
| C1-24 |
96.2 |
1.85 |
1.03 |
27.2 |
7.8 |
35 |
| C2-24 |
109.7 |
1.89 |
1.22 |
23.9 |
11.1 |
35 |
| E1-48 |
226.4 |
1.91 |
1.46 |
11.6 |
23.4 |
35 |
| E2-48 |
290.9 |
1.92 |
1.6 |
9.0 |
26.0 |
35 |
| C1-48 |
92.1 |
1.89 |
1.1 |
28.4 |
6.6 |
35 |
| C2-48 |
74.8 |
1.86 |
1.07 |
n/a |
n/a |
35 |
Next Step:
RT for cDNA
Reverse Transcription:
Summary:Reverse transcribe RNA to cDNA
Procedure:
Make cDNA:
Turn thermocycler on and get ice bucket ready
Mix stock RNA by inverting tube
Put 5 ul (374 ng total for this sample) of RNA sample into fresh PCR tube
Incubate tube at 75C for 5 min in thermocycler
Immediately after the incubation, place tube on ice for at least 5 min
Make Master Mix:
Recipe per Reaction:
4 ul 5x Buffer (MMLV-RT Buffer)
8 ul dNTPs (10mM total)
1 ul MMLV-RTranscriptase
1 ul Random Primer (prokaryotes do not have Poly A tail) 500ug/ml
1 ul RNase free water
Total = 15 ul
Three reactions per sample for a total of 60 ul of cDNA per sample.
Add MM components above to the PCR tube with the diluted RNA sample (total vol should be 20ul)
Vortex briefly and spin down quickly
Incubate at RT for 10 min
Place in thermocycler and incubate at 37C for 1 hr
Heat inactivate at 95C for 3 min
Spin down briefly
Store at -20C or on ice until ready to PCR
I made 3 rxns of each sample for a total volume of 60ul of cDNA. This should be enough to run 3+ primer sets with ~6 samples in each.
Next step:
Double check primer sets using genomic DNA to make sure they work.
11-26-09
Summary: Primer check before running qPCR
Procedure:
6 Primer sets tested:
rpoS (set#2): (F:tgccttacgtggtgatgaag; R: tggtgtaggttcgtggtcaa)
Product size: 299 bp
rseA (F: gtatcactcgctgtgatcttaggc, R:gaagaacgtgtcaagctcactggt)
Product size: ? bp (I need to find the sequence that this primer was developed off of)
tdh-1: (F: tccaaacgggtcgaaaatag; R: tgggatgagtggtcacaaaa)
Product size: 315 bp
tdh-2: (F: CACATCAAAACCTTCGGTCA; R: TCTATTTTCGACCCGTTTGG)
Product size: 273 bp
tdh-3:(F: GGTCGAAAATAGACGCCAGA; R: TGGGATGAGTGGTCACAAAA)
Product size: 307 bp
tcpA (set #2) Forward: AGTTGCACTGACACAGACATACC; Reverse: TGGGAACATATCACCGACAC)
Product size: 265 bp
Master Mix Recipe:
Vol (ul) per rxn x 4 rxns
| PCR H20 |
4.75 |
x4 = |
19.00 |
|
| 5xBuffer |
5.00 |
20.00 |
||
| MgCl2 |
2.00 |
8.00 |
||
| BSA |
1.00 |
4.00 |
||
| dNTPs |
0.50 |
2.00 |
||
| F Primer (10mM) |
0.75 |
3.00 |
||
| R Primer (10mM) |
0.75 |
3.00 |
||
| Taq |
0.25 |
1.00 |
per rxn |
|
| Template |
10.00 |
Add individually |
15 ul |
|
| TOTAL |
25.00 |
ul |
Thermocycler Parameters:
95 for 3 min
95 for 15 sec
57 for 1 min } Steps 2-4 for 36 cycles
72 for 30 sec
72 for 10 min
Run on a 1% agarose gel at 100v for 45 min. Order of samples on gel: rpoS, rseA, tdh1, tdh2, tdh3, tcpA.
Results:


rpoS: Band size looks approx right on both lanes. Blank lane was negative. This is the only primer set I've seen work on this gene. I am going to run with this and see what happens on qPCR. I may up the Tm a little on the qPCR parameters to get a higher specificity for the gene (ie try to eliminate some of the extra bands there). Overall, pretty happy with the result.
rseA: Nice bright bands on both lanes, approx 200 bp in length. Blank lane was negative. Need the product size for this gene. SR gave me these primers to work with and I don't have the original sequence that it was developed off of. If the bands are the appropriate size, the primers work well with this strain. Again, faint band on the second lane. Higher temp may eliminate that.
tdh1: Nice bands of appropriate size. Blank lane was negative.Again, specificity needs to be tightened up. Move ahead with qPCR on this primer set.
tdh2: Nice bands, but I am seeing 2 bands on each lane. One is the right size, the other is approx 600bp. Blank lane was negative.
tdh3: I've got 2 bands here, but they look like they are the wrong product size - too big (~450bp). Blank lane was negative.
tcpA: Looks like I have one band that's approx the right size (265bp) but the first lane there's a very faint band (I think). Blank lane was negative. I will pass on this primer set. Maybe useful in the future though.
Next Steps and Conclusions:
Run rpoS, rseA and tdh1 with qPCR. These genes had pretty good results on cPCR using genomic Vt RE22 strain (to be used on the SK grant). These may be of interest in future research.
I will run qPCR on 12-1-09 and use a Tm of 58C, slightly higher than the Tm used in the cPCR above.
12-1-09
Summary: qPCR
Procedure: qPCR using SYBR green chemistry
Develop Master Mix recipe
Prepare mix
Dilute RNA 1:4 (add 2 ul of RNA into 6 ul water)
Devy out mix and put the cap on for running qPCR
Master Mix Recipe:
For 25 ul Reaction:
12.5 uL Immomix 2X
1.0 ul Syto-13 dye (50uM)
0.5 ul F Primer (10 uM)
0.5 ul R Primer (10 uM)
9.5 ul pH2O
1.0 ul cDNA
For 6 x 10 samples = 60 rxns (for 1st primer set: rseA - 2 cDNA, 2 blank, 1 RNA)
750 ul Immomix 2x
60 ul Syto-13 dye (50uM)
30 ul F Primer (10 uM)
30 ul R Primer (10 uM)
570 ul pH2O
then 5 x 10 samples = 50 rxns for primers tdh and rpoS (2 cDNA, 2 blank)
625 ul Immomix 2x
50 ul Syto-13 dye (50uM)
25 ul F Primer (10 uM)
25 ul R Primer (10 uM)
475 ul pH2O
Load the plate
Set parameters for the run:
qPCR Cycling Parameters:
Rxn volume: 25 ul
95C - 10 min
95C - 30 sec
55C - 1 min
72C - 1 min
40 cycles steps #2-4
95C - 1 min
55C - 30 sec
95C - 30 sec
Place plate in machine and run!
Results:



tdh - SAMPLES 3-10:

tdh - SAMPLES 1 and 2:

Ran the samples with the highest Ct values for rpoS and tdh on 1% agarose gel to look at product size. I was trying to see if the small amount of amplification I see in the graph is the gene product or primer dimer. Each sample was run with 10 ul of qPCR product in duplicate. rpoS on lanes 2-3 and tdh on lanes 4-5 and 100 bp ladder in lane one.
Here's the gel pic:

I am expecting a product size of ~ 300 bp for both genes. Looks like rpoS is nothing but dimer, but tdh looks like the right size and may be amplification of the gene. Small, but it's something. Shows the primers work at least.
Ct values and arbitrary gene expression results:
Expression Results and Ct Values_ED
Next Steps and Conclusions:
Conclusions about the qPCR results above:
Good results for rseA.
Not so good results for rpoS - looks like the peak is due to primer dimer, not any gene expression.
tdh - the cPCR above shows that the product is ~300 bp in length and that the peak I saw on qPCR was not primer dimer, but expression of tdh. There is very little expressed, but exciting nonetheless.
Genomic carryover: it looks like there's a bit of genomic carryover on samples 5-10, although the expression is pretty small. Not really sure what this means for my data. I need to talk with Mac about possible implications.
Next steps:
Analyze the data for any differences in expression of control and experimental samples for genes rseA and tdh, if any.
I plan on running genes chiA (chitinase) and vtpA (metalloprotease) to see if there's any expression since I have the cDNA. Primers for these two genes have been tested in the past.
Gel pictures:
12-3-09
Re-DNasing samples C1-24 thru C2-48 due to genomic carryover, re reverse transcribing and qPCR do-over.
DNase Procedure:
Add to PCR tube:
2.5 ul DNase Buffer
1 ul turbo DNase
15 ul RNA sample
5.5 ul of pH20 (I didn't have enough RNA from last DNase to make the usual 20.5 ul volume)
TOTAL - 24 ul
Incubate samples for 30min at 37C
After 30 min, add 1 ul of turbo DNase
Incubate samples for 30min at 37C
After 30 min add 2.5 ul Inactivation Reagent
Leave at RT for 2 min, mixing occasionally
Spin down at 10000 ref for 1.5 min
Transfer supernatant to new tube.
Quant and normalize to the lowest RNA value.
Results:
Quanting and Normalizing results:
| Second Dnase |
||||||
| Samples |
ng/ul |
260/280 |
260/230 |
Normalizing Calculation |
uL Water Added |
Total Volume (ul) |
| T0-1 |
76.1 |
2.02 |
1.78 |
5.3 |
4.7 |
10 |
| T0-2 |
75.8 |
2 |
1.85 |
5.3 |
4.7 |
10 |
| E1-24 |
77.3 |
1.84 |
1.18 |
5.2 |
4.8 |
10 |
| E2-24 |
75 |
1.85 |
1.18 |
5.4 |
4.6 |
10 |
| C1-24 |
41.4 |
1.68 |
0.95 |
9.8 |
0.2 |
10 |
| C2-24 |
41 |
1.7 |
1.08 |
9.9 |
0.1 |
10 |
| E1-48 |
42.3 |
1.73 |
42.3 |
9.6 |
0.4 |
10 |
| E2-48 |
40.5 |
1.75 |
1.46 |
10.0 |
0.0 |
10 |
| C1-48 |
42.2 |
1.68 |
0.97 |
9.6 |
0.4 |
10 |
| C2-48 |
41.8 |
1.76 |
1.03 |
9.7 |
0.3 |
10 |
Reverse Transcription:
Summary:Reverse transcribe RNA to cDNA
Procedure:
Make cDNA:
Turn thermocycler on and get ice bucket ready
Mix stock RNA by inverting tube
Put 5 ul (374 ng total for this sample) of RNA sample into fresh PCR tube
Incubate tube at 75C for 5 min in thermocycler
Immediately after the incubation, place tube on ice for at least 5 min
Make Master Mix:
Recipe per Reaction:
4 ul 5x Buffer (MMLV-RT Buffer)
8 ul dNTPs (10mM total)
1 ul MMLV-RTranscriptase
1 ul Random Primer (prokaryotes do not have Poly A tail) 500ug/ml
1 ul RNase free water
Total = 15 ul
For 11 rxns:
44 ul 5x Buffer
88 ul dNTPs
11 ul MMLV traanscriptase
11 ul Random Primer
11 ul RNase free water
Add MM components above to the PCR tube with the diluted RNA sample (total vol should be 20ul)
Vortex briefly and spin down quickly
Incubate at RT for 10 min
Place in thermocycler and incubate at 37C for 1 hr
Heat inactivate at 95C for 3 min
Spin down briefly
Store at -20C or on ice until ready to PCR
Next up:
Re-run qPCR with diluted RNA samples to double check any carryover in the morning.
12-4-09
Summary: qPCR
Procedure: qPCR using SYBR green chemistry
Develop Master Mix recipe
Prepare mix
Dilute RNA 1:4 (add 2 ul of RNA into 6 ul water)
Devy out mix and put the cap on for running qPCR
Master Mix Recipe:
12.5 ul Sensimix
0.8 ul F Primer (10uM)
0.8 ul R Primer (10uM)
0.5 ul SYBR
9.4 ul PCR water
= 24 ul
For 55 rxns : rseA primers (Duplicate samples and 2 blanks + 10 RNA samples to test for carry over)
687.5 ul Sensimix
44.0 ul F Primer (10uM)
44.0 ul R Primer (10uM)
27.5 ul SYBR
517 ul PCR water
For 45 rxns: chiA and tdh primers
562.5 ul Sensimix
36.0 ul F Primer (10uM)
36.0 ul R Primer (10uM)
22.5 ul SYBR
423 ul PCR water
+ 1ul cDNA template = 25 ul rxns
qPCR Parameters: (note increase in temp to increase specificity of primer sets)
Rxn volume: 25 ul
95C - 10 min
95C - 30 sec
58C - 1 min
72C - 1 min
40 cycles steps #2-4
95C - 1 min
55C - 30 sec
95C - 30 sec
Place plate in machine and run!
Results:
No genomic carryover!! Hooray!
Looks like better amplification with SYBR in general. There are good peaks for rseA and chiA. The gene tdh wasn't so good.
Disassociation Curves:



Ct Values and Arbitrary Gene Expression:
Expression Results and Ct Values_ED
Next Steps/Conlusions:
- SYBR worked better for this experiment in general
- Second DNase was necessary. Next time DNase twice, DNase carefully, and make sure to run RNA after DNasing onto gel or qPCR before running any samples. That should save time/$ in the future.
Next up: Data analysis...