Secondary stress: proteomics
I played around a bit with protein-protein interaction network stuff today. I tried Nascent and something called ENT. It looks like a limitation for a lot of these packages is that my data is annotated from multiple model species.
For Nascent I tried to make C. gigas both the source (annotation) and target (networked) animal, basing the calculation on sequence similarity. That returned empty results. Same thing when I tried to make M. musculus the source animal. Seems like this might be a good bioinformatics project.
Moving on to iPath, I made 3 separate plots for 2-fold differentially expressed proteins from each treatment condition. Red = expressed higher in stress condition and blue = lower, line width = 20.
I also made blots for all proteins expressed higher across all 3 stress conditions and all expressed lower. blue = OA only, yellow = mech stress only, red = OA + mech stress, green = protein changes for both OA only and Mech stress only (i.e. for proteins shared across responses color = combination of the single response colors, brown = across all 3). These plots are harder to understand by just quickly looking at them, so I think the 3-plot approach is more useful.
December 19, 2013
Secondary Stress: Proteomics
Made heat maps for each treatment comparison with proteins at least 2-fold differentially expressed. If a protein had no annotation, it was marked as "Unannotated #". If two proteins had the same annotation a letter was added (i.e. a, b,...). Data were log-transformed and clustering was done for columns and rows using euclidean distance. For each heat map treatment groups clustered together.
Playing around with Cytoscape...The user manual is not that awesome (i.e. major terms are not defined). I figured out that I need to create a network of protein-protein interactions. There's a R package for that! WGCNA :
http://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/#WGCNAIntro
First will use GO Terms as trait data. Annotated 2-fold differentially expressed proteins with GO terms.
SELECT * FROM [emmats@washington.edu].[2-fold diff proteins for venn]
LEFT JOIN [dhalperi@washington.edu].[SPID_GOnumber.txt]
ON [emmats@washington.edu].[2-fold diff proteins for venn].SPID=[dhalperi@washington.edu].[SPID_GOnumber.txt].A0A000
Ran Select Distinct for file to get rid of redundant entries. Created file with diff expressed proteins for OA only associated with GO terms (removed proteins that had no associated GO term).
There are too many 0 values for WGCNA to work. I tried it with oysters with at least 4, 5, and 6 oysters required to have expression data for each protein but it was still too many 0s. I've contacted one of the package developers to see if he might be able to give me a hint.
December 18, 2013
Secondary Stress: Proteomics
Analysis of proteomics data to include proteins that are at least 2-fold differentially expressed. Compiled a list of proteins with at least a 2-fold change (up or down) from the file NSAF avg SpC. The list is called 2-fold protein list.csv and I made sure that all proteins with q-value < 0.1 were included (only 1 from 2800 mech stress comparison was not in the list because of <2-fold difference in expression). Uploaded list to SQL and joined with file of NSAF and SPID annotations.
SELECT * FROM [emmats@washington.edu].[2-fold_protein_list.csv]
LEFT JOIN [emmats@washington.edu].[NSAF based on avg SpC with SPIDs]
ON [emmats@washington.edu].[2-fold_protein_list.csv].Protein=[emmats@washington.edu].[NSAF based on avg SpC with SPIDs].[All Proteins]
In Excel, removed annotations that were >1E-10. Separated proteins by treatment comparison and for each treatment removed proteins that were expressed in <2 oysters within a single treatment (i.e. if the protein was expressed in just 1 oyster in each treatment it was removed).
In response to OA + mechanical stress, 137 proteins were expressed lower or not at all compared to 2800 µatm only and 149 were elevated under the dual stress. 138 proteins were expressed less in the mechanical stress alone compared to 400 µatm and 107 were expressed higher. In response to OA only, 136 proteins are expressed less at elevated pCO2 and 148 are expressed more.
A venn diagram was executed in eulerAPE for the at least 2-fold differentially expressed proteins among treatment comparisons. A non-redundant list of all 2-fold proteins was generated and then lists of diff exp proteins from individual treatment comparisons were joined to the master list.
SELECT * FROM [emmats@washington.edu].[all_2-fold_proteins.txt]
LEFT JOIN [emmats@washington.edu].[OA_2-fold.txt]
ON [emmats@washington.edu].[all_2-fold_proteins.txt].Protein=[emmats@washington.edu].[OA_2-fold.txt].Protein
LEFT JOIN [emmats@washington.edu].[400MechS_2-fold.txt]
ON [emmats@washington.edu].[all_2-fold_proteins.txt].Protein=[emmats@washington.edu].[400MechS_2-fold.txt].Protein
LEFT JOIN [emmats@washington.edu].[2800MechS_2-fold.txt]
ON [emmats@washington.edu].[all_2-fold_proteins.txt].Protein=[emmats@washington.edu].[2800MechS_2-fold.txt].Protein
DAVID enrichment analysis on 2-fold differentially expressed proteins. Non-redundant lists of SPIDs were created for each treatment comparison (3 lists) and for the entire gill proteome (1347 SPIDs). With the significance cutoff of p<0.075, the significantly enriched GO terms for the treatment comparisons are the following:
OA - vitamin metabolic process, visual perception, sensory perception of light stimulus, dorsal closure, cell-cell junction organization, transcription, regulation of transcription DNA-dependent, homophilic cell adhesion, regulation of transcription
400MechS - regulation of RNA Metabolic process, regulation of transcription DNA-dependent, gamete generation
2800MechS - cellular polysaccharide biosynthetic process, transcription, polysaccharide metabolic process, cellular polysaccharide metabolic process
December 17, 2013
Proteomics: Brest
I could see enough in the development of the films from the western blot to see that the antibody worked well and made a nice band. However, I messed up something with the revelation and cannot do an expression comparison.
December 16, 2013
Proteomics: Brest
Western blot with primary antibody phosphorylated site for MAPKAPK-2 (Thr222), CST #3316. Product is 49 kDa, antibody made in rabbit. Samples are the same used for Nov 26 and gel layouts are the same except there is no control and I included samples 41 and 44 on both gels (although on gel 2 there is only 20 µl of 41 and 15 µl of 44, on gel 1 there is 25 µl of each). 41 and 44 are in the last 2 wells of each gel. I made new SDS 10% and new 10x electrophoresis buffer this morning.
Primary antibody was diluted 1:1000 in PBS-1% tween-5% BSA.
December 13, 2013
Proteomics: Brest
The blots did not turn out. I did very long exposure times (up to 1/2 hour) with the film. I can see that there are a couple of very faint bands. Charlotte thinks that the problem is the primary antibody that was used once before. We are going to order new antibody and she may redo the blot for me in January.
Next step: redo one of the blots that I previously did (probably antibody 2793) with the samples prepared yesterday.
December 12, 2013
Proteomics: Brest
The blots that I did Dec 3-4 and Dec 11-12 both failed to show expression of AMPKa (the antibody bound to the dye front on the membranes from 12/3-4). I am re-preparing samples to do the blot today with antibody #2603 (otherwise known as 23A3). Samples are diluted in lysis buffer to 2.24 mg/ml (to 50 µl) and I prepared new loading buffer (16.65 µl added to each sample).
The well for sample 266 had a little blip at the bottom, which may affect the shape of the band.
The primary antibody diluted in PBS-tween-BSA was used once before.
December 11, 2013
Proteomics: Brest
Day 2 of western blot. This particular form of AMPKa does not seem to be expressed in oyster gill tissue. I left films to develop in the cassette for 4 and 8 minutes and the band for the ampk-treated control cells turned out very well, but there was no expression in any of my samples. Tomorrow I will use the other antibody that is supposed to be similar to this one (#2603) to see if I get the same results.
(Re)did quantifications for previous 2 blots. I think there is a chemiluminescence problem with membrane 2 from Dec 4 and with sample 41 on membrane 1. I will redo membrane 2, but no need to redo 41 since it is a mech stress sample and I'm not including those in this analysis. Based on ANOVA, there is no difference in expression among treatments (excluding 2800 MS) of AMPKa (antibody 2793), phosphorylation site of AMPKa for full-length protein, or for phosphorylation site of truncated protein (p > 0.05).
The ladder I've been using is a BioRad precision plus protein dual color standard, #151-0374.
December 10, 2013
Proteomics: Brest
Day 1 of western blot using AMPKa primary antibody (CST #5832). The first gels I made this morning did not polymerize so I had to make new ones, which seem to have worked out fine. I used the same samples that were prepared for the Nov 26 western blot (still enough left for one more blot!) and the CST AMPK control cells, treated.
December 9, 2013
Proteomics: Brest
I met with Catherine to discuss my SOD results. She thought the best interpretation would be to choose a time point where all the samples had a linear slope and do a single point calculation. I chose the time point 15 min, 10s (only 1 sample had to be excluded due to non-linearity at this time point, 59.2). I calculated the coefficient of variation for the sample replicates and all were <20%. I plotted the standard curve and the linear version (by taking LN of the SOD U/ml). % inhibition was calculated for all samples based on the equation: ([abs blank 1]-[abs sample])/[abs blank 1] *100.
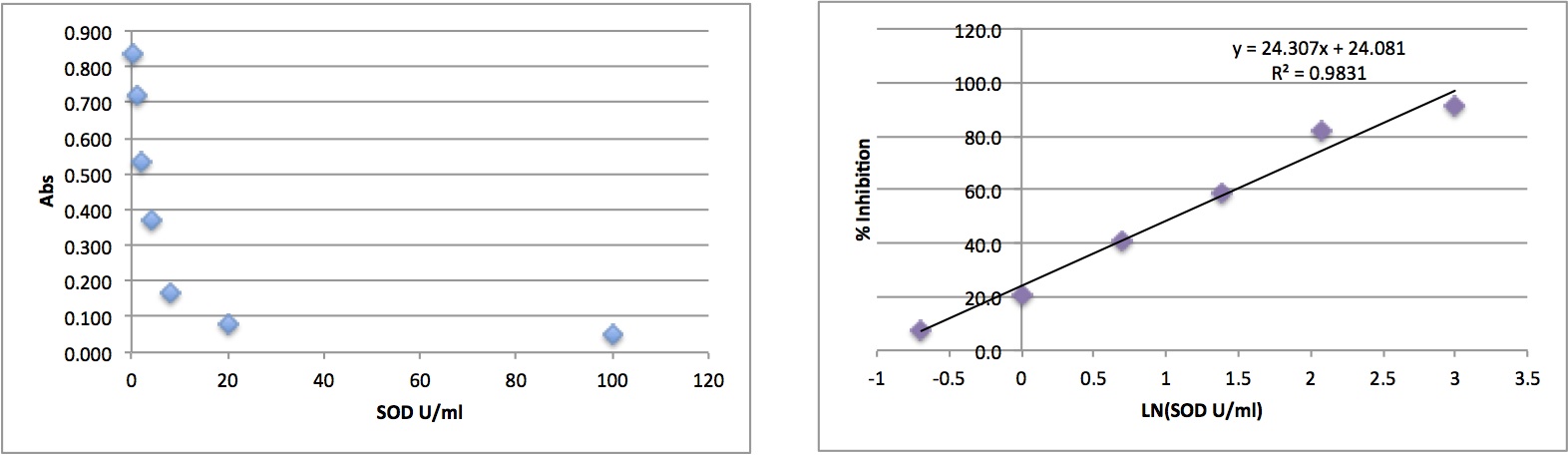
To achieve linearity, the 100 standard was removed from the linear curve before calculation of the best fit line (in excel). The equation of the best fit line was used to calculate SOD U/ml for each sample based on its % inhibition. SOD U/mL were multiplied by 2 (dilution factor) and then divided by the average concentration of the protein sample to get SOD U/mg. These values were used in R to calculate differences among the treatment groups.
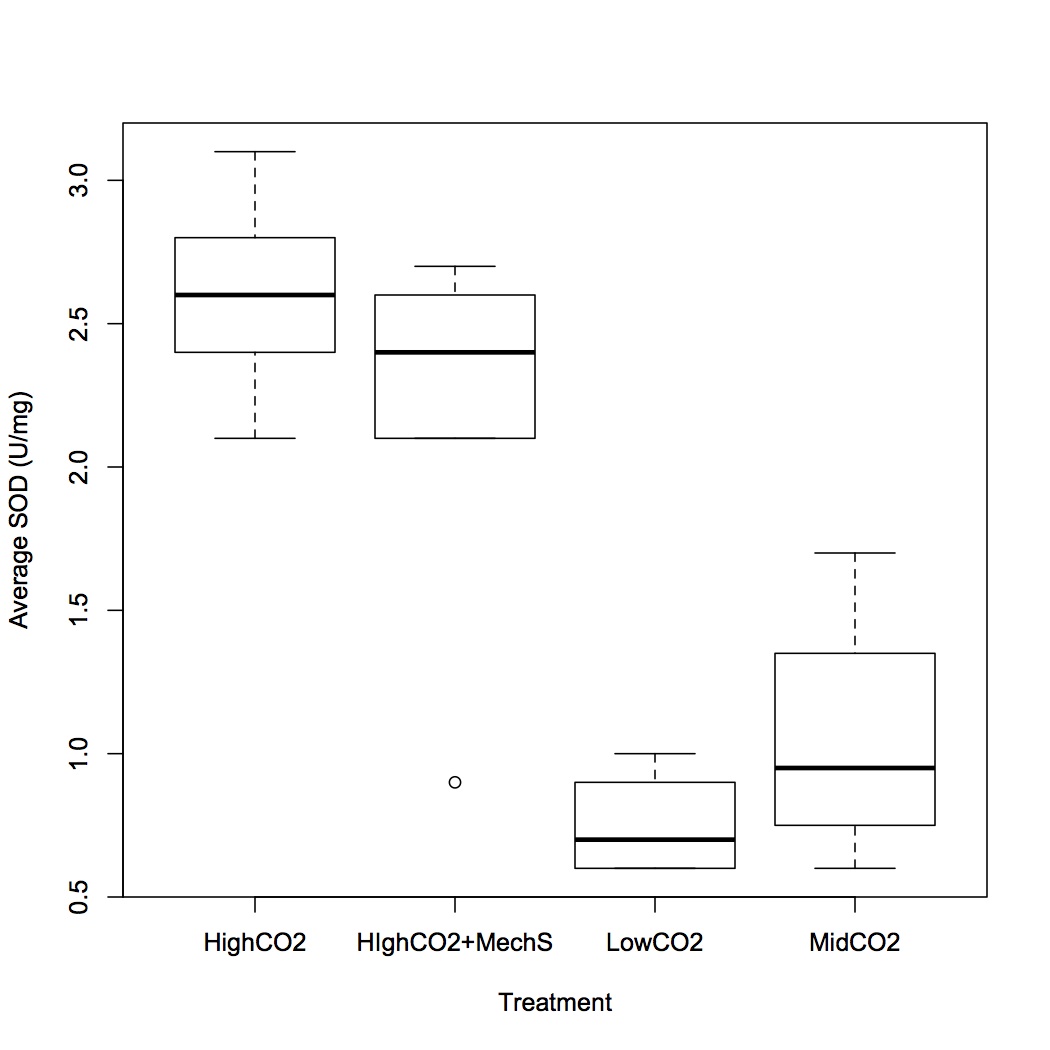
SOD activity is significantly greater at 2800 and 2800 + mech stress compared to the other two treatments (400 and 1000), but are not different from each other. 1000 µatm is not different from the control.
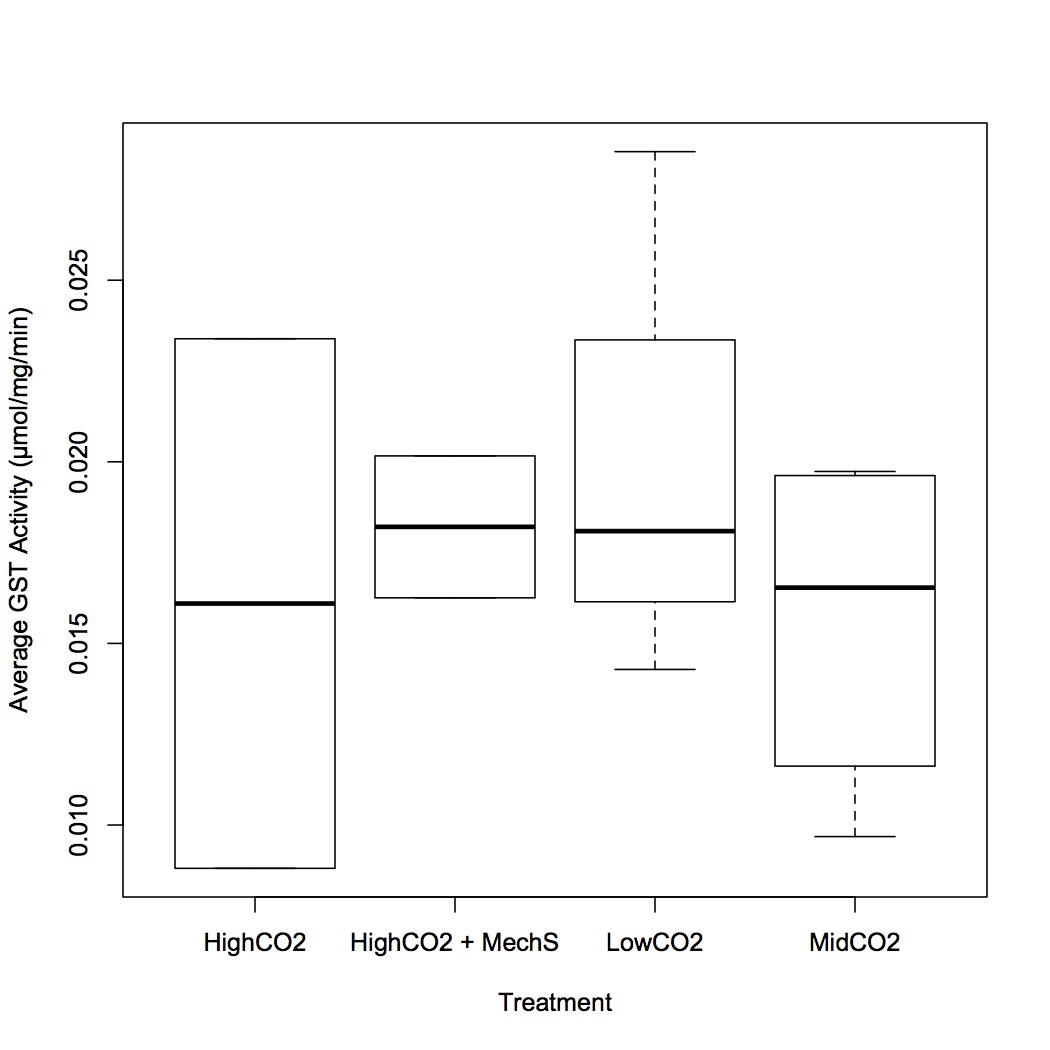
These results are in contrast to the previous no difference in GST activity among all 4 treatments.
December 4, 2013
Proteomics: Brest
Day 2 of western blot. Secondary antibody is goat anti-mouse. Blot seems to have worked well, but the oyster AMPKa is much smaller than the one in the control. It looks like all samples express AMPK at about the same level, but I will do quantifications next week.
Charlotte says that it is the truncated form that reacted with the antibody, not the total protein (that's why it's smaller). The control is ~55kDa and the oyster truncated form is 10-15 kDa. (This is in fact much much smaller than the truncated form that Eric saw in his work, which seems weird...)
December 3, 2013
Proteomics: Brest
Day 1 of western blot. Same samples used (and in same order) as Nov 26. Gel 1 tore a little bit, but will hopefully turn out ok. Since samples had already been prepped on 11/26, I just reheated them for 5 minutes at 100°C before leaving on bench for 10 min and then storing on ice. Primary antibody = CST #2793, anti-AMPK alpha in mouse. Antibody is diluted 1/1000 (50 µl in 50 ml of PBS-tween-5% BSA).
Did quantifications of western blot from 11/26-11/27. I quantified 2 different images for each membrane. Global background was set based on the image background for each membrane photo.
December 2, 2013
Proteomics: Brest
SOD activity assay of samples extraction in lysis buffer (TL). All samples run in triplicate and diluted 1:2 in dilution buffer from kit. Manufacturer's protocol followed except ddH2O was not put in the blank 1 and 3 wells. Used standard curve but also ran kinetic protocol, reading plate ever 1 min 10s over 20 minutes. 20 µl of lysis buffer was put in wells for blank 2.
Some of the reaction curves for the SOD assay had data points that were obviously incorrect (i.e. were scatter randomly about the plot instead of in a linear relationship). If no part of the plot was linear, the data will not be used. If the plot became linear after the random scattering, then only the linear portion of the plot will be used. Here is a list of samples that had to be completely deleted (sample number.replicate): 14.2, 28.3, 34.2, 47.1, 278.1, 376.1. The 2 lowest concentration standards (0.5 and 0.25) needed to have points removed because the reaction curve plateaued after a certain point. Most of the samples that had points removed had them removed from the beginning of the kinetic reaction. The samples that had points deleted were: 34.1, 38.2, 38.3, 41.1, 44.3, 56.1, 56.3, 59.2, 236.3, 266.1, 266.2, 278.2, 278.3, 367.1, 367.2, 367.3, 373.2, 373.3, 376.3, 379.1, 379.3. Overall, I think it was a good call to do the kinetic reaction curves since so many of the samples had inaccurate starting absorbance values.
Slopes of the linear curves were calculated for each sample, blank, and standard. Slopes for blanks 2 and 3 were 0. Calculated coefficient of variation (standard dev/mean) of the triplicate slopes for each sample. If CV>20%, the outlier slope was excluded from further calculations. Sample 47 shows too much variability to be included. I calculated the inhibition rate according to the equation in the manufacturer's protocol, which ends up being ([slope for blank 1]-[slope for sample])/[slope for blank 1] *100 since slopes for other blanks = 0. However, this resulted in inhibition rates ranging from >100% to >2000%, which doesn't make any sense.
I plotted the standard concentrations against the average slope values for each standard (excluded 0.5 and 0.25) and fitted the logarithmic curve in Excel. I used the logarithmic eqn to calculate concentrations based on slope values for the other samples. I also plotted the linear relationship between LN(standard concentration)~slope average and derived sample concentrations. I think the latter is more accurate because the former results in negative sample concentrations.
November 27, 2013
Proteomics: Brest
Second day of western blot started Nov 26. Photos were successfully developed and the blots worked really well - I don't need to redo them! I will quantify them tomorrow.
The control is ~65 kDa, the larger band is ~75 and the smaller bad is ~37.
November 26, 2013
Proteomics: Brest
Western blot using AMPK phosphorylated Thr172 as previously described. Samples used are those extracted in tampon de lyse Nov 13 and 18. Samples were prepared by diluting all of them to 2.235 mg/ml (based on least concentrated sample's concentration) in tampon de lyse to make 100 µl. 33.3 µl of loading buffer (475 µl loading dye + 25 µl beta mercaptoethanol) was added to each sample, vortexed, heated at ~100°C for 10 minutes, incubated at RT for 10 min, vortexed, spun down, and stored on ice. Samples were almost immediately loaded into prepared gels. Order of gels are left to right when looking at the gel from behind. 5 µl of ladder was loaded, 10 µl of control, and 25 µl of each sample. On each gel, there are 2 protein samples from 400 µatm, 2 from 1000, 3 from 2800, and 1 from 2800 + mechanical stress. 2 additional 2800 + MS samples were prepared and not loaded (47 and 28) and samples 38 and 34 were not prepared. Upper corner near ladder is cut on gel 1, all other 3 corners are cut on gel 2.
Gel 1 order: ladder, AMPK treated control (cell signaling technologies), 278, 236, 373, 367, 17, 56, 59, 41
Gel 2 order: ladder, control, 233, 266, 379, 376, 53, 14, 50, 44
November 25, 2013
Proteomics: Brest
SOD activity assay using kit of samples extracted in lysis buffer, diluted 1:10 in kit dilution buffer. With 1:10 dilution, sample concentrations were at the very end of the standard curve. Additionally, on reading the protocol more carefully, apparently the enzyme working solution is not good after 3 weeks at 4°C. Ours has definitely been around longer than this so I need to follow up with Catherine. I'm also having trouble with post-assay calculations...I will redo the assay with less dilute samples and may try the kinetic method instead of the standard curve.
November 22, 2013
Proteomics: Brest
Ran GST assay in triplicate for all the samples extracted in Triton buffer. Activities were calculated using the equation in the manufacturer's protocol. Activities were corrected for different starting concentrations by dividing the activity (µmol/ml/min) by the sample concentration (mg/ml). The entire 20 minutes of the reaction was used to calculate activity since the entire curve was linear. Using ANOVA and binomial GLM, there are no significant differences among the treatment groups. If you squint, there is a slight suppression of activity at 1000 and 2800 µatm compared to control and then activity becomes elevated again at 2800 + mechanical stress. However, due to low sample sizes and some inter-individual variability it's hard to tell if the trends are "real".
November 21, 2013
Proteomics: Brest
Test of GST and SOD enzymatic assays with most recently extracted samples. I chose 2 samples that have the lowest concentrations and one with a higher concentration for each test (n=3 samples tested total). For SOD this was 278, 38, and 50 and for GST 382, 272, and 361. All samples run in duplicate.
For SOD, followed manufacturer's protocol but did not do a standard curve because Catherine was gone and I couldn't find the SOD standard. Samples were diluted both 1:2 and 1:10. It seems that the assay will still work with samples diluted 1:10, based on the values from October 9.
Followed the manufacturer's protocol for GST. For the GST control, used 4 µl of GST diluted 1:10 or 1:5. Used 20 µl undiluted sample. Assay was run for 20 minutes with reads every 50s. It looks like the 1:5 dilution of the control gives a good, linear curve. The 3 samples all showed a linear response, although 361 had by far the steepest curve (being the more concentrated sample). But even the least concentrated (382 at 1.7 mg/ml) had a linear kinetic response.
November 20, 2013
Proteomics: Brest
NMDS of antioxidant enzymes from proteomics dataset (see 11/5/13). There was one redundant protein that has been removed from the dataset so that there are 30 proteins. NSAF data were Log(x+1) transformed and Bray-Curtis dissimilarity matrix was used. In plot, orange points represent 2800 µatm, 400 µatm are in blue, and mechanically stressed oysters are the closed circles. There is no difference among treatment groups for expression of antioxidant enzymes.
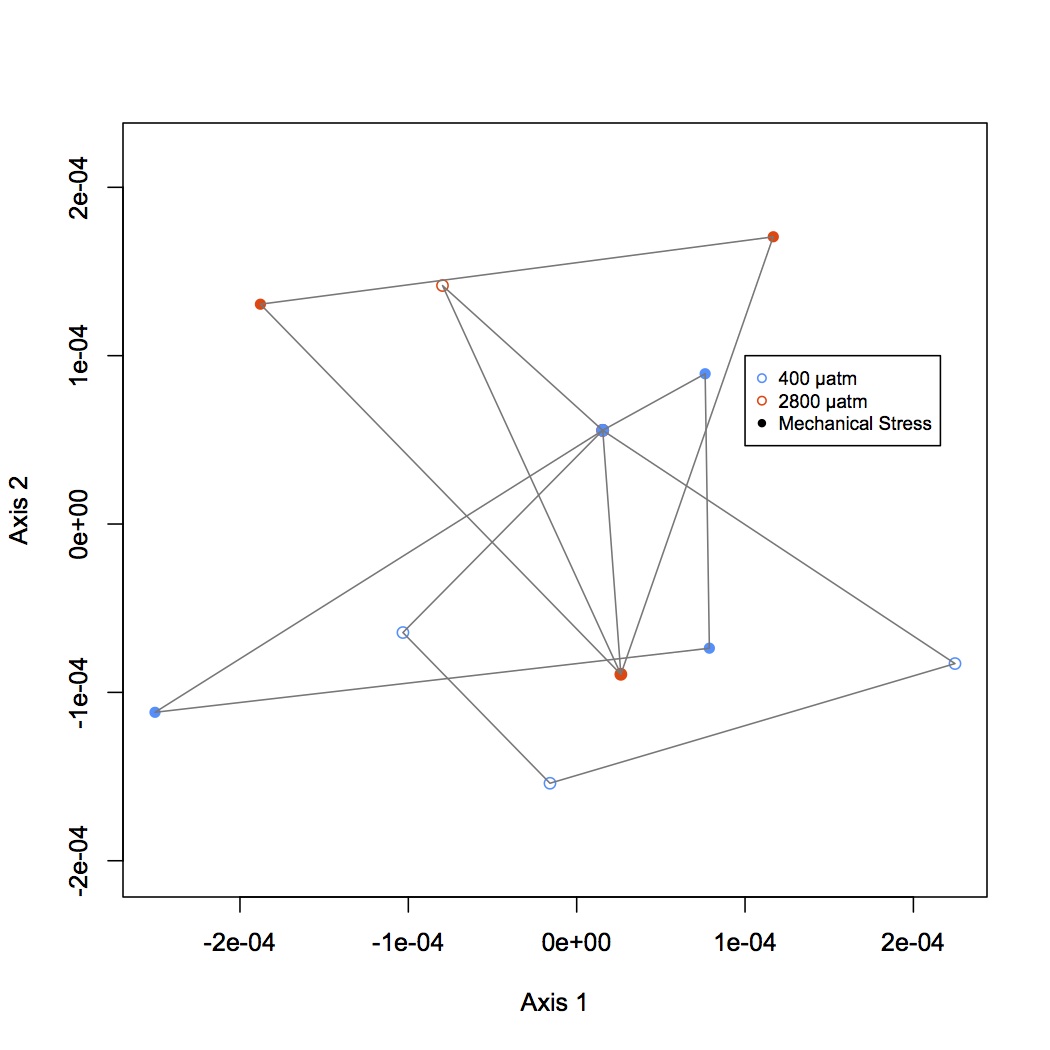
November 18, 2013
Proteomics: Brest
Extraction of samples 14, 17, 20, 23, 50, 53, 56, 59, 25, 28, 31, 34, 38, 41, 44, 47. Some are extracted in lysis buffer and some in triton (see excel spreadsheet for details).
The concentration values were weird for the first plate read, so I read it again and they seemed more consistent. I saved both outputs (the second good one is appended with "bon").
Protein concentrations ranged from 2.38-9.396 mg/ml.
There is a weak correlation between mass of tissue extracted and protein sample concentration (R^2 = 0.53), but this relationship is stronger when R^2 is calculated individually for each extraction method (0.77 for lysis buffer, 0.72 for triton).
November 13, 2013
Proteomics: Brest
All samples are well below 100 mg (0.1 g) so I cannot divide the tissues between the 2 extraction buffers. I am doing each sample with one extraction method and Sam will send more samples to round out the sample sizes. Prepared new Tampon de Lyse (TL) this morning by adding to 50 mL of lysis buffer 2 tables protease, 500 µl phosphatase, and 1 mL NaPPi. No inhibitors were added to triton (TR). Samples are extracted in batches of 8 and details can be found in excel worksheet titled "protein extractions". Both extraction types were done following the TL protocol exactly. 2 µl of each protein sample was diluted 1:10 in 18 µl of water to find concentrations. Samples were stored at -80°C.
Protein concentration was determined using the BioRad DC assay. Samples were all diluted 1:10 and manufacturer's protocol was followed. All samples run in triplicate. If the %CV was >20, the outlier concentration was excluded from the mean. Concentrations ranged from 1.6-7.5 mg/ml.
November 12, 2013
Proteomics: Brest
To test whether the inhibition of the GST assay in the lysis buffer extraction is caused by the buffer itself or the inhibitors that are added, prepared some triton with inhibitors. To 12.5 ml of triton prepared on 10/3/13, added 1/2 tablet protease inhibitors, 250 µl NaPPi (decrystalized), and 125 µl phosphatase inhibitors. For some of the triton assays, will added 10 µl of sample and 10 µl of triton-extracted sample. For the GST control, did 2 wells of 2 µl of undiluted enzyme and 2 wells of 4 µl of 10x diluted enzyme. Assay was read at 340 nm ever 50 s for 20 minutes.
Ground gill samples with mortar and pestle in liquid nitrogen. The following samples were ground. * = posterior gill, others are anterior.
control (400 µatm): 233*, 236*, 239*, 266*, 269*, 272*, 275*, 278*
1000 µatm: 361, 364, 367, 370, 373, 376, 379, 382
2800 µatm: 14*, 17*, 20*, 23*, 50*, 53*, 56*, 59*2800 + mech stress: 25, 28, 31, 34, 38*, 41*, 44*, 47*367 fell on the floor but was still processedhalf of 41 was flung across the room so i will probably need a replacement
These samples were not mailed with the previous batch and need to be sent
1000 + mechanical stress: 385, 389, 392, 395, 398, 401, 404, 407
November 8, 2013
Proteomics: Brest
Received GST kit and did assay of samples extracted Oct 3 and concentrations done Oct 9. Followed manufacturer's protocol for 96-well format. GST control was diluted 10x in sample buffer. Samples were run undiluted (4 µl) and diluted 10x in milliQ water (4 µl). Activity was read at 340 nm every 50 s for 10 min.
At these concentrations it looks like the triton extractions work much better than the lysis buffer extractions. Overall, activity values were pretty weak (2-3 times what was measured in the blanks). I'm going to redo this on Tuesday using greater concentrations of the GST control (undiluted, diluted 5x, 10x, 20x), and greater concentrations of the different samples (undiluted for all, 4 µl, 10 µl, 20 µl).
Note for future assays: aliquot samples into wells first and add enzyme solution last because plate is supposed to be loaded immediately after all is put together.
November 5, 2013
Proteomics: Brest
Created list of antioxidant proteins that were sequenced in proteomics (from supp table S3 from manuscript). Searched for the following terms to make list: superoxide, catalase, glutathione, dual oxidase, peroxidase, peroxiredoxin, thioredoxin, glutathione reductase. This resulted in 31 proteins (some CGI numbers have identical annotations - need to do alignments to determine if really different proteins). In response to OA, glutathione S-transferase omega-1 and dual oxidase 2 were expressed >2-fold in high pCO2 vs low pCO2 and mitochondrial glutathione reductase (n=2), glutathione S-transferase 3 (n=2), another glutathione S-transferase omega-1, and thioredoxin domain-containing protein 16 were expressed only at high pCO2. A GST omega-1, GST mu 3, and dual oxidase 2 were downregulated at elevated pCO2. For response to mechanical stress alone GST omega-1 was upreg at least 2-fold and glutathione reductase (n=2), GST 3 (n=2), GST omega-1, dual oxidase 2, thioredoxin domain-containing protein 16 were all expressed only after mech stress. GST P2, GST omega-1, dual oxidase 2, and thioredoxin domain-containing protein 17 were all downreg after mech stress. In response to OA + mech stress GST A, GST 3, and thioredoxin domain-containing protein 17 were upreg at least 2-fold while GST omega-1, GST 3, GST Mu 3, dual oxidase 2 (n=2), and thioredoxin domain-containing protein were expressed only after mech stress. GST A and GST omega-1 were at least 2-fold down reg.
October 29, 2013
Proteomics: Brest
Day 2 of western blot. I made new PBS + tween because Charlotte thinks that one source of poor images for the blots is bacteria growing in the PBS. The images turned out well, although there is still some background. Yanouk thinks this is due to using the older (small) bottles of chemiluminescent reagent.
October 28, 2013
Proteomics: Brest
Charlotte finished up the blots started 10/24 on the 25th and they turned out well.
Day 1 of western blot for samples 39, 42, 45, 48, 51, 54 (gel 1) and 94, 97, 100, 103, 106, 109 (gel 2). corner near the ladder is cut for gel 1 and all 3 other corners are cut for gel 2. The control A1 (see 9/25) was used for both gels. Samples were diluted (see 9/18) and prepared in loading buffer before loading on gels.
A new primary antibody solution was used.
October 24, 2013
Proteomics: Brest
Day 1 of western blot (same samples as 10/22 but in numerical order and only 1 ladder on each gel). Prepared new témoin (positive control A1). To 25 µl of protein added 8.3 µl of loading buffer. Mixed, heated at ~100°C for 10 minutes, cooled at RT, spun down and stored on ice until gel was loaded (13.3 µl of control loaded onto gel).
Samples were previously prepared in loading buffer and so were denatured at 100°C for just 5 minutes.
for gel 1 (samples 38-53), upper corner on ladder side (right when facing gel) is cut. for gel 2 (samples 93-107) all 3 corners except upper corner on ladder side are cut.
October 23, 2013
Proteomics: Brest
Day 2 of western blot. I had forgotten to add SDS to the electrophoresis buffer yesterday. When I noticed, I added some and finished running the gel but it ended up making the bands kind of smeary. At least the revelation of the gels worked ( made new developing reagents), but the blots need to be redone.
October 22, 2013
Proteomics: Brest
Day 1 of western blot (previously described). Gel 1 samples (in order): ladder, témoin, 38, 41, 44, 50, 47, 53. Gel 2 samples (in order): ladder, ladder, témoin, 93, 96, 99, 102, 105, 107. Primary antibody had been used once before. Membrane 1 upper corner near ladder is cut, membrane 2 2 corners opposite ladder are cut.
Charlotte gave me samples from different tissues from an experiment done in 2008-2009 to test the new MAPKAP-2 antibody. Found concentrations using BioRad assay. All samples were diluted to 4.3 mg/ml using tampon de lyse and then put in -80°C. to load 45 µg of sample will be 13.9 µl of protein diluted in loading buffer.
| Sample |
conc (mg/ml) |
| mantle |
11.1 |
| mantle border |
6.3 |
| gill |
4.3 |
| smooth muscle |
5.3 |
| striated muscle |
10.5 |
| palps |
7.0 |
| dig gland |
6.2 |
October 9, 2013
Proteomics: Brest
Measured concentrations (using biorad assay kit and plate reader) of extractions done 10/3/13. Samples were only diluted 10x. Also found average expression for only 10x dilution for concentrations done 9/30/13. TL = extracted in tampon de lyse, TR = extracted in Triton.
| Sample |
Conc (mg/ml) |
| TL1 |
7.59 |
| TL2 |
6.41 |
| TL3 |
5.62 |
| TL4 |
9.43 |
| TR1 |
9.57 |
| TR2 |
8.56 |
SOD (total) enzyme activity using Sigma kit (manufacturer's protocol can be found here: http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Datasheet/6/19160dat.Par.0001.File.tmp/19160dat.pdf) For the most part I followed the manufacturer's protocol (exceptions noted with "^"). I diluted the samples 1:10 in the buffer that they were extracted with (tampon de lyse or triton mix.), I realized after that samples are actually supposed to be diluted in the kit provided dilution buffer. Prepared WST working solution and enzyme working solution. ^Prepared standard curve as follows^:
| final SOD U/ml |
vol SOD |
SOD to be diluted |
vol dilution buffer |
| 100 |
10 |
stock |
1490 |
| 20 |
80 |
100 |
320 |
| 8 |
160 |
20 |
240 |
| 4 |
200 |
8 |
200 |
| 2 |
200 |
4 |
200 |
| 1 |
200 |
2 |
200 |
| 0.5 |
200 |
1 |
200 |
| 0.25 |
200 |
0.5 |
200 |
All samples were done in triplicate and aliquoted with buffers according to manufacturer's protocol. For some reason, there was not enough of the Triton samples to put 20 µl in all 3 wells but there was enough of the T de L samples ( I made them at the same time so I'm not sure what I did wrong here). As a result, the 3rd triton wells were left out of calculations. Incubated plate at 37°C for 20 min, then placed in plate reader and read at 450 nm (NB: make sure plate reader is warmed up to 37°C before it's time to read the plate).
The activity assay is actually a measure of SOD inhibiting a reaction (the less the inhibition, the darker the yellow color in the well). For the standard curve, absorbance values must be log-transformed to make a linear curve. The 100 standard can be excluded from the curve to increase linearity. Activity of SOD in the tampon de lyse-extracted samples was about 4X > activity in the triton-extracted samples. For the tampon de lyse samples, there was good within and between sample consistency of SOD activity.
This is an efficient assay because with the concentrations of these samples (5-9 mg/ml), only 7 µl was needed to make a working solution of 70 µl.
October 3, 2013
Proteomics: Brest
Finished Day 2 of western blot with Fiz. With >4 minutes of development time, there are clear AMPKa phosphorylation bands visible. Expression does seem to change with the different diet regimes the larvae experienced.
extractions of samples from 9/27 using triton to compare the enzymatic assay results for both extraction methods. Made 50 mL of Triton extraction buffer: 50 mL PBS 10 mM, 0.0185 g EDTA 1 mM, 50 µl Tritonx100. Added 100 mg of tissue to 300 µl triton buffer. Only extracting 2 samples: 1 = mix of oysters 1,3,and 4 and 2 = oyster 2. Homogenized tissue in triton and rinsed off blades with 100 µl triton. Kept at RT for ~7 min then spun 1 hour, 4000 rpm, 4°C.
October 2, 2013
Proteomics: Brest
Today I helped Fiz with day 1 of the western blot protocol. I realized that one mistake I made the other day was that I did not equilibrate the gels in transfer buffer before assembling the transfer sandwich.
October 1, 2013
Proteomics: Brest
Day 2 of western blot. Recommended PBS+tween washes at 1 pm (see 9/25/13). Saved and froze primary antibody solution.
The blots didn't work out very well. And I think I probably mixed up the gels, although neither really looks like the one I did 9/25. The membrane I ended up calling membrane 1 had a very faint signal - most of the bands couldn't be seen with an exposure <4 minutes. The other membrane has a dark shadow in front of it and cannot be clearly viewed.
September 30, 2013
Proteomics: Brest
Diluted samples extracted 9/27/13 1:5, 1:10, and 1:20 in water. Followed protocol outlined 9/18/13 for finding concentration of samples using standard curve. Best dilution seemed to be 1:10. Samples are ~6 mg/ml. Also found concentration of new tube of control (A1, now called T blot). Its concentration was ~4 mg/ml. For 20 µg of T blot, load 13.3 µl into gel.
Day 1 of western blot (see 9/24/13). Redid samples from 9/24, but used same dilutions in loading buffer previously made. To denature, heated 5 min at 100°C. The second gel had 6 new samples: 92, 95, 98, 101, 104, 107. Primary antibody = AMPKa phosphorylation sites. *I am 90% sure that I didn't mix the gels up and that the one with 1 corner cut off is gel 1 (with samples from 9/24) and the one with 2 corners cut off is the new samples. Membranes left to incubate with primary antibody in cold room at 6:30 pm.
September 27, 2013
Proteomics: Brest
Went to Argenton this morning and sampled gill tissue from 4 oysters. Dissected gill and placed directly in liquid N2. Brought back to lab and ground frozen tissue into powder using metal thing with ball. Weighed 100 mg of tissue into a tube containing 300 µl lysis buffer (with added protease, etc.). Homogenized (on ice), rinsed off homogenizer with 100 µl additional lysis buffer, and then let sit 40 minutes at RT.
Spun samples for 1 hour at 4°C, 4000 rpm.
Removed middle layer to new tube. Spun at 10,000rpm, 45 min, 4°C.
September 25, 2013
Proteomics Brest
Day 2 of western blot
Removed primary antibody wash and incubated membranes on rocker with PBS+tween for 10 minutes and then for 2*5 minutes. Incubated with rabbit anti-goat antibody for 1 hour (in PBS+tween+BSA, 20 µl antibody in 50 mL - dilution = 1/5000).
After second antibody, incubated 10 min with PBS+tween and then 5 minutes. Left membranes in PBS+tween until development of films (on bench top).
For each membrane, made 5 mL of reagent that binds to antibody and emits light: 2.5 mL Immun-star HRP luminol/enhancer + 2.5 mL Immun-star HRP peroxide buffer (keep away from light). In dark room, placed membranes in dish and poured reagent over, covering membrane surface. Incubated 5 min. Transferred membranes into casette lined with plastic (no wrinkles!) and then folded to cover membranes with plastic. Placed film on top of the plastic-coated membranes and close casette (best time seems to be about 3.5 minutes). Transfer film to realizing reagent (made ahead of time), dipping it in a few times and checking band darkness against red led. Then put film in fixative (made ahead of time) for a minute or so. Transfer to water.
The top row is loaded with 10 µl of product (45 µg) and the bottom has 20 µl (90 µg). It seems that we could probably load about 25 and still have plenty of signal. A1 is a control (undifferentiated oyster gonad) and 10 µl were loaded.
https://www.evernote.com/shard/s242/sh/85d68e07-749f-410f-827c-ca59f94013a8/a254033e3bfe19e381796b26afd8f985
Secondary stress: Proteomics
Edited xy plots showing differentially expressed proteins by color to highlight those proteins that have q-value < 0.1.
Analysis comparing proteomic response to mechanical stress at 2 different pCO2 - this is a direct comparison of expressed proteins at 400 + Mech stress and 2800 + Mech stress. Did q-value in R and all q-values = 1 (no sig qvalues). Created dataset of proteins that are at least 5-fold different between the 2 treatments. Made xy plot, heat map, and did enrichment analysis.
Annotated these proteins with SPIDs.
SELECT * FROM [emmats@washington.edu].[mech_stress_diff_exp_for_annotation.txt]
LEFT JOIN [emmats@washington.edu].[table_TJGR_Gene_SPID_evalue_Description.txt]
ON [emmats@washington.edu].[mech_stress_diff_exp_for_annotation.txt].[All Proteins]=[emmats@washington.edu].[table_TJGR_Gene_SPID_evalue_Description.txt].[CGI Protein]
Made sure dataset only includes proteins that are expressed in at least 2 oysters across all 8 for the comparison. Enrichment analysis showed that RNA splicing, mRNA processing, chordate embryonic development, and mRNA metabolic process are enriched.
Redid q-value for all treatments because realized that I had included p-values for comparisons of expression = 0 in both treatments being compared. The results were qualitatively the same (the same proteins were significant) but q-values were slightly different. For the comparison of just mechanically stressed oysters, one protein was sig diff: 26S protease regulatory subunit 8, which is expressed only in oysters exposed to OA before mech stress.
September 24, 2013
Proteomics: Brest
Test of Western Blot using phosphorylated AMPK antibody. Samples tested (from microtraces): 37, 40, 43, 46, 49, 52. All water used is milliQ.
Made 2 gels de séparation (10%): 4.05 ml H2O, 2.5 ml Tris lower, 100 µl SDS 10%, 3.3 ml acrylamide. Stirred for a few minutes then added 50 µl 10% APS, 5 µl TEMED. Stirred briefly and pipetted 4.7 ml into 2 gel molds. Added about 100 µl of water to the left and right of the gel to make a straight line at the top. Let solidify 45 minutes.
Made gel de concentration (4%): 6.1 ml H2O, 2.5 ml Tris upper, 100 µl SDS 10%, 1.33 ml acrylamide. Poured off water from top of gels de séparation. Stirred for a few minutes then added 50 µl 10% APS, 15 µl TEMED. Filled gel molds to the top with gel de concentration. Insert combs (refilled with gel de concentration). Let solidify 40 min.
Yanouk prepared 1X electrophoresis buffer and 1X transfer buffer (the latter stored at 4°C).
Prepared samples: diluted all samples to 6 mg/ml in tampon de lyse to a total volume of 100 µl. Added 33.3 µl of loading buffer (prepared ahead of time). Heated at 100°C for 10 minutes, cooled at RT 10 minutes, spun down and stored on ice.
Removed gel combs and added water to top to get rid of bubbles. Poured off water and emptied wells using a thin strip of whatman paper. Secured gels in electrophoresis rig, wells facing in. Filled with electrophoresis buffer. In far left well for each gel put in 5 µl ladder and in the next well put in 10 µl of A1 control. In the first gel (upper corner opposite latter cut off) put in 10 µl of each sample. In second gel (both corners opposite ladder cut off) put in 20 µl of sample. Fil rig halfway with electrophoresis buffer and ran gels for 10 min 40 mA constant/100V then 45 min 80 mA/200V. When gels are run, remove from molds and cut off the gel de concentration.
Cut 2 membranes to correct size and equilibrated: 15 s in MeOH, 1 min in H2O, >10 min transfer buffer on rotating platform. Soaked scotch brite (4) and whatman paper (4) in transfer buffer. Before sandwich assembly, equilibrated gels in transfer buffer (<10 min). For plastic sandwich holder, the black part is the bottom. Layer 1 scotch brite, 1 whatman paper, reversed gel (ladder on right, up is up), membrane, whatman, scotch brite. For the last 2 steps, roll out bubbles.
Assemble rig with black sides of sandwiches facing black part of rig. Add ice pack and stir bar. Fill with transfer buffer and place on stir plate. Run for 1 hour at 100 V/250 mA constant.
Unmake membrane sandwiches and cut membrane corners as gel corners are cut. Place membranes in PBS on rocker for 5 minutes. Then 2 successive 5 minute incubations of PBS with 1% Tween 20. Incubate with PBS + tween + 3% BSA at ~40°C for 1 hour on a rocker.
Wash with PBS+tween: 10 minutes then 2 times, 5 minutes each.
Incubated over night at 4°C with primary antibody (diluted 1/1000, 50 µl in 50 mL PBS+tween+BSA). The primary antibody is AMPK alpha phospho Thr 172 - binds to phosphorylated threonine in AMPKa.
Secondary stress: proteomics
Preparing abstract for Ocean Sciences 2014. Joined proteins used for analysis with GO terms:
SELECT * FROM [emmats@washington.edu].[NSAF with averages, 5-fold, SPIDs, enrichment]
LEFT JOIN [dhalperi@washington.edu].[SPID_GOnumber.txt]
ON [emmats@washington.edu].[NSAF with averages, 5-fold, SPIDs, enrichment].SPID=[dhalperi@washington.edu].[SPID_GOnumber.txt].A0A000
Then joined with GO Annotations/GO Slim
SELECT * FROM [emmats@washington.edu].[Proteins with NSAF and GO]
LEFT JOIN [sr320@washington.edu].[GO_to_GOslim]
ON [emmats@washington.edu].[Proteins with NSAF and GO].[GO:0003824]=[sr320@washington.edu].[GO_to_GOslim].GO_id
Made a list of proteins corresponding to immune-related GO terms: MAPKKK cascade, activation of MAPK activity, response to reactive oxygen species, response to superoxides, age-dependent response to oxidative stress, age-dep response to ROS, toll-like receptor signaling pathway, antigen processing and presentation of peptide antigen via MHC class I, antigen processing and presentation of exogenous peptide antigen via MCH class I TAP-dependent, mast cell chemotaxis, MyD88-dep toll-like receptor signaling pathway (and independent), negative regulation of inflammatory response to antigenic stimulus, superoxide metabolic process, xenobiotic met. process, phagocytosis, phagocytosis engulfment,autophagy, apoptosis, anti-apoptosis, induction of apoptosis, activation of caspase activity, cell structure and disassembly during apoptosis, response to stress, defense response, inflammatory response, immune response, response to oxidative stress, leukocyte adhesion, I-kappaB kinase/NF-kappaB cascade, JNK cascade, toll signaling pathway, cell death, induction of apoptosis by extracellular (& intracellular) signals, activation of caspase activity by cytochrome c, pathogenesis, response to virus, response to bacterium, positive regulation of necrotic cell death, pos reg of cell death, viral reproduction, detection of bacterium, immunoglobulin mediated immune response, reactivation of latent virus, viral infectious cycle, intracellular transport of viral proteins in host cell, viral assembly maturation egress and release, viral transcription, cytokine-mediated signaling pathway, removal of superoxide radicals, antimicrobial humoral response, apoptotic nuclear changes, neg reg of NF-kappaB transcription factor activity, interleukin-10 production, interleukin-12 production, reg of interleukin-6 prod, reg of tumor necrosis factor prod, neg reg of TNF prod, pos reg of interleukin-1 beta prod, pos reg of superoxide release, TNF-mediated signaling pathway,response to cytokine stimulus, toll-like receptor 1 (2,3,4) signaling pathway, cellular response to oxidative stress, cell. response to ROS, hemocyte proliferation, wound healing, cytokine biosynthetic process, B cell proliferation, positive regulation of T cell prolif., T cell activation, B cell activation, neutrophil activation, xenobiotic catabolic process, natural killer cell mediated cytotoxicity, pos reg of TNF biosyn process, response to H2O2, superoxide anion generation, antigen processing and presentation of exogenous peptide antigen via MHC class I, defense response to bacterium, H2O2 metabolic process, H2O2 catabolic process, xenobiotic transport, regulation of apoptosis, positive reg of apoptosis, negative reg of apoptosis, neg reg of PCD, regulation of I-kappaB kinase/NF-kappaB cascade, pos reg of I-kB kinase/NFkB cascade, regulation of MAPKKK cascade, reg of JUN kinase activity, pos reg of JUN kinase activity, neg reg of JUN kinase activity, neg reg of neuron apoptosis, innate immune response, reg of innate immune response, reg of anti-apoptosis, reg of JNK cascade, pos reg of JNK cascade, neg reg of JNK cascade, viral genome transport in host cell, cytokine secretion, H2O2 biosynthetic process, reg of inflammatory response, neg reg of inflammatory response, neg reg of immune response, T cell receptor signaling pathway, B cell receptor signaling pathway, reg of T cell activation, pos reg of NFkB transcription factor activity, stress activated MAPK cascade, defense response to virus, neg reg of cell death, cellular response to H2O2, response to interleukin-1, response to interleukin-15
Removed all annotations with e-value <1E-10. Removed redundancies. 189 proteins are annotated with one of 117 immune-related GO term. 13 of these proteins are expressed at least 5-fold different in 2800 vs. 400 µatm: universal stress protein MSMEG_3950, universal stress protein SII1388, coactosin, lymphocyte cytosolic protein 2, glutathione S-transferase omega-1, allograft inflammatory factor 1, neurogenic locus notch homolog protein 1, 60S ribosomal protein L21, 40S ribosomal protein S9, quinone oxidoreductase, 26S proteasome non-ATPase regulatory subunit 14, host cell factor 1, tubulin alpha-1 chain, 26S non-ATPase regulatory subunit 14.
7 immune-related proteins are at least 5-fold different in response to mechanical stress at 400 µatm and 15 change in response to mech stress after 1 month at elevated pCO2.
Revisions of data for manuscript. Verified that proteins differentially expressed according to q-value are included in the 5-fold data sets for OA and mech stress at 400. 2 proteins are not included in the mech stress at 2800 data set, so I am adding them in (CGI#s 10023513 and 10005784) and redoing enrichment, heat map, and venn diagram for this treatment comparison.
September 18, 2013
Proteomics: Brest
Total protein concentration using the BioRad DC assay. First needed to determine the dilution factor for the pooled whole body samples. I diluted 3 samples (95, 38, 47) 1:2 to make further dilutions 1:5 (40 µl 1:2 + 60 µl H2O), 10 (20 1:2 + 80 H2O), 20 (10 1:2 + 90 H2O) and 40 (2.5 1:2 + 97.5 H2O). Standards are 0 µg BSA/ml, 250, 500, 750, 1000, and 1500. All samples are measured in triplicate. 5 µl of each standard or sample is aliquoted into 3 wells in a welled plate to go on the plate reader. 25 µl of solution A' is added to each well (A' = 1 ml solution A + 20 µl solution S). 200 µl of solution B is added to each well. Plate is gently agitated for 5 s on desktop and then left to incubate at RT for 15 minutes. Plate is then loaded into reader and KC4 software is used to analyze.
KC4 instructions:
click "new"
click "wizard"
enter plate layout - sample designator, dilution factor
read plate
after plate is read, click "curves" and choose M750
save as
export to excel
load standard curve, table with curve fit, and table with concentrations x dil.
the samples diluted 20x fell in the middle of the curve. Diluted the rest of the extracted samples 1:20 by putting 5 µl of extracted protein in 95 µl water. Repeated same steps as above (had to use 2 plates to accommodate all the samples). A few samples had one well (out of 3) that did not change color yellow -> blue after 15 minutes so did not enter them as samples in the plate wizard. NB: the software automatically does not include replicates in the mean that are way far off the range of the other 2. Plate data saved as microtraces 1 & 2 18092013.
Secondary stress: proteomics
Query from yesterday never finished running so I killed it. Steven has a perl script that will do the same thing and he is going to fix table S6.
Working on pathway representation for manuscript. I'm playing around with ipath now. I've made a file of all the proteins expressed at least 5-fold different between treatments. Proteins that change in response to OA are in red, in response to mech stress at 400 µatm are blue, and in response to mech stress at 2800 µatm in yellow. If proteins change in response to more than one stress, they are the combination of those colors: OA + mech at 400 = purple, OA + mech at 2800 = orange, both mech = green, all 3 = black. If the proteins are expressed higher in the stress treatment(s), the lines are thickest. If they are expressed lower, the lines are thinnest. If they do not change in the same direction the lines are an intermediate weight.
Made another version of the same plot, but this time all proteins have same line weight and lighter lines represent down-reg and darker lines represent up-reg. If down vs. up is not consistent for proteins responding to >1 treatment, then color is dark.
September 17, 2013
Proteomics: Brest
Extracted 6 more of Yanouk's samples as described 9/12/13: 95, 96, 97, 98, 100, 107. Samples were thawed overnight at 4°C. After second spin, samples were left for ~1 hour on ice (layers were still separated).
Claudie showed me how to use the BIO Rad DC protein assay kit for measuring protein concentration.
Secondary Stress: proteomics
Editing supplementary table S6. Uploaded table successfully to new SQL share. I'm trying to put each spid in its own row. I found code to do this here: http://stackoverflow.com/questions/5493510/turning-a-comma-separated-string-into-individual-rows
I'm not sure if this actually works yet because it is still running...
;with tmp(Comparison, [Enriched GO Term], PValue, [Fold Enrichment], FDR, Proteins, DataItem) as (
SELECT Comparison, [Enriched GO Term], PValue, [Fold Enrichment], FDR, LEFT(Proteins, CHARINDEX(',', Proteins+',')-1),
STUFF(Proteins, 1, CHARINDEX(',', Proteins+','),'')
FROM [emmats@washington.edu].[Supplementary_Table_S6.txt]
UNION ALL
SELECT Comparison, [Enriched GO Term], PValue, [Fold Enrichment], FDR, LEFT(Proteins, CHARINDEX(',', Proteins+',')-1),
STUFF(Proteins, 1, CHARINDEX(',', Proteins+','),'')
FROM tmp
WHERE Proteins > ''
)
SELECT Comparison, [Enriched GO Term], PValue, [Fold Enrichment], FDR, DataItem
FROM tmp
ORDER BY Comparison
OPTION (maxrecursion 0)
September 16, 2013
Proteomics: Brest
I extracted 10 more of Yanouk's samples as described 9/12/13. Sample numbers were: 40, 44, 47, 93, 99, 92, 101, 103, 104, 109.
September 12, 2013
Proteomics: Brest
Extraction of Yanouk's samples from 15 days of pesticide exposure (16 pooled spat per sample): 41, 42, 38, 39, 51, 54, 45, 50,104, 106 (104 and 106 were digested in lysis buffer today by YE)
Previously, Yanouk homogenized the tissue samples and incubated them in lysis buffer with protease inhibitors, phosphatase inhibitors, and NaPPi (sodium pyrophosphate, another phosphatase inhibitor).
Thawed samples on ice. Transferred to new tubes. Spun 1 hour, 3000xg, 4°C. Removed middle layer into 2 eppendorfs (except for 104 and 106, which had smaller volumes and were aliquoted into one tube). Centrifuged at 10,000xg, 45 minutes, 4°C. (Before centrifugations, centrifuges were precooled to 4°C.)
After centrifugation, if samples were in separate tubes they were combined into 1 5-mL tube, vortexed, and then aliquoted into 2 eppendorfs. Stored at -80°C.
Downloaded MKK2 sequences from multiple species/taxa. Exported as fasta to do alignment in ClustalX. Made tree in Geneious: cost matrix=Blosum62, gap open penalty = 12, gap extension penalty = 3, alignment type = global alignment with free end gaps, genetic distance model = jukes-cantor, tree build method = neighbor-joining, outgroup = none.
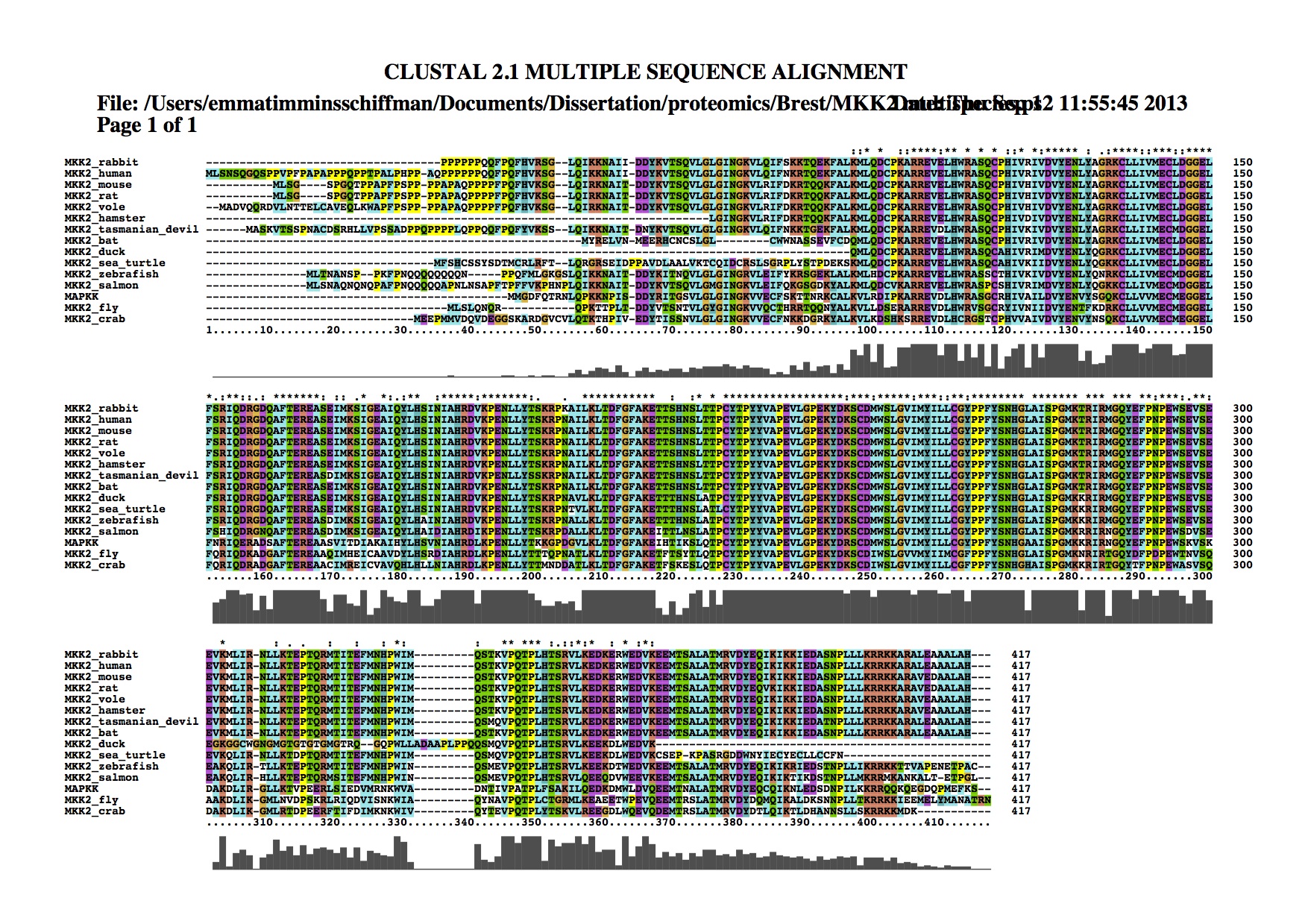
Tree can be found here: https://www.evernote.com/shard/s242/sh/d16f1229-e12d-446d-bfb5-e9c0489e185f/4f889303abb956068fbe8f51e8e4cee0 Invertebrates cluster together, as do mammals and fish. The one species each from birds (duck) and reptiles (sea turtle) cluster together probably because there aren't enough representatives of each group to give good support.
September 11, 2013
Proteomics: Brest
We have decided to investigate GST and MAP kinase-activated protein kinase. Used pearson's correlation coefficient to determine which proteins change expression similarly to MAPKK. With a cutoff of |0.70|, 9 proteins are positively correlated with MAPKK and 0 are negatively correlated. Correlated proteins are: 3 unknown, aminoacylase 1, ras-related protein rab-10, annexin a7, tubulin beta chain, uncharacterized oxidoreductase YajO, lysosomal alpha-glucosidase.
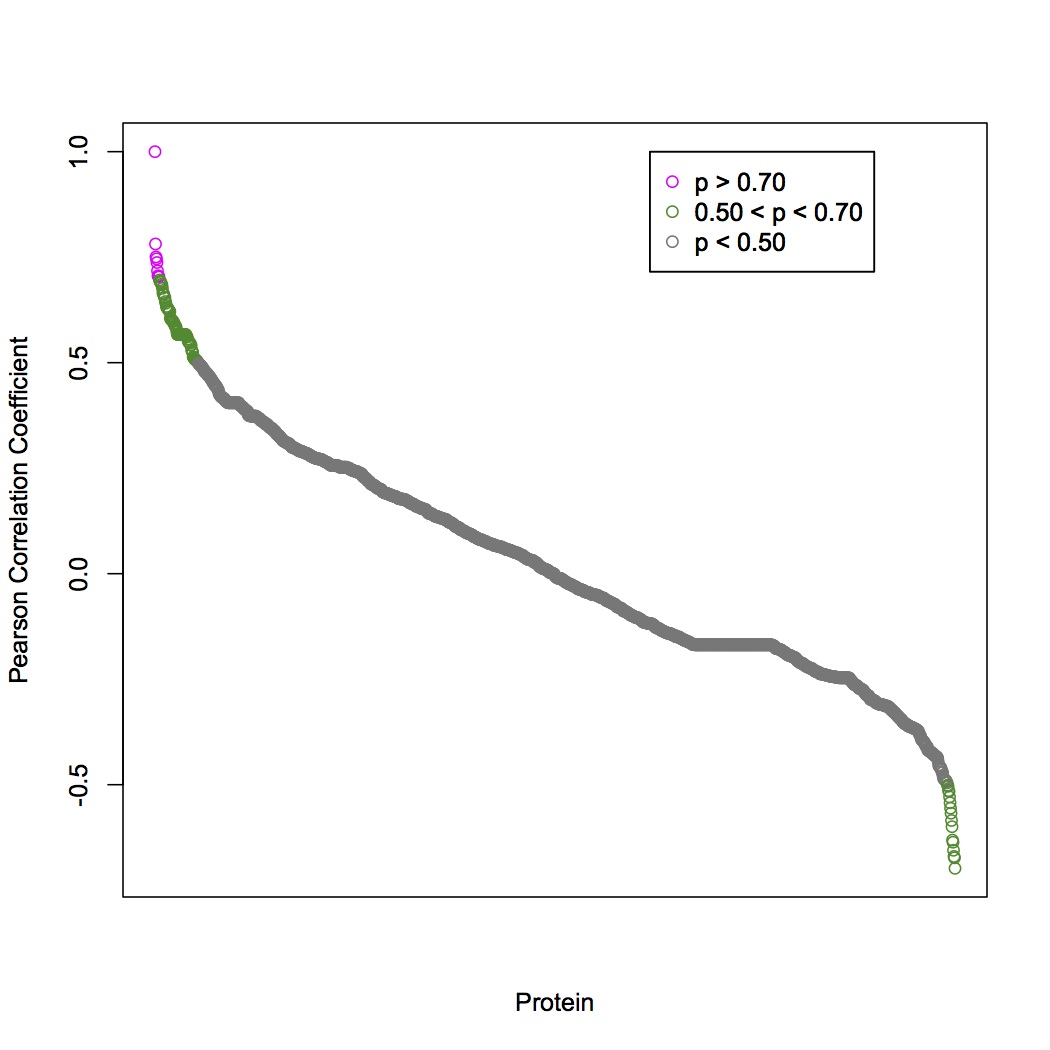
To find MAPKK isoforms, uploaded blast output for entire proteome (see 9/9/13) to SQL and renamed columns. Selected data that only corresponded to MAPKK as query.
SELECT * FROM [emmats@washington.edu].[Brest_proteins_blastpout]
WHERE Query='CGI_10003308'
4 proteins have >40% identification with MAPKK (this is an arbitrary cut-off that I have selected, it seems pretty liberal). In Geneious, aligned these protein sequences with the MAPKK sequence. Only 2 of them really shared sequence similarity with MAPKK in its active regions (the first 200 aa). Neither of these proteins were identified in the experiment as being expressed. CGI_10026336 is a map kinase-activated protein kinase 5 (the one we have identified is MAPKK 2) and CGI_10012098 is calcium/calmodulin protein kinase 1.
September 10, 2013
Proteomics: Brest
I've taken stock of the remaining samples from the OA/mech stress experiment. 3 tissue samples were taken for each oyster: anterior gill, posterior gill, whole body (i.e. remaining body tissues).
| Treatment |
#AntGill |
#PostGill |
#WholeBody |
Tank |
| 400 |
0 |
11 |
8 |
103B |
| 400+mech |
8 |
4 |
8 |
103B |
| 600 |
16 |
16 |
16 |
102B |
| 600+mech |
8 |
8 |
8 |
102B |
| 800 |
16 |
16 |
16 |
104A |
| 800+mech |
8 |
8 |
8 |
104A |
| 1000 |
16 |
16 |
8 |
104B |
| 100+mech |
8 |
8 |
8 |
104B |
| 1200 |
16 |
16 |
16 |
103A |
| 1200+mech |
8 |
8 |
8 |
103A |
| 2800 |
1 |
12 |
8 |
101B |
| 2800+mech |
8 |
4 |
8 |
101B |
September 9, 2013
Proteomics: Brest
Installed new version of ncbi blast (2.2.28+) on my computer. Navigated to bin folder within blast.
./makeblastdb -in /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/oyster_v9_aa_format1.fasta -dbtype prot -out /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/proteomeDB
./blastp -num_threads 2 -out /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/Brest_proteins_blastpout -db /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/proteomeDB -outfmt 6 -evalue 1 -max_target_seqs 100 -query /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/oyster_v9_aa_format1.fasta
./blastp -num_threads 2 -out /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/Brest_proteins_blastpout_12prot -db /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/proteomeDB -outfmt 6 -evalue 1 -max_target_seqs 100 -query /Users/emmatimminsschiffman/Documents/Dissertation/proteomics/Brest/12proteins.fasta
Renamed column names for blastp output
SELECT
[Column1] AS [Query],
[Column2] AS [Subject],
[Column3] AS [perc ID],
[Column4] AS [align lengths],
[Column5] AS [mismatches],
[Column6] AS [gap openings],
[Column7] AS [query start],
[Column8] AS [query end],
[Column9] AS [subject start],
[Column10] AS [subject end],
[Column11] AS [e-value],
[Column12] AS [bit score]
FROM [emmats@washington.edu].[table_Brest_proteins_blastpout_12prot]
September 6, 2013
Proteomics: Brest
Created list of proteins that show differences between treatment groups from OA/mech stress experiment (https://docs.google.com/spreadsheet/ccc?key=0An4PXFyBBnDEdC1VZWNGaGhIbmZjQnFrX3dhaHB0N0E#gid=0). These proteins have a good e-value (low), are expressed across multiple oysters, and are interesting physiologically. Entered entire protein sequences into Geneious (proteomics > Brest) and created a database of the C. gigas proteome (v9). Searched each protein sequence agains the entire proteome to look for isoforms with the following paramters: blastp, max hits = 20, max e-value = 1e-10, word size = 3, matrix = BLOSUM62, gap cost = 11 1, # CPUs = 1.
The first search of alpha L-fucosidase returned 13 results. The first 3 include the actual sequence and 2 very similar sequences (e-value =0). However, the 10 other sequences look pretty different from the query and I'm having a hard time deciding what a "true" isoform is. I'm going to think about this....
Aligned alpha L-fucosidase with the entire proteome using geneious alignment: cost matrix = Blosum62, gap open penalty = 12, gap extension penalty = 3, global slignment with free end gaps, refinement iterations = 2. This didn't work because there was not enough memory.