Mukilteo water chem
Sammi made new dye this morning. I did spec pH of incoming water and 1 tank from lab, GHA and GHB. Also did dye correction for new dye.
March 21, 2012
Secondary stress
Met with Peter Westley (Quinn lab) to talk about using ImageJ to quantify shell corrosion on the inside of shells. He thinks I should use the polygon function to measure the total shell pixel area and the area of the white spots. To take better pictures, he suggests using a regular digital camera attached to a stand so that it is parallel to the bench top. Put the oyster shell on a white background (probably use only the flat valve to minimize problems with glare), include a scale of some sort, and light with a couple of regular desk lamps. Try to take a picture with the maximum resolution - raw if possible, or tiff, try to make the pictures 4 MB.
March 20, 2012
Secondary stress: proteomics
Retrieved blastall file from 3/15/12 (saved as Galaxy 36 blastall results.xlsx in proteomics folder). Edited file so that there are only 3 columns: Sigenae ID, Swissprot ID, and e-value and uploaded into Galaxy (file 43). Joined this file with GO annotations using Swissprot IDs (Galaxy 44) and then joined with GO Slim terms (Galaxy 45). This file was exported and is called: Galaxy 45 - annotated proteins from blastall.xlsx. Sorted by GO type so that only biological processes were left and then sorted by e-value (cut off =1e-5). Made pivot table from GO slim terms.
Mukilteo water chem
Did spec pH of 9 Muk water samples: incoming water and 2 tanks for each GHA, GHB, and lab.
Also did dye correction of dye made 3/19/12.
March 19, 2012
Mukilteo water chem
Made new m-cresol purple with 0.032 g dye in 40 mL nanopure water. Adjusted pH so that A1/A2 (578/434) ~1.6 (really about 1.43). Next, time start with just 1 drop of 5N NaOH.
Did spec pH of 3 Muk water samples: incoming water for GHA, GHB, and lab.
March 16, 2012
Mukilteo water chem
Spec pH of incoming water for Greenhouse A (GHA), Greenhouse B (GHB), and lab, and one tank from each location as well (total = 6). Sammi had made the dye and done dye correction 3/6/12. Files are saved on Friedman lab computer in 236.
March 15, 2012
Secondary stress
Analysis of size data (t0 and t 1 month) for Experiment 2, 1 month OA exposure.
Did a linear regression of shell weight on buoyant weight since for t0 we only have BW and for t1mo we have both weights. BW (adjusted for forcep mass) is a decent predictor of shell weight via the eqn: y=0.8329(x)+0.2581. Buoyant weight and total weight are also highly correlated, as are shell weight and total weight. Lenght, width, and LxW are not well correlated to BW.
Based on a one-way ANOVA with pCO2 as the fixed factor, there is no difference in shell weight (using BW as a proxy) within treatments at either time point or across time points (t-test was used for the latter).
Secondary stress: proteomics
From file Galaxy 36-annotated proteomics, took all Sigenae IDs that had a nsp-adjusted probability of at least 0.9 and retrieved contig sequences from Sigenae, this file is contigs for galaxy 36 proteins. Sigenae only returned 571 contigs out of 604 total. Did a blastall in wetgenes (blastx against swissprot db).
In Galaxy, joined the Sigenae ID columns in the Galaxy 36 dataset with GO annotations (Galaxy 41). Then joined with GO Slim terms (Galaxy 42) and exported file (Galaxy 42- Go annotated proteins.xlsx). These data were filtered for nsp-adjusted probability of at least 0.9 and biological process GO category. The resulting GO Slim terms were put into a pivot table.
March 13, 2012
Secondary stress: proteomics
Explanation of how ProteinProphet produces a protein probability - from Nesvizhskii et al. 2003.
Background of protein sequencing and identification: Proteins are cleaved (by trypsin) and the complexity of the peptide mixture is reduced by chromatography separation. After this separation, the mixture is subjected to reverse-phase chromatography coupled with mass spec. Peptides are ionized and ions are subjected to fragmentation to produce tandem mass spectra. The spectra are entered into a database search (e.g. SEQUEST). Such programs create a theoretical database of spectra based on the protein sequences so that the data (spectra) from the mass spec are actually assigned to these theoretical spectra that correspond to an accession number in the db. SEQUEST creates a Qscore (kind of like an e-value) that takes into account the total number of identified peptides in the data set and the number of peptides that correspond to each protein. This QScore and the peptide sequence are the inputs into ProteinProphet.
degenerate peptide = sequence is present in more than a single entry in the database, in ProteinProphet degenerate peptides are assigned to all of their hits (degeneracy is usually due to db redundancy)
probability of observed data being correct (p(+|D)) = the model learns to distinguish correct and incorrect peptide assignments based on the particular db and so computes a probability for each peptide assignment being correct. By using the data itself to "learn", the method is robust to sample purity, mass spectral quality, proteolytic digest efficiency, etc.
Number of sibling peptides = if a peptide is part of a "multihit" protein (high coverage) then it has a high NSP. NSP is the expected number of other correctly identified peptides corresponding to the same protein.
Combining p(+|D) and NSP: If a single hit peptide has a p(+|D) close to 1, then it is penalized less for its low NSP.
Example data using ProteinProphet: all probabilities of at least 0.99 were correctly assigned and only 3 were incorrect at >0.9. 92% of correct peptide assignments have NSP > 5, majority of incorrect assignments have NSP < 0.25. More MS runs of the same data increases coverage so that more correctly identified proteins are multihit (have a large NSP).
March 12, 2012
Secondary stress: proteomics
Explanations of protein output data (also found at ).
weight: contribution of peptide among each of multiple sequence entries, * = unique entry
nsp adjusted probability: probability is adjusted based on number of sibling peptides found for that protein (the greater the number of sibling peptides, the greater the probability)
Initial probability: probability assigned by ProteinProphet without regard to corresponding protein
NTT (number of tolerable termini): 0, 1, or 2 expected cleavage termini
nsp bin: 0-7, # of sibling peptides after discretization
total: number of instances peptide was identified in dataset
peptide group indicator: independent evidence of peptide identity if found in different charge states (also indicated in column)
group probability:
protein probability:
percent coverage:
number unique peptides:
percent share of spectrum IDs:
description:
nondegenerate evidence:
precursor ion charge:
Made a list of just the Sigenae contig numbers that corresponded to the sequenced proteins and joined this list with a file of Sigenae contig numbers with their top blast hit to a protein (Galaxy 5 Cg Sigenae 8 best hits). This essentially annotates the list of proteins that were sequenced. Exported the file from Galaxy and removed all proteins that didn't have a clear annotation - i.e. deleted all that were simply "hypothetical" proteins or predicted proteins without a function. This file saved in excel workbook Galaxy 32 - annotated proteins 031212.
Took ProteinProphet output file (proteomics output first gill sample.xlsx) and edited it so that each protein was on one row. An extra column was added so that Sigenae ID has only the first ID and all IDs are in a separate column. Saved as a text file and joined with Sigenae 8 best hits in Galaxy. Saved as Galaxy 36-annotated proteomics.xlsx.
February 29, 2012
Secondary stress
Brought over one sample (Exp2.218, posterior gill from control, tank 103B1) to Byron Gallis at the mass spec facility in Health Sciences (School of Medicinal Chemistry). He is going to prep and run the sample on the MS for proteomic sequencing.
February 28, 2012
Secondary stress
Sorted tubes from Experiment 1 today. Exp.1-200 were divided between 2 boxes so that odd number tubes are in one box and even in the other (NB: Exp1.199 is out of order because one spot was skipped). This should divide up duplicate samples for oysters since 2 gill samples were taken in consecutive tubes for each animal. Heat shocked samples (collected 1/20/12) are in their own box and LHT are in one half with SLT+LHT in the other half (except for 103B LHT2, which is in the overflow box). Exp1.201-249 are in a fourth box.
I also organized all the samples from Experiment 2 (1 month exposure). Samples are divided between boxes by tissue type: anterior gill, posterior gill, and whole body. It appears that there never was a tube Exp2.387 so the sample sheet will be adjusted accordingly.
February 18, 2012
Secondary stress
Labeled tubes and made sample sheets for last day of experiment, 2/19/12, when Carolyn and Steven are going to sample the oysters.
Took spec pH of source water for all treatments. Spec pH is corrected for temperature (13 or 20°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.04 |
29.6 |
8.032 |
| 102A |
7.64 |
29.6 |
7.593 |
| 102B |
8.02 |
29.6 |
8.014 |
| 103B |
8.01 |
29.6 |
8.035 |
| 104A |
8.05 |
29.7 |
8.013 |
| 104B |
7.70 |
29.7 |
7.684 |
February 17, 2012
Secondary stress and Multispecies OA
Counted morts of geoduck, small C. gigas, and large C. gigas.
Took spec pH of all source water. Messed up the 101A sample so do not have pH data for it. Also somehow forgot to record the salinity and Durafet pH data, so salinity is estimated to be 29.7 ppt. Spec pH is corrected for temperature (either 13 or 20°C) using a TA of 2095 µmol/kg.
| Tank |
spec_pH |
| 102A |
7.612 |
| 102B |
8.045 |
| 103A |
8.012 |
| 103B |
8.057 |
| 104A |
8.030 |
| 104B |
7.683 |
February 16, 2012
Bioinformatics
The focus of my analysis is now going to be completely on gene discovery. I am going to do a comparison of gene discovery using a de novo assembly vs. a backbone assembled from available H. asinina reads.
De novo assembly of trimmed reads: reads assembled to create 9,301 reference sequences. Exported pdf of mapping summary report. Opened de novo and selected all contigs and chose "open consensus". Exported FASTA file of consensus sequences.
Reads mapped back to H. asi backbone: did the same as above - extracted consensus sequences from 1122 references and exported FASTA file. (NB: these are still the untrimed reads)
For both files of consensus sequences, imported into the SAFS Inquiry portal to do a blastall. Blastall run with same parameters as Jan 9, 2012: blastx against swissprot db, etc.
Workflow for results from H.asi backbone:
- opened blastall file in excel and separated columns by "|"; made sure there were no gaps in the data (saved as blastall Hasi backbone.txt)
- uploaded into galaxy and joined with the uniprot swissprot IDs file based on swissprot IDs (1st column in uniprot file, 5th in blastall file) = job 20; kept unmantching and incomplete lines
- joined resulting table (column 5) with swissprot associations file (column 2) - couldn't keep unmatching and incomplete lines because kept on getting an error = job 23
- joined resulting table (column 25) with GO to GO Slim file column 1, keeping unmatching and incomplete lines = job 24
- exported into excel and saved as Galaxy Hasi backbone.xlsx
- Made a tab with the data filtered for only biological processes
- filtered the biological processes to those with evalues less than or equal to 1e-5 in a new tab
- Used the GO Slim terms to make a pivot table and pie chart of the processes discovered (see below)
- exported unique swissprot IDs from the blastall file to DAVID as the background list and exported the filtered (biol process, e-value cut-off) unique list of IDs as the gene list. Used the functional annotation tool, selected Gene Ontology, and exported the GO_TERM_BP FAT file. (see below)
- Pasted GO terms with associated p-values into revigo.
Workflow results for de novo assembly (same as above but differences are noted):
- blastall file - blastall de novo.txt
- join with uniprot/swissprot IDs= job26
- join with swissprot associations=job 27
- join with GO to GO slim file = job 28
- saved as Galaxy de novo.xlsx
Secondary stress
Cleaned tanks and counted mortalities.
From 43°C HS (tank 102B) the following are dead: hot pink (102B3), yellow and dark pink (102B4), and red (102B5). From 44°C HS the following are dead: hot pink and dark pink (103A1), hot pink (103A2), seafoam and turquoise (103A3), turquoise and yellow (103A5), dark pink (103A7). All oysters dead from yesterday and today were measured (length and width), shucked, and shell weight was taken.
Durafet probes were calibrated for temperature. Spec pH was taken of all source water and is corrected for temperature (13°C) with a TA of 2090 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.01 |
29.6 |
8.043 |
| 102A |
7.68 |
29.7 |
7.654 |
| 102B |
8.02 |
29.6 |
8.035 |
| 103A |
8.03 |
29.7 |
8.034 |
| 103B |
8.04 |
29.7 |
8.080 |
| 104A |
8.05 |
29.9 |
8.037 |
| 104B |
7.69 |
29.8 |
7.689 |
Multispecies OA
All tanks were cleaned (I cleaned Oly tanks, Dave cleaned multispecies tanks). Geoduck morts were counted and photos put in dropbox: 3 in 102A3 (1 crushed), 2 in 102A6, and 1 in 102A4. Small C. gigas morts were 1 each in 101A5 and 102A6. See above for spec pH (Oly pH is corrected for temp of 20°C).
February 15, 2012
Bioinformatics
The RNA-Seq says that it "ended abnormally" so I guess that means it didn't work. Mapping reads (both libraries) back to reference (H. asi backbone) to make sure that there are actually sequences in common between them.
Started work flow for trimmed sequences (that SR had trimmed). Summary of SR trimming:
| Name |
Numberofreads |
AVg.length |
Numb.reads.after.trim |
percentag.trimmed |
avg.length.after.trim |
| high CO2 |
41,027,557 |
36.0 |
40,816,572 |
99.59% |
35.9 |
| ambient |
54,101,070 |
36.0 |
53,961,227 |
99.74% |
36.0 |
removal of ambiguous nucleotides: maximal 2 nucleotides allowed
removal of sequences on lenght: min length 25 nucleotides
De novo assembly of 2 libraries of trimmed reads (all saved within "SR" directory under trimmed sequences): mismatch cost = 2, limit = 8, fast ungapped alignment, vote conflict resolution, random non-specific matches, min contig length = 200, map reads back to contigs (create summary report).
Secondary stressor and Multispecies OA
Took spec pH of all source water and containers 5 and 6 for every treatment. Took TA of all source water and of containers 5 and 6 for 104 A and B (Olys) and 101A and 102A (Multispecies OA). Spec pH in the table is corrected for temperature (13°C for all except 20°C for 104A and 104B) using the given TA.
| Tank |
Durafet_pH |
salinity |
spec_pH |
TA |
| 101A |
8.00 |
29.7 |
8.027 |
2093.01 |
| 102A |
7.66 |
29.7 |
7.604 |
2091.32 |
| 102B |
8.02 |
29.7 |
8.046 |
2091.92 |
| 103A |
8.04 |
29.9 |
8.032 |
2090.77 |
| 103B |
8.03 |
29.9 |
8.067 |
2091.02 |
| 104A |
8.05 |
29.9 |
8.042 |
2092.84 |
| 104B |
7.69 |
30 |
7.69 |
2093.12 |
Small C. gigas mortality: 101A5 (1), 101A6 (1).
Secondary stress C. gigas mortality for 44°C:
| Tank |
101B(hot_pink) |
102B(turquoise) |
103A(yellow) |
103B(dark_pink) |
104A(sea_foam) |
104B(red) |
| 103A1 |
x |
x |
x |
x |
||
| 103A2 |
x |
x |
x |
x |
||
| 103A3 |
x |
x |
x |
x |
||
| 103A4 |
x |
x |
x |
x |
x |
x |
| 103A5 |
x |
x |
x |
x |
||
| 103A6 |
x |
x |
x |
x |
x |
x |
| 103A7 |
x |
x |
x |
x |
x |
|
| 103A8 |
x |
x |
x |
x |
x |
Saved all the morts in the freezer to get measurements and shell weights tomorrow.
February 14, 2012
Bioinformatics
RNA-Seq from 2/7/12 seems to have not worked so repeated steps to do RNA-seq for both libraries agains the H.asi backbone.
Secondary stressor
The electricity was turned off this afternoon for about 15 minutes around 1:30 pm. When it came back on all the controllers were in manual mode and so the pH went up for all the treatments to 8.1-8.2. They were set back to auto around 3:30 pm.
All tanks were cleaned. There were no mortalities although many of the oysters from the 44°C HS were slow to close their valves upon emersion (lethargic).
Took spec pH of all source water. Spec pH is corrected for temperature (13°C except for 104 A and B which are at 20°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.99 |
29.7 |
7.990 |
| 102A |
7.63 |
29.9 |
7.551 |
| 102B |
8.01 |
29.8 |
8.034 |
| 103A |
8.00 |
29.7 |
7.999 |
| 103B |
8.03 |
29.9 |
8.081 |
| 104A |
8.02 |
29.9 |
8.009 |
| 104B |
7.65 |
29.8 |
7.668 |
Multispecies OA
Did geoduck mortality checks and sent photos of dead ducks to Carolyn. Also did C. gigas mortality checks. There was 1 C. gigas dead in the following containers: 101A5, 102A6, 102A2, 102A5.
20°C set points for 104A (400 ppm) and 104B (1000 ppm) using TA of 2095 µmol/kg and s of 29.8 ppt: 104B = 7.683, 104A = 8.037.
February 13, 2012
Multispecies OA
One mortality of C. gigas: 400 ppm, 43°C bag 5
New set points for Oly tanks (104A and 104B) at T of 17°C, s = 29.9 ppt, TA=2095 µmol/kg: 400 ppm = 8.036, 1000 ppm = 7.678 (adjusted pH set points accordingly). Put tanks at 18°C in the evening.
Calibrated new batch of acid that I made last night. New concentration is 0.09608 N.
Did spec pH of source water for all tanks. Spec pH is corrected for temperature (13°C for all except for 104 A and B, which are 15°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.99 |
29.7 |
7.998 |
| 102A |
7.64 |
29.8 |
7.582 |
| 102B |
8.06 |
29.8 |
8.043 |
| 103A |
8.02 |
29.7 |
7.992 |
| 103B |
8.02 |
29.9 |
8.021 |
| 104A |
8.01 |
29.9 |
7.997 |
| 104B |
7.65 |
29.9 |
7.637 |
Calibrated Durafet probes for temperature on all tanks.
Secondary stressor
No mortality from HS.
February 12, 2012
Multispecies OA
No mortality from HS 2/12/12 for small C. gigas.
Secondary stressor
Heat shocked oysters - 1 oyster from each container (n = 8) per treatment at each temperature: 42, 43, and 44°C. Oysters were randomly taken from each container measured (length, width), weighed (total weight and buoyant weight) and labeled with nail polish corresponding to the treatment from which they came. HS was comprised of a 10 minute warm-up followed by 1 hour in 800 mL of heated seawater. The same water was used for 42 and 43°C, but the water was changed for 44°C. After HS, the oysters were all returned to ambient conditions (pH = 8.03, T = 13°C) for observation over 1 week. In each container, there is one oyster from each treatment. Oysters HS'd at 42°C are in 103B, 43°C are in 102B and 44°C are in 103A. Time that HS occurred was also recorded.
Put all histo samples in 70% EtOH.
February 11, 2012
Secondary stressor and Multispecies OA
Sampling day! Sampling began around 8 am and ended around 3 pm. From each of the 6 pCO2 treatments, oysters were sacrificed in 3 groups of 8 (1 from each container in each group of 8). The first group was sampled as controls, the second group underwent mechanical stress and the third group was another control. After being removed from their containers, each oyster was measured (length and width) and weighed (total weight and buoyant weight). Oysters were then shucked by Mac who also sampled the anterior gill and poster gill lamellae (in 2 separate tubes). Steven took a cross-section that included digestive gland and gill and a slice of the adductors muscle, which were immediately fixed in Invertebrate Davidson's solution (oysters from 101B and 102B sections were not fixed right away, all 8 were stockpiled until the end and then all put in the fixative at the same time). SR then put the remaining body in a third tube. I removed excess tissue from the shell, dried the shell, labeled both valves, and weighed the shell. The shells were saved in large weigh boats to air dry. For the control oysters, after measurements were taken the oysters were left sitting on the lab bench for 5 minutes before sampling. For the mechanically stressed oysters, after measurements all 8 oysters were spun together in a salad spinner (by Ro) for 5 minutes, after which they were immediately sampled. All samples were flash frozen in liquid nitrogen and stored at -80°C.
February 10, 2012
Secondary stressor and Multispecies OA
Cleaned feeding lines by flushing with fresh (DI) water and then salt water.
Calibrated temperature probes.
Small C. gigas HS mortality: 3 at 43°C (90% mortality).
Heat stressed small C. gigas: randomly picked one from each container and heat shocked 2 groups at each temperature per pCO2 treatment at 41, 42, and 43°C.
Took spec pH of all source water. Spec pH is corrected for temperature (13°C) using a TA of 2090 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.99 |
29.5 |
7.989 |
| 101B |
7.36 |
29.4 |
7.400 |
| 102A |
7.65 |
29.5 |
7.571 |
| 102B |
7.85 |
29.5 |
7.871 |
| 103A |
7.60 |
29.6 |
7.631 |
| 103B |
8.03 |
29.6 |
8.029 |
| 104A |
7.74 |
29.6 |
7.778 |
| 104B |
7.65 |
29.6 |
7.645 |
February 9, 2012
Secondary stressor and Multispecies OA
Sampled C. gigas for Carolyn's experiment: 1 oyster from each container, sampled mantle, gill and whole body and measured length, width, total weight, buoyant weight, and shell weight.
Took spec pH of all source water from each of the treatments. Spec pH is corrected for temperature (13°C) using a TA of 2070 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.95 |
29.4 |
7.942 |
| 101B |
7.51 |
29.3 |
7.512 |
| 102A |
7.58 |
29.3 |
7.600 |
| 102B |
7.84 |
29.3 |
7.875 |
| 103A |
7.59 |
29.3 |
7.632 |
| 103B |
8.05 |
29.4 |
8.044 |
| 104A |
7.74 |
29.3 |
7.769 |
| 104B |
7.64 |
29.4 |
7.643 |
Calibrated Durafet probes for temperature.
Turned off feeding for 101A and 102A in the morning so that the animals would not be fed right before their feeding trial. Turned the pumps back on for evening feeding.
Geoduck HS mortality: 1 at 22°C
Small C. gigas HS mortality: 4 at 43°C, 2 at 44, 4 at 45.
Cleaned all containers for the secondary stress experiment (i.e. did not clean 101A or 102A).
February 8, 2012
Secondary stressor and Multispecies OA
The breaker is getting fixed today so the electricity shouldn't short out anymore.
Ran TA and spec pH for all source water and containers 1 and 2 for each treatment (container 2 was poisoned and archived for later TA analysis). Spec pH is corrected for temperature (13°C) using the TA that was measured for the source water of each cooler.
| Tank |
Durafet_pH |
salinity |
TA |
Spec_pH |
| 101A |
8.03 |
29.5 |
2069.72 |
8.015 |
| 101B |
7.40 |
29.5 |
2069.96 |
7.419 |
| 102A |
7.69 |
29.5 |
2071.4 |
7.629 |
| 102B |
7.87 |
29.6 |
2070.99 |
7.892 |
| 103A |
7.57 |
29.5 |
2072.1 |
7.638 |
| 103B |
8.04 |
29.6 |
2071.87 |
8.047 |
| 104A |
7.74 |
29.6 |
2076.24 |
7.775 |
| 104B |
7.64 |
29.7 |
2074.87 |
7.670 |
Geoduck mortality (1 each): 101A1, 101A6, 102A2, 102A7. 102A2 appeared to be crushed and 102A7 had a chipped shell.
Geoduck HS mortality: 2 at 28°C
Small C. gigas HS mortality: 2 at 43°C, 8 at 44°C, and 6 at 45°C.
February 7, 2012
Secondary stressor and Multispecies OA
When I went down to the lab this morning the power had tripped to the 101 and 102 tanks again (it had been off for about 2 hours). I turned it back on. Heaters didn't seem to be working correctly in 101 tanks so I set them to autotune.
Took spec pH of all source water. Spec pH is corrected for temperature (13°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.01 |
29.6 |
8.012 |
| 101B |
7.14 |
29.6 |
7.136 |
| 102A |
8.00 |
29.6 |
7.926 |
| 102B |
8.03 |
29.6 |
8.045 |
| 103A |
7.60 |
29.6 |
7.588 |
| 103B |
8.03 |
29.6 |
7.974 |
| 104A |
7.73 |
29.6 |
7.726 |
| 104B |
7.66 |
29.6 |
7.661 |
Cleaned all tanks.
Geoduck mortality: 101A3 (1), 102A2 (2). Photo sent to CSF and shells stored in 95% EtOH.
Geoduck HS mortality: 1 at 29°C
Small C. gigas HS mortality: 0
HS'd small C. gigas (n=10) at 44 (blue zip tie) and 45°C (white bag) for 1 hour, with 10 minute warm up.
Made new m-cresol purple. Intercept = 0.023052 and slope = -0.0491.
Bioinformatics
Did RNA-Seq for both libraries (untrimmed) against consensus sequences from H. asi backbone (1176 references). See 1/9/12 for details on method.
February 6, 2012
Secondary stressor and Multispecies OA
Matt fixed the relay in the controller for 102B in the afternoon so it is now working correctly. However, in the evening the heater for 101B was staying on even though the temperature was 13.6°C (and rising). By 9:30 pm, the temp was at 13.9°C and I unplugged the heater for the night. Also, during the day the power kept on shorting out for tanks 101 and 102. When this happens, the controller for 102 for some reason automatically goes to manual mode and makes the pH creep up towards 8 (there were a lot of pH fluctuations for 102A and B today).
Did spec pH of source water and containers 3 and 4 for all treatments. Spec pH is corrected for temperature (13°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.98 |
29.5 |
7.984 |
| 101B |
7.17 |
29.5 |
7.177 |
| 102A |
7.98 |
29.5 |
7.885 |
| 102B |
7.86 |
29.5 |
7.932 |
| 103A |
7.61 |
29.4 |
7.695 |
| 103B |
8.04 |
29.5 |
8.003 |
| 104A |
7.73 |
29.4 |
7.742 |
| 104B |
7.65 |
29.6 |
7.665 |
Geoduck mortality (1 each): 101A1, 102A4, 102A5, 102A7. Photo sent to CSF and shells saved in EtOH.
Geoduck HS mortality: 1 at 28°C (probably crushed), 4 at 29, and 2 at 30.
Small C. gigas HS mortality: 0.
February 5, 2012
Secondary stress and Multispecies OA
Cleaned all containers.
Fed animals in sea tables.
Geoduck mortality from HS: 0 dead from 22 and 25°C, 6 dead in 28, 5 dead in 29, 3 dead in 30, and 1 dead in 31.
small C. gigas mortality from HS: none
Geoduck mortality from OA: 101A4 (1), 101A6 (1, crushed), 101A7 (2), 102A2 (2, 1 crushed), 102A4 (1, lost the shell), 102A5 (1). Sent the photo to CSF and saved the shells in 95% EtOH.
I never heard back from Matt about fixing 102B so the temperature has remained steady around 11.8-11.9°C.
Calibrated Durafet probes for temperature.
February 4, 2012
Secondary stress and Multispecies OA
Took spec pH of all cooler water. Spec pH is corrected for temperature (13°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
7.98 |
29.5 |
7.963 |
| 101B |
7.34 |
29.6 |
7.326 |
| 102A |
7.66 |
29.6 |
7.598 |
| 102B |
7.88 |
29.5 |
7.854 |
| 103A |
7.58 |
29.6 |
7.593 |
| 103B |
8.04 |
29.6 |
7.975 |
| 104A |
7.73 |
29.6 |
7.770 |
| 104B |
7.63 |
29.7 |
7.653 |
The temperature in 102B is about 11.9°C and the heater is not going on, meaning that there is something wrong with the relay. I'm not sure how to fix this, so I texted Matt about the problem with no response (it is a weekend).
February 3, 2012
Secondary stress and Multispecies OA
The water pressure was still bad in the 102 tanks this morning. I had removed the feeding coil and other parts so that it was just the one tube carrying water leading to the drippers (I put the feeding apparatus back together before the morning feeding but removed it soon after the pumps finished delivering food to maintain water flow to the tanks). Matt came and looked at it and realized that somehow the air had not been turned on enough after he had fixed the solenoid yesterday. He turned the air up and the water pressure problem was fixed. This means that from yesterday afternoon until this morning 102A and 102B were less aerated (had less O2) than the other treatment tanks.
Took spec pH of all cooler water and adjusted for temperature (13°C) using a TA of 2095 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.08 |
29.8 |
8.061 |
| 101B |
7.17 |
29.8 |
7.217 |
| 102A |
7.68 |
29.7 |
7.609 |
| 102B |
7.85 |
29.8 |
7.830 |
| 103A |
7.61 |
29.9 |
7.597 |
| 103B |
8.05 |
29.8 |
8.000 |
| 104A |
7.72 |
29.9 |
7.751 |
| 104B |
7.64 |
29.9 |
7.650 |
- 101B: 2 dead, 0 left alive
- 102B: 0 dead, 1 alive
- 103A: 2 dead, 2 alive
- 103B: 1 dead, 1 alive
- 104A: 0 dead, 1 alive
- 104B: 2 dead, 2 alive
Mortality from geoduck HS: 0 morts from 22, 25, 28, and 29°C. 5 morts at 30 °C and 1 mort at 31°C (1 geoduck is missing since there is only one left).
Mortality from small C. gigas HS: 0 mortalities at all temperatures.
Cleaned feeding lines going to tanks. Emptied all salt water and algae from the buckets and refilled ~1/3 with DI water. Used this same DI water to wipe down the sides of the bucket and flushed out the lines with this DI water/old algae mixture. Once buckets were empty of this liquid, refilled all the way with new DI water and flushed the lines. After the DI water was flushed through, Filled buckets to ~2/3 with salt water and ran through the lines. At the end, refilled the buckets with salt water and algae for the next feeding.
February 2, 2012
Secondary stress and Multispecies OA
Did total alkalinity and spec pH of all source water and container 7s. Poisoned 600 mL of water from container 8s for later analysis (with 75 µL HgCl2). Spec pH is corrected for temperature (13°C) using the given TA. The containers in 102A are a higher pH than the source water, which is unusual. I think this is because there was a lot of pH fluctuation in 102A today (see below) and there's a lag in what the containers experience relative to the source water during pH variation.
| TANK |
salinity |
Durafet_pH |
spec_pH |
TA |
| 101A |
29.9 |
8.07 |
8.041 |
2093.75 |
| 101B |
30 |
7.18 |
7.196 |
2095.42 |
| 102A |
30 |
7.48 |
7.401 |
2094.81 |
| 102B |
30.1 |
7.83 |
7.823 |
2095.16 |
| 103A |
29.9 |
7.61 |
7.633 |
2094.84 |
| 103B |
30 |
8.03 |
8.004 |
2094.14 |
| 104A |
30 |
7.73 |
7.776 |
2094.52 |
| 104B |
30.2 |
7.60 |
7.669 |
2095.96 |
The solenoid in tank 102 had to be replaced because it was not working properly (CO2 was not being dispensed appropriately to meet and maintain the set point pH). After it was replaced, I re-calibrated the probes using spec pH and ran accutune.
The am feeding seemed to have clogged some of the tubing in 102A. I replaced the part that was clogged and it is working fine now.
All the animals in the sea tables were fed.
Mortality from HS 1/27/12: 1 in 103A and 1 in 102B.
Geoduck mortality: 1 in 101A7 and 1 in 102A7.
Heat shocked geoduck at 29 and 30°C for 1 hour each. Mortality from yesterday's 'duck HS: 7 dead from 31°C and 1 dead from 28°C, although it looks to have died from being crushed. The three "live" geoduck from the 31 HS may not really be alive, but it was hard to tell if they were truly dead or just extremely lethargic from the stress.
Heat shocked 10 small Pacific oysters each at 40, 41, 42, and 43°C (10 minute warm up followed by 1 hour HS). Zip tie colors correspond to the following temperatures: pink = 40, purple = 41, yellow = 42, red = 43.
Calibrated Durafet probes for temperature.
February 1, 2012
Secondary stress and Multispecies OA
Took spec pH of source water for all coolers. Spec pH is corrected for temperature (13°C) at a TA of 2099 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.07 |
29.9 |
8.056 |
| 101B |
7.22 |
29.9 |
7.273 |
| 102A |
7.79 |
30 |
7.759 |
| 102B |
8.20 |
30 |
8.174 |
| 103A |
7.61 |
29.9 |
7.645 |
| 103B |
7.99 |
30 |
7.958 |
| 104A |
7.74 |
29.9 |
7.767 |
| 104B |
7.77 |
30 |
7.767 |
Calibrated temperature in all coolers.
Had to manually adjust CO2 flow in 102A and B and 104B this morning because they were not reaching the correct set points. The pH had settled by the afternoon so I ran accutune in 102A and B (don't think it is needed in 104B).
Cleaned all containers. Fed animals in sea tables.
Mortality from HS 1/27/12: 1 in 104B, 3 in 103A, 1 in 102B, 1 in 104A, 1 in 101B, 1 in 103B.
Today I helped to sample the oysters in Carolyn's experiment - took samples of mantle, gill, and the rest of the body for 1 C. gigas from each of her containers in 101A and 102A.
We are trying to find the lethal temperature for the geoduck seed. We are doing HS starting at 21°C at every 3 degrees. Today, HS was done at 21, 25, 28, and 31°C. The geoduck were not looking good after the 31°C - gaping, lethargic, siphons unresponsive to stimulation.
January 31, 2012
Secondary stress and Multispecies OA
Calibrated the new batch of acid from 1/30/12 with CRM 113. Ran samples that were taken previously and poisoned on 1/18 and 1/24/12.
Moose and Matt fixed an air flow problem to the system, but it made all the controllers go crazy. They ran accutune so everything should be better by this evening.
Mortality from HS 1/27/12: 1 each in 101B and 104A
Made new m-cresol purple.
Fed animals in sea table.
Geoduck mortalities (1 each): 101A1, 101A8, 102A8. Photo sent to CSF and shells preserved.
102B is having trouble getting up to its set point pH (7.87). I turned the CO2 flow down so there is less CO2 entering the Venturi.
Bioinformatics
downloaded all H. asinina ESTs (8,355) from GenBank and uploaded FASTA into CLC. This is the abalone with the greatest number of ESTs. Did de novo assembly of these ESTs: mismatch cost 2, insertion cost 3, deletion cost 3, length fraction 0.5, similarity 0.8, vote for conflict resolution, random non-specific matches, min contig length 200, map reads back to contigs.
This will be the Hasi backbone. Mapped both libraries back to the Hasi backbone. Made consensus sequences of Hasi assembly. Mapped both Pinto libraries back to Hasi backbone: mismatch cost 2, limit 8, fast ungapped alignment, vote conflict resolution, random non-specific matches.
January 30, 2012
Bioinformatics
Did some research on abalone phylogeny to find out if there is a closely related species to pinto abalone that may have a lot of data in GenBank to use as a backbone for assembly. In Coleman and Vacquier (2002), pinto abalone clusters the most closely to Haliotis sorenseni (white abalone) but is also in the same clade as H. rufescens (red abalone) and H. walallensis (flat abalone) based on intergenic spacer regions of ribosomal DNA (ITS) markers. In general, these markers showed very little variation within the North Pacific clade (shown below).
Number of sequences in NCBI:
H. sorenseni: no ESTs, 71 nucleotide
H. rufescens: 225 nucleotide, 358 ESTs
H. walallensis: 21 nucleotide, no ESTs
H. corrugata: 467 nucleotid, no ESTs
H. cracherodii: no ESTs, 87 nucleotide
H. fulgens: 136 nucleotide, no ESTs
Downloaded all H. rufescens ESTs from NCBI and uploaded FASTA file into CLC. Ran de novo assembly: mismatch cost 2, insertion cost 3, deletion cost 3, length fraction 0.5, similarity 0.8, conflict resolution vote, non-specific matches random, minimum contig length 200, map reads back to contigs. Also did a regular sequence assembly: did not trime seqs before assembly, min aligned read length 50, medium alignment stringency, vote for conflicts, create full contigs including trace data for output. Did the same assembly but this time created only consensus sequences. The de novo assembly resulted in 41 references; the first regular assembly resulted in 56 contigs.
Downloaded all EST and mRNA sequences for the species listed above.
Sequence assembly of H. rufescens ESTs and mRNA from NCBI: 50 min aligned read lengths, medium alignment stringency, vote for conflicts, create only consensus seqs.
Sequence assembly of all abalone ESTs and mRNA from NCBI: same parameters as above.
There are now 2 backbones: H. rufescens (Hruf backbone) and all North Pacific abalone species (allab backbone).
Mapped the untrimmed reads from sequencing (air and high CO2 together) back to the Hruf and allab backbones. Parameters: mismatch cost 2, limit 8, fast ungapped alignment, vote for conflict resolution, random non-specific matches. The mapped reads are saved within the directories that contain the contigs that make up the backbones in a file called "seqs mapped to ref".
Secondary stress and Multispecies OA
Ran Elene's TA samples. Cory made a new batch of acid and I tried to calibrate it with CRM 113 but couldn't get a consistent enough TA value so I will try again tomorrow.
Did spec pH on all source water. Spec pH is corrected for temperature (13°C) at a TA of 2099 µmol/kg. Matt fixed the controllers in tanks 104 so I calibrated the Durafets, reset the set points, and ran accutune.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.06 |
29.9 |
8.047 |
| 101B |
7.28 |
29.9 |
7.336 |
| 102A |
7.71 |
29.8 |
7.659 |
| 102B |
7.87 |
29.8 |
7.873 |
| 103A |
7.59 |
29.8 |
7.618 |
| 103B |
8.00 |
30 |
7.964 |
| 104A |
7.93 |
30 |
7.879 |
| 104B |
7.82 |
30.1 |
7.778 |
Cleaned all containers and did mortality checks. In 101A4, there was a polychaete-type animal in the bag with the geoduck (I killed it). Geoduck mortality (1 each): 101A1, 101A8, 102A1, 102A4, 102A8. Photo was sent to CSF and shells were preserved.
Mortality from HS 1/27/12: None
January 29, 2012
Secondary stress and Mulispecies OA
Took spec pH of source water from all tanks. pH is corrected for temperature (13° ) with a TA of 2099 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.05 |
29.8 |
8.042 |
| 101B |
7.27 |
29.9 |
7.335 |
| 102A |
7.70 |
29.9 |
7.658 |
| 102B |
7.85 |
30 |
7.861 |
| 103A |
7.60 |
29.9 |
7.593 |
| 103B |
8.02 |
30 |
8.010 |
| 104A |
7.78 |
29.9 |
7.795 |
| 104B |
7.66 |
30.1 |
7.723 |
Mortality from HS 1/27/12: None
Animals in sea tables fed.
The stopcock between the water coming the containers and incoming water had been shut off and some point and not put back on so there was no new water circulating in the tanks. Food had gotten in from the peristaltic pumps. Opened the stopcock so water could start circulating.
January 28, 2012
Secondary stress and Multispecies OA
Cleaned tanks and did mortality checks. There were 3 geoduck mortalities: 1 in 102A1, 1 in 102A2, and 1 in 102A5. Photo was sent to CSF and shells were preserved. One of the clams in 102A3 was discovered to be an empty shell and was discarded (measurements = 12 x 16 mm). There is a slight chance that the oysters in 101A5 and 101A6 were switched and are now in the other container (the incorrect lid may have been put on the containers). One of the manila clams in 101A4 has a large crack in its shell but still appears to be alive.
The feeding lines were flushed with DI water. The buckets were filled with DI water and left to run into empty buckets until empty. The buckets were then filled halfway with salt water and that was run through the lines. The sides of the buckets were wiped down to remove residual algae. After cleaning, the buckets were refilled with salt water and 19 mL of algae and the animals were fed. During the cleaning process, the amount of liquid that comes out of the lines in 30 s was measured with a graduated cylinder for each tank. The amounts were pretty much the same between tanks (28 or 30 mL per 30 s). In the morning, ~1 mL of water from the drippers was taken during feeding for algal cell counts as described 1/27/12.
| Tank |
Numb_mL/30s |
| 101A |
30 |
| 101B |
30 |
| 102A |
28 |
| 102B |
28 |
| 103A |
30 |
| 103B |
30 |
| 104A |
28 |
| 104B |
30 |
| Tank |
salinity |
Durafet_pH |
spec_pH |
| 101A |
30 |
8.02 |
8.007 |
| 101B |
30 |
7.20 |
7.235 |
| 102A |
30.1 |
7.63 |
7.602 |
| 102B |
30.1 |
7.87 |
7.861 |
| 103A |
30.1 |
7.62 |
7.584 |
| 103B |
30.2 |
8.01 |
7.961 |
| 104A |
30.1 |
7.76 |
7.757 |
| 104B |
30.1 |
7.70 |
7.676 |
Mortalities from HS 1/27/12: 1 in 104B, 1 in 103B, and 2 in 104A.
Animals in sea tables fed.
January 27, 2012
Secondary stress and Multispecies OA
Algae counts: During the morning feeding, around 9:50 am, collected ~1 mL of water + algae directly from the drippers from each container #1. Counted the algae cells in a hemocytometer. I will repeat this for another container this evening or tomorrow morning. Also counted the cells from 1 mL of water taken directly from a feeding bucket. Samples were shaken before being loaded onto the hemocytometer. All counts are for the entire center square of the hemocytometer (i.e. 1 square millimeter). The counts were done in duplicate for each sample.
| Tank |
Algae_count1 |
Algae_count2 |
| 101A |
3 |
4 |
| 101B |
4 |
7 |
| 102A |
2 |
3 |
| 102B |
5 |
4 |
| 103A |
15 |
7 |
| 103B |
5 |
5 |
| 104A |
1 |
4 |
| 104B |
3 |
8 |
Took spec pH of the source water from all treatments and from containers 7 and 8. Spec pH is corrected for temperature (13°C) and calculated using a TA of 2099 µmol/kg.
| Tank |
Durafet_pH |
salinity |
spec_pH |
| 101A |
8.02 |
29.9 |
7.977 |
| 101B |
7.18 |
30 |
7.218 |
| 102A |
7.62 |
30.1 |
7.612 |
| 102B |
7.87 |
30 |
7.854 |
| 103A |
7.60 |
30.1 |
7.608 |
| 103B |
8.02 |
30.1 |
7.982 |
| 104A |
7.76 |
30 |
7.746 |
| 104B |
7.68 |
30.1 |
7.661 |
Sea table animals fed.
Heat shock: Did LHT HS of oysters that were HS'd at SLT on 1/13/12. Oysters were warmed at 44°C for 10 minutes before HS at 44°C for 1 hour (monitored constantly by the Fluke temperature probe). Checked mortality at 6 pm (about 6 hours post-HS): 3 dead in 104A, 2 dead in 104B, 4 dead in 101B, 5 dead in 102B, and 4 dead in 103B.
2 geoduck mortalities: 1 in 101A7 and 1 in 102A7. Photo sent to CSF and shells preserved in 95% EtOH.
January 26, 2012
Secondary stress
Mortality from HS 1/19/12: 1 was dead from the 44°C treatment (100 % mortality). This is the new LHT.
Had to reset the relay in 104A.
Calibrated temperature for the Durafet probes.
All tanks were cleaned.
Algae counts: Collected 1 mL of water from the corner of each container #1 that is the farthest from the dripper during feeding (around 9:50 pm). Shook the tube and put 10 µL on a hemocytometer. The solution was too dilute to effectively count cells.
Fed all animals in sea tables.
Multispecies OA
replaced the durafet probe in tank 101A because it wasn't working. Also had to reset the relay because it wasn't communicating well with the heater. Calibrated the probe using spec pH.
Both tanks were cleaned.
Geoduck mortalities: 101A4 and 102A8 (1 in each).
Bioinformatics
The previous quality trimming that was done resulted in 0 sequences after trimming, meaning that none of the sequences were good enough to pass the filters. To test the quality of the sequences, I am going to map all the reads (untrimmed) back to reference sequences from GenBank.
Downloaded all pinto abalone (Haliotis kamtschatkana) nucleotide sequences from GenBank (43 sequences in all, these are the same as the ESTs in NCBI). Uploaded the FASTA file to CLC and from the high throughput sequencing tools, mapped the reads of each library back to the reference (the 43 sequences). Default parameters used and results saved to Pinto Ab file.
From DAVID results on 1/19/12, entered the GO Terms and p-values into revigo to get visualization. Exported the resulting table that goes with the figure and saved as REVIGO 012612.
January 25, 2012
Secondary stress
Mortality from HS 1/19/12: 2 were dead from the 44°C treatment and 2 were dead from the 43°C.
Power outage at midnight last night be most everything was righted by morning. Moose had to turn the CO2 scrubbers off for a while so there were some dips in the higher pH treatments.
Multispecies OA
3 geoduck were dead, 1 each in 101A1 101A7 and 101A8.
January 24, 2012
Secondary stress and Multispecies OA
Did TA for source water and from container 5 from all treatments. Took 600 mL sample from container 6, poisoned with 75µL HgCl2 and stored for later analysis. Also did spec pH of all source water and containers 5 and 6. Feeding occurred at 9:30 so I did not take any samples from containers between 9:30 and 11:00 (feeding ended at 10 and it takes about an hour for the water to turnover in the containers), although I continued to sample and analyze the source water. Matt cleaned all the containers in coolers 103A, 103B, 104A, and 104B and I cleaned all the 101s and 102s.
All animals in the sea table were fed.
Mortality from HS 1/19/12: None
Multispecies OA
Today was Carolyn's sampling day. She and Dave were not able to finish sampling before they had to catch the ferry so I sampled C. gigas from containers 101A7 and 8 and 102A2-8: length, width, total weight, buoyant weight, shell weight, gill, mantle, and rest of body (the last three were flash frozen in liquid N2 and stored at -80°C). Also, I took 1 geoduck from each container, took a picture of all 8 in each treatment, and then put 8 into a tube (1 tube for each treatment) and flash froze for transcriptomics.
January 23, 2012
Secondary stress and Multispecies OA
Texted Matt about broken heater but have not heard back from him. Looked at other heaters in other tanks, but they cannot simply be unplugged and swapped out so have to wait for Matt...
Mortality from HS 1/19/12: None
Spec pH was done for all source water and is corrected for temperature (13°C) at TA of 2076 µmol/kg.
| Tank |
salinity |
Durafet_pH |
Spec_pH |
| 101A |
29.4 |
8.04 |
7.997 |
| 101B |
29.8 |
7.10 |
7.158 |
| 102A |
29.7 |
7.64 |
7.637 |
| 102B |
29.8 |
7.87 |
7.846 |
| 103A |
29.8 |
7.58 |
7.573 |
| 103B |
29.9 |
8.03 |
7.993 |
| 104A |
29.7 |
7.72 |
7.710 |
| 104B |
29.8 |
7.68 |
7.587 |
January 22, 2012
Secondary stress and Multispecies OA
All containers were cleaned. One geoduck was dead in each of 101A3 and 102A6 (photo emailed to CSF and shells stored in EtOH).
Mortality from HS 1/19/12: 2 oysters were dead from the 45°C treatment and 3 were dead from the 44°C treatment.
The heater in 101A seems to be broken and the temperature is around 11.5°C. I told Moose about it but he had no suggestions. Matt is out of town still but will get in touch with him tomorrow.
January 20, 2012
Secondary stress
Sampled gill tissue from 96 oysters that were heat shocked last Friday (1/13/12) at either 43°C for one hour (LHT) or 38°C for 1 hour (SLT) followed immediately by LHT. Each oyster was shucked and the anterior portion of all lamellae was sampled and flash frozen in liquid N2. The samples were stored at -80°C. Oysters were measured to match them to the measurements taken pre-HS last Friday and shell weight was recorded.
Spec pH was taken of all source water and from containers 3 and 4 in each treatment. In the table, spec pH is corrected for temperature (13°C) using a TA of 2076 µmol/kg.
All containers were cleaned.
| Tank |
Durafet_pH |
Spec_pH |
Salinity |
| 101A |
8.04 |
8.012 |
29.7 |
| 101B |
7.21 |
7.247 |
29.8 |
| 102A |
7.71 |
7.713 |
29.9 |
| 102B |
7.86 |
7.870 |
29.6 |
| 103A |
7.61 |
7.637 |
29.8 |
| 103B |
8.06 |
8.026 |
29.8 |
| 104A |
7.73 |
7.704 |
30 |
| 104B |
7.64 |
7.653 |
29.9 |
Mortality from HS 1/19/12: 5 oysters were dead from the 45°C treatment and 2 were dead from the 44°C treatment. Heat shocked oysters, along with all other animals in the sea tables, were fed.
Multispecies OA
2 geoduck were dead in 101A8 (1 appeared to be dead from a crushed shell) and 1 was dead in 102A8. Geoduck were photographed and the shells were stored in 95% EtOH. In 101A6, 1 manila clam was discovered to be an empty shell so the sample size is only 9 for this container. The clam's measurements are 13x17 mm. One of the Olympia oysters from 102A1 fell into the 102A2 container. The oysters in 102A2 all need to be measured and the correct oyster returned to 102A1.
As described for secondary stress, spec pH was taken of the source water and containers 3 and 4. All containers were clean and animals examined for mortality.
January 19, 2012
Multispecies OA
Tank 102A was still at a high pH (7.78) this morning. I set the control to manual and opened the CO2 flow valve to let more CO2 into the water (I think that it wasn't open enough to allow for that much lowering of the pH given that the incoming water is about 8.14). Once the pH got down to about 7.70 I turned it back to auto. At pH of 7.65 I lowered the CO2 flow meter back down and set accutune.
Took spec pH of source water from both coolers (see table below).
Set up feeding system. Used Matt's spreadsheet to calculate how much Shellfish Diet 1800 to dilute in 15,000 mL of seawater (19 mL). The animals will be fed twice a day with the pumps on for 30 minutes each time. The leftmost bucket sources the food for tanks 101A and 102A; the middle bucket is for 101B, 102B, and 103A; and the rightmost bucket is for 103B, 104A and 104B. Cycle time is set for 12 hours and % on is 6%.
2 geoduck were dead in container 101A4 (one of them seemed to have a crushed shell). One geoduck was dead in 102A4 and it's shell was so brittle that it crushed when I touched it. The geoduck shells were preserved in 95% EtOH and a photograph of them was sent to CSF.
Secondary stress
Tank 104B had crashed to a pH of <7 so I emptied most of the water out to refill. The pH should be about 7.67. I also noticed that when the controller is on auto, there's some sort of offset and its setpoint pH is actually about 0.2 units below what is entered. I raised the setpoint to about 7.8 and I'll ask Matt about this when he is back next week.
Took spec pH of source water from all coolers. Spec pH is corrected for temperature (13°C) and based on a TA of 2076 µmol/kg. Durafet probes were calibrated for temperature as needed. See above for feeding information.
| Tank |
Durafet_Temp |
Fluke_Temp |
Durafet_pH |
Spec_pH |
Salinity |
| 101A |
12.6 |
12.8 |
8.06 |
8.022 |
29.6 |
| 101B |
11.5 |
11.7 |
7.22 |
7.231 |
29.5 |
| 102A |
12.1 |
12.7 |
7.67 |
7.694 |
29.6 |
| 102B |
12.1 |
12.8 |
7.86 |
7.846 |
29.7 |
| 103A |
12.9 |
13.5 |
7.58 |
7.589 |
29.9 |
| 103B |
12.8 |
13.2 |
8.05 |
8.021 |
29.7 |
| 104A |
12.6 |
13.3 |
7.71 |
7.729 |
29.6 |
| 104B |
12.7 |
13.3 |
7.53 |
7.530 |
29.5 |
Re-establishing LHT: heat shock of 8 oysters per temperature at 42 (purple bag), 43 (white zip tie), 44 (pink zip tie), and 45°C (purple zip tie). The oysters are given a 10 minute warm-up and then HS'd for 1 hour. It seems that there was a problem with thermometer accuracy last time and that the thermometer used for the LHT read 43°C when the temp was really only 40.6°C. The Fluke digital thermometer will be used for all future HS. The oysters that were supposed to be HS'd tomorrow (7 days post SLT) will instead be HS'd next week after the LHT has been determined with the Fluke.
Bioinformatics
Removed spaces from the first column of the RNA-Seq file exported from Galaxy so that the contig indicators match the blastall file (e.g. "Consensusfromcontig1"). Saved file as tab delimited and uploaded into Galaxy. Got rid of header information in blastall file and separated columns by "|". Made sure there were no gaps in the data in this file or in the blastall file. Contig number is now in the first column for both datasets.
Under "Join, Subtract, and Group" chose "Join 2 datasets". For job 12, did not keep lines that didn't match and did not fill empty columns. For job 13, kept unmatching and incomplete lines and filled empty columns. Continued analysis with job 13.
Joined resulting table from job 13 with uniprot swissprot IDs (22nd column of the former and 1st column of the latter). This is job 14. Joined this resulting table with swissprot associations based on swissprot ID (column 22 with column 2): job 15.
Joined job 15 table with GO to GOslim table based on GO ID (column 42 with column 1): job 16. Exported this table to Excel. Removed redundant and unnecessary columns and saved file as "Galaxy Job 16 joined tables 011912". Sorted file based on e-value and removed all e-values less than 1e-5. Filtered the file again based on Fold change and deleted all rows with fold change between -2 and 2 (i.e. fold change less than or equal to 2). Saved this file as Galaxy Job 16 joined tabled 011912 fold change. Deleted all information except for column of SPIDs and saved as SPIDs for DAVID 011912. Uploaded to DAVID as gene list.
Edited the original SwissProt ID table uploaded into Galaxy by doing advanced filter in Excel and copying just the unique values into new columns to make a file of unique SPIDs (file is saved with this name). This file contains just one column of SPIDs. Uploaded as the background file into the DAVID Gene Functional Classification Tool indicating that it was Uniprot Accession numbers.
This analysis didn't yield any results (0 clusters, whatever that means) so maybe this isn't the tool that I want. Am going to try Functional Annotation Clustering. Chose Gene Ontology from the list of options.
Watched SR's video on DAVID and I was using the wrong background. Went back to the original exported Galaxy file (job 16) and deleted all columns except for SPID. I then copied unique values only into a new column and saved these as DAVID background 011912. I uploaded this file into DAVID. I then uploaded the SPIDs for DAVID 011912 as my gene list and did functional annotation clustering. Chose Gene Ontology and looked at chart for GOTERM_BP_FAT (see screen shot below). Downloaded the file as DAVID GO table 011912.
January 18, 2012
Secondary stress and Multispecies OA
Ran total alkalinity for all source water and for container 1 from each treatment (8 coolers). Took alkalinity sample (600 mL) from container 2 and poisoned with 75 µL HgCl2 for later analysis. The alkalinity has dropped about 20 units to approximately 2076 µmol/kg seawater. I recalculated setpoints for the coolers based on this new TA and on the desired pCO2 levels and the pH does not change considerably for any of the treatments (i.e. this 20 unit change in TA results in changes to pH in the thousandth place). Took 2 replicate samples of source water from 101A, 101B, and 102A in beer bottles to send off to test 2 companies that sell titrators.
Did spec pH of all source water and from containers 1 and 2 from all 8 coolers. Spec pH of source water corroborated what the Durafet pH probes read. pH from containers 1 and 2 was not more than 0.05 units different from source water pH.
Since the weather has gotten colder, the water in the treatments has been closer to 12°C than 13°C so I turned the chillers up from 11 to 12°C so that the heaters have less work to do.
The pH in 102A will not go down to 7.67. I tried to set it below the setpoint, but it still hovers around 7.78-7.80 (the probe is properly calibrated). I ran accutune overnight to try and fix the problem.
Fed all animals and cleaned containers. Did mortality checks. 3 geoduck were dead in the higher pCO2 treatment (102A): 1 each from containers 1, 3, and 4. Photographed the geoduck to send to CSF and preserved the shells in 95% EtOH for later SEM analysis.
There were no mortalities of HS'd oysters. Tomorrow I will do another HS with n = 8 oysters per temperature at 42, 43, 44, and 45°C.
January 17, 2012
Secondary stress
Made new m-cresol purple. Took spec pH of all coolers and did 2nd dye addition to do a correction.
The slope of the new dye correction curve is-0.03482 and the intercept is 0.018166. Spec pH is corrected for temperature based on a TA of 2095 µmol/kg.
| Tank |
salinity |
Durafet_pH |
Spec_pH |
| 101A |
29.5 |
8.03 |
8.011 |
| 101B |
29.6 |
7.27 |
7.294 |
| 102A |
29.8 |
7.69 |
7.789 |
| 102B |
29.6 |
7.86 |
7.859 |
| 103A |
29.6 |
7.57 |
7.586 |
| 103B |
29.8 |
8.02 |
8.002 |
| 104A |
29.5 |
7.74 |
7.714 |
| 104B |
29.4 |
7.66 |
7.673 |
Animals were fed. No mortalities from HS.
Matt showed me the set up for the automated feeding and I think I will work on that Thursday. The algae delivered to the containers ends up being at 5% concentration of the algae in the source buckets. The buckets are marked with a fill line at 20,000 mL for calculating how much algae to add to a bucket of seawater.
Multispecies OA
Took spec pH of both coolers (see above table).
Animals were fed.
January 16, 2012
Secondary stress
Ran total alkalinity samples that were taken on 1/7/12 and 1/10/12. Also ran 3 of the Mukilteo samples that Carolyn brought up. Did spec pH on the source water for all 8 coolers. NB: salinity was not taken at the exact time of spec pH, but was taken about 1 hour later. Spec pH has been corrected for temperature based on a TA of 2095 µmol/kg.
| Tanks |
salinity |
spec_pH |
| 103A |
29.4 |
7.638 |
| 103B |
29.5 |
8.014 |
| 104A |
29.5 |
7.731 |
| 104B |
29.6 |
7.664 |
| 101A |
28.9 |
8.037 |
| 101B |
29.1 |
7.401 |
| 102A |
29.4 |
7.780 |
| 102B |
29.4 |
7.853 |
Fed oysters and cleaned containers after feeding.
There was no mortality in the HS'd oysters.
Cleaned spec pH cuvettes overnight.
Multispecies OA
Throughout the day adjusted the pH in cooler 102A to about 7.67 (pCO2 of 1000 ppm).
Fed bivalves and cleaned containers. Checked all animals for mortality. 4 geoduck were dead, 1 in each of containers 101A1, 101A3, 102A3, and 102A6. Photographed the dead animals with a scale for size.
NB: When cleaning these containers make sure that none of the geoduck have fallen out of their bag to the bottom of the container.
January 15, 2012
Secondary stress
There is no mortality from HS.
Oysters were fed 120,000 cells per mL (water circulation turned off for 1 hour).
Multispecies OA
Animals were fed 120,000 cells per mL.
January 14, 2012
Secondary stress
Fixed samples were moved from Davidson's fixative into jars of 70% EtOH.
There was no mortality from HS.
The oysters for the 1 month OA exposure were measured, weighed individually for buoyant weight, and weighed for total mass of all the animals in a container together. 6 oysters were put in each container at pCO2 of 400, 600, 800, 1000, 1200, or 1400.
Multispecies OA
Animals were measured before being put into one of 8 containers in each of 2 coolers. The coolers are both currently set at a pH of >8.0 (pCO2 ~400 ppm).
Pacific oysters - 25 animals were measured lengthwise and weighed individually for buoyant weight to get an average. There are 10 C. gigas in each container and these 10 animals were massed for buoyant weight together before being placed in pillow bags in the container.
Geoduck - There are 20 geoduck in soft mesh bags in each of the containers. Before being placed in the containers the geoduck (in groups of 10) were photographed for measurements and weighed for total weight.
Olympia oysters - There are 3-4 Olys per container. Before being placed loose in the containers Olys were measured (length and width) and buoyant weight was taken. Epibionts were scraped off. Of the three containers that have only 3 Olys, 2 of them have Olys with smaller Olys settled on them and 1 has a particularly large oyster.
January 13, 2012
Secondary stress
Today was the end of the 1 week OA exposure experiment and the big sampling/heat shock day. I started at 7 am and finished at 11:30 pm. 4 different heat shock (HS) regimes were performed on the oysters and for each one 1 oyster from each container per treatment (n = 8 per treatment) were used. HS1 was 38°C for 1 hour and in one week (1/20) these oysters will be HS'd at 43°C for 1 hour and then monitored for mortality. HS2 was 38°C for 1 hour followed immediately by shucking and sampling of gill tissue for NGS. HS3 was 43°C for 1 hour and monitoring throughout the week for mortality. HS4 was 38°C for 1 hour followed immediately by 43°C for 1 hour and monitoring for mortality. After HS, all oysters were put in labeled mesh bags in a sea table where they will remain throughout monitoring.
Sampling was done on all other oysters. Oysters were measured (length and width) and weighed before sampling. Oysters were shucked and separate gill tissue samples were taken for gene and protein expression and immediately flash frozen in liquid nitrogen. A section was taken for histology that included the DG and gill as well as a separate section of part of the adductor muscle. These sections were fixed for 24 hours in invertebrate Davidson's fixative.
January 12, 2012
Bioinformatics
Exported RNA-Seq files from CLC into "Emma's data" folder. Also began new RNA-Seq on de novo assembly 2.
Uploaded blastall results into Galaxy. Tried to upload RNA-Seq files, but they wouldn't upload.
Ran experiment on RNA-Seq files to do comparison of gene up- and down-regulation and exported as csv files. This file was uploaded successfully into Galaxy.
Secondary stress
Fed all animals and cleaned containers in acclimation coolers.
January 11, 2012
Secondary stress
Took spec pH of all experimental tanks and calibrated probes for temperature and pH. pH is corrected for temperature.
| Tank |
salinity |
Durafet_T |
Fluke_T |
Durafet_pH |
spec_pH |
| 101B |
29.9 |
13.1 |
13.1 |
7.31 |
7.366 |
| 102B |
29.9 |
12.9 |
13 |
7.86 |
7.864 |
| 103A |
29.9 |
12.9 |
13 |
7.55 |
7.60 |
| 103B |
30.1 |
12.9 |
12.9 |
8.03 |
8.013 |
| 104A |
30.1 |
12.9 |
13.1 |
7.78 |
7.750 |
| 104B |
30.1 |
13.2 |
13.2 |
7.63 |
7.634 |
Fed all the oysters and cleaned experimental tanks.
Multispecies OA
Received manila clams and put in sea table. Fed all animals.
January 10, 2012
Secondary stress
Did full TA and spec pH for all experimental coolers. Ran titrations for all source water and samples from all containers number 3. Took and poisoned samples for all containers number 4. Did spec pH on samples from all source water and all containers 3 and 4. Spec pH was calculated based on the TA that was found for the source water in each cooler. The TA for 104B is a little off becuase it was run with a salinity of 29.7 rather than 30.2. Spec pH has been corrected for temperature.
| Tank |
salinity |
Durafet_pH |
spec_pH |
TA |
| 101B |
29.4 |
7.26 |
7.274 |
2091.87 |
| 102B |
29.9 |
7.86 |
7.857 |
2092.27 |
| 103A |
29.6 |
7.54 |
7.552 |
2097.14 |
| 103B |
29.7 |
8.03 |
7.968 |
2093.44 |
| 104A |
29.7 |
7.76 |
7.664 |
2094.94 |
| 104B |
30.2 |
7.68 |
7.596 |
2092.69 |
Did spec pH on source water for acclimation coolers. Calibrated the Durafet probes for temperature. Spec pH has been corrected for temperature based on a TA of 2095 µmol/kg.
| Tank |
salinity |
Durafet_T |
Fluke_T |
Durafet_pH |
spec_pH |
| 105A |
29.5 |
12.9 |
12.8 |
7.96 |
7.913 |
| 105B |
29.7 |
13.6 |
13.5 |
7.97 |
7.958 |
| 106A |
29.9 |
12.9 |
12.8 |
7.94 |
7.926 |
| 106B |
30 |
13.1 |
13 |
7.92 |
7.841 |
| 102A |
30 |
12.7 |
12.5 |
7.94 |
7.917 |
| 101A |
30 |
12.9 |
12.8 |
7.97 |
8.092 |
Multispecies OA
Carolyn brought up mussels, geoduck, olympia oysters, and pacific oysters for a multispecies OA experiment. The Pacifics are small, about the size of a thumbnail. The Olys are larger, about 1 inch long. The mussels look like they are just post-larval and the geoduck are also small seed, but larger than the mussels. Manila clams are arriving tomorrow. All animals were put in sea tables. All animals were fed shellfish diet 1800.
January 9, 2012
Secondary stress
Made new m-cresol purple (see 1/2/12). Took spec pH from all experimental coolers and used them to do the dye correction. The intercept for the dye correction is 0.013592 and the slope is -0.03138. Calculated the temperature-corrected pH from spec pH at 25°C with an assumed TA of 2095 µmol/kg.
| Tank |
salinity |
Durafet_pH |
spec_pH |
| 101B |
29.7 |
7.29 |
7.265 |
| 102B |
29.8 |
7.86 |
7.867 |
| 103A |
29.9 |
7.49 |
7.428 |
| 103B |
29.8 |
7.93 |
7.894 |
| 104A |
29.9 |
7.70 |
7.729 |
| 204B |
30 |
7.62 |
7.650 |
Changed set points on a couple of the coolers that are consistently below their set points.
Fed all the oysters and cleaned the containers for the experimental oysters.
Bioinformatics
Started a new de novo assembly (Emma de novo 2) on pinto ab OA data. The parameters for the new de novo are all default, except the mismatch costs were changed to 3 and the min contig length was decreased to 150.
Began RNA-Seq analysis to find differentially expressed genes between data sets. The consensus sequences from the de novo assembly were used as a reference backbone (8911 sequences). The analysis was done on a reference without annotations and with default assembly settings, except the unmapped sequences box was unchecked. RNA-Seq was done for both data sets.
Detailed instructions:
- Open file of consensus sequences in CLC so you can view it and select all sequences by clicking on the first one, scrollingn down, hold shift and click on last one
- Click "open consensus" at the bottom of the window
- Resave the file into a folder in your CLC directory - this file will be your reference for RNA-Seq
- Right click on your raw data file and select toolbox -> CLCserver -> RNAseq analysis
- Use default options on the RNA-Seq with the following changes: 1) reference without annotations, 2) uncheck unmapped sequences
- Repeat RNA-seq with your second sequence file
- To do a comparison of your RNA-seq for your datasets, go to high throughput sequencing in your toolbox and select "Run an experiment". Keep your default settings
Quality trimmed raw data, discarding lengths below 20 and saving the trimmed sequences in the trimmed sequence folder.
Annotated the consensus sequences using the wetgenes INQuiry portal. Used the blastall tool with the following parameters: blastx against swissprot database, short description and number of alignments changed from 500 and 250 (respectively) to 1, tabular output.
January 8, 2012
Secondary stress
Calibrated Durafet probes in acclimation tanks for temperature and pH as described 1/7/12. Saw that 103B2 was not attached to its dripper so no clean water was circulating through (dripper was reattached).
| Tank |
salinity |
Durafet_pH |
Durafet_Temp |
Fluke_Temp |
spec_pH |
| 105A |
29.9 |
7.97 |
12.9 |
13.4 |
7.908 |
| 105B |
29.9 |
7.96 |
13.5 |
13.9 |
7.867 |
| 106A |
30 |
7.95 |
11.6 |
12.3 |
7.762 |
| 106B |
30 |
8.06 |
11.4 |
12.1 |
7.707 |
| 102A |
30 |
7.97 |
13 |
12.8 |
7.939 |
| 101A |
30 |
7.96 |
13 |
13 |
7.947 |
| Tank |
salinity |
Durafet_pH |
Durafet_Temp |
Fluke_Temp |
spec_pH |
| 101B |
29.7 |
7.27 |
13.1 |
12.9 |
7.290 |
| 102B |
29.8 |
7.86 |
13.1 |
13.1 |
7.860 |
| 103A |
29.9 |
7.52 |
13 |
12.9 |
7.508 |
| 103B |
29.9 |
7.95 |
13 |
12.8 |
7.924 |
| 104A |
30 |
7.71 |
13 |
13.1 |
7.729 |
| 104B |
30 |
7.65 |
13.1 |
13.1 |
7.687 |
In the evening did mortality checks and fed all the oysters. Cleaned all the containers in the acclimation coolers. The process is slightly different from the experimental coolers: all oysters are removed and put in a common bucket while the containers are cleaned. After all containers are clean, oysters are re-assorted into containers randomly.
January 7, 2012
Secondary stress
For the most part, the pH seems to be stable at about the right value for all of the coolers except for 101B. Continue to empty water from this cooler and refill with new (high pH) water to counterbalance the low pH of about 7.
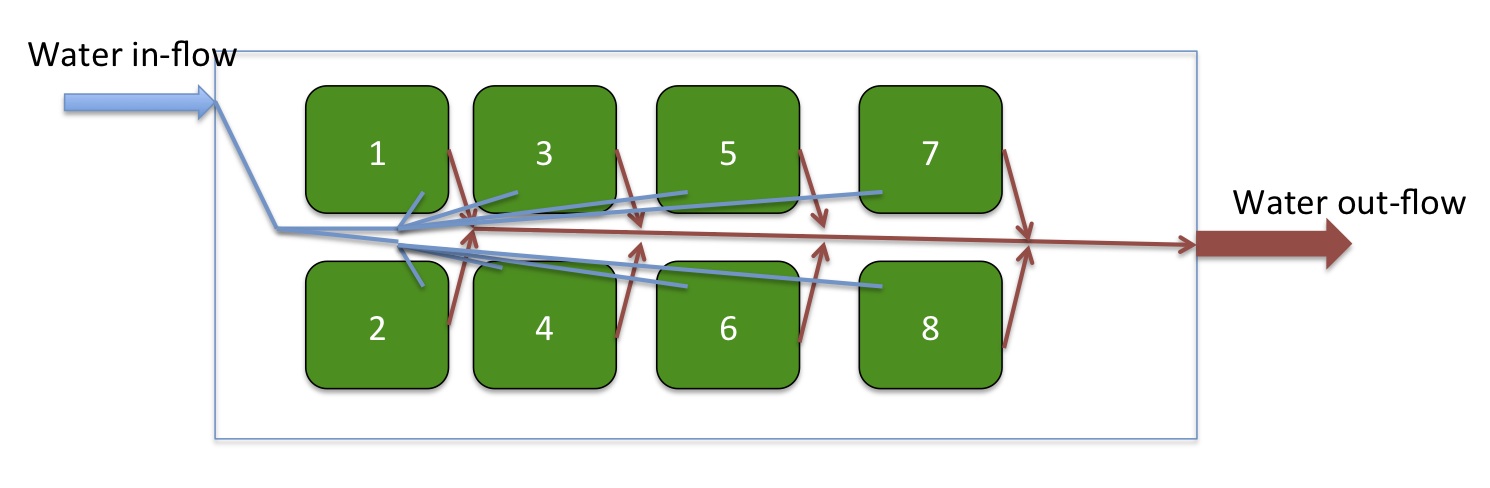
Did chemistry for all the coolers. Took spec pH and TA samples from the source water and from containers 1 and 2 (see diagram below for container numbers). The pH in the containers was not more than 0.05 pH units less than the source water pH. Used the spec pH values to calibrate the Durafet probes and also calibrated probes for temperature using the Fluke probe (see table). All probes were pretty accurate, except for 103A, which is the control treatment. The probe read a pH of about 8, but it was actually around 7. I verified this with a second spec pH. I calibrated the probe, drained the water and it quickly righted itself but the oysters were exposed to low pH water for about 36 hours. All the treatments equilibrated to the new pH much better after the probes had been calibrated. I also ran TA titrations for the source water and for one of the containers for all the treatments. In general, the source water TA was around 2095 µmol/kg seawater and the container TA was about 10-20 units lower. TA samples were taken for all container 2s by rinsing out sample jars with 60 mL of seawater, filling them with 300 mL of seawater from the container and poisoning with 75 µL HgCl2. I ran CRMs at the beginning, middle, and end of the titrations. I also got TA values for Liza's 4 samples from her pinto abalone experiment done 9/29/11.
I also calibrated the Durafet monitoring the incoming water using 2 spec pH samples. The probe read pH 8.1 and, assuming a salinity of 29.7 and temperature of 10°C, the actual pH is about 8.1.
In the evening, I fed all the oysters (experimental and acclimation) by diluting 19 mL of shellfish diet in 941 mL of ambient seawater and pipetting 9 mL of this mixture into each of the containers (flow turned off). I let the oysters sit for about an hour so that they could feed. After an hour, I turned the flow back on and cleaned all the experimental containers. It takes about 15 minutes to clean all 8 containers in each treatment and I do 2 at a time, so the oysters are only out of water for about 2 minutes. I put all the oysters back in the container that I had taken them from.
| Tank |
salinity |
Durafet_pH |
Durafet_Temp |
Fluke_Temp |
spec_pH |
| 104A |
29.5 |
7.70 |
13 |
12.4 |
7.56 |
| 104B |
29.7 |
7.59 |
13 |
12.4 |
7.49 |
| 102B |
30.1 |
7.86 |
13 |
12.4 |
7.67 |
| 103B |
30 |
7.91 |
12.8 |
13 |
6.84 |
| 103A_2pm |
29.9 |
12.9 |
13.2 |
||
| 103A_4pm |
29.7 |
7.52 |
7.31 |
||
| 101B |
30 |
7.08 |
13 |
12.8 |
7.03 |
January 6, 2012
Secondary stress
Set the pH set points at 7 am. Some of the oysters got switched to different tanks if I was having trouble with the controlers. The tanks and their set points are in the table below. Changed the PID tune parameters to the numbers from cooler 103B since it has been the most consistent in keeping the correct pH: Prop bank 0.911, Rate 1.446, Reset 0.173. When pH gets too low (>0.2 units below the set point), I drain most of the water from the tank and refill it with fresh water. Accutune is run once the pH has reached a level close to the set point.
| Tank |
pCO2(ppm) |
pHsetpoint |
| 101B |
1400 |
7.533 |
| 102B |
600 |
7.876 |
| 104A |
800 |
7.762 |
| 104B |
1000 |
7.671 |
| 103A |
1200 |
7.597 |
| 103B |
400 |
8.034 |
Continued to monitor and adjust coolers throughout the day to get the pH to stabilize at the correct value. Having trouble with coolers 104A and B and 101B and a little bit with 103A. 101B is especially troublesome because I'm having trouble getting it to stay above a pH of 7, let alone get to 7.53.
In the evening, fed the oysters in each of the experimental tanks and the oysters in the sea tables. After feeding, moved oysters from see tables into containers in acclimation coolers (6 coolers, 8 3-L containers per cooler). There are 8 oysters per container. pH remains steady at about 8 in all. There were no mortalities.
January 5, 2012
Secondary stress and Total Alkalinity
Cleaned out tanks 103A and 103B as described 1/1/12.
Calibrated Durafet pH probe in tank 101A using spec pH.
Ran total alkalinity for 2 more beer bottle samples. Began by running replicate junk samples until the TA values were within 2 units of each other (this took 3 samples). Ran a CRM (batch 113 bottle 0780) but the first run did not yield a TA close enough to the value: the experimental value was 2219.14 and it needs to be 2224.65. Ran another CRM, which was also not the right number. Ran 4 junk samples to check precision of titrator and got the same number for each sample. Ran a CRM which gave a TA of 2220.5. Decided to run allt he beer bottle samples for 1/5/12 - the data can be found in the spreadsheet chemistry 010512.
Calculated set points for each of the tanks based on a presumed TA of 2100 µmol/kg and a temperature of 13°C.
| Tank |
pCO2(ppm) |
pHsetpoint |
| 101A |
800 |
7.762 |
| 101B |
400 |
8.034 |
| 102A |
1400 |
7.533 |
| 102B |
600 |
7.876 |
| 104A |
1200 |
7.597 |
| 104B |
1000 |
7.671 |
Fed the oysters as previously described and cleaned the containers. There was one dead oyster in 101B and I replaced it with an oyster from the sea table.
Bioinformatics
Pinto abalone larvae NGS data were uploaded into CLC genomics workbench. There are 2 experimental groups: ambient (control) and high pCO2. Ran 2 different de novo assemblies on them.
I. default parameters: mismatch cost 2; limit 8; fast ungapped alignment; vote ACGT; non-specific matches random; min contig length 200; map reads back to contigs; result handling open; make log
II. same parameters except turned global alignment on
January 4, 2012
Secondary stress
Checked pH of source water and 5 containers to see if pH had changed over night after the water change. There was an approximate 0.05 pH unit change between all the containers and the source water.
Calibrated all the Durafet probes for temperature and pH using the Fluke probe and spec pH, respectively.
Fed all the oysters as previously described.
January 3, 2012
Secondary stress
Calibrated the acid for the total alkalinity titrator by running 3 CRMs within 1 unit of each other (they were all less than 1 unit different) and using those numbers to back calculate the acid concentration since the CRM TA is known.
Did a dye correction for the spec pH using 2 samples from 2 difference coolers and then 2 other samples to which either NaOH or HCl was added. The slope for this dye batch (1/2/12) is -0.02313 an the intercept is 0.009991.
Measured pH from the source water and 1 container each from 101A and 104B (see table below). pH values are corrected for dye addition, but not for temperature at which they were taken. Since there seems to be a difference between source and container water, took spec pH of 102B source water and 5 containers (table below). Also did TA titrations of 102B source water and 2 containers.
| Sample |
TA(µmol/kg) |
pH |
pHdifferencefromsource |
| 101A source (s) |
7.53 |
||
| 101A container(c) |
7.49 |
0.036 |
|
| 104B s |
7.51 |
||
| 104B c |
7.39 |
0.12 |
|
| 102B s |
2111.97 |
7.53 |
|
| 102B c1 |
2068.51 |
7.45 |
0.081 |
| 102B c2 |
7.46 |
0.065 |
|
| 102B c3 |
7.46 |
0.066 |
|
| 102B c4 |
7.47 |
0.055 |
|
| 102B c5 |
2075.44 |
7.47 |
0.057 |
Fed all the oysters as described 1/2/12. Cleaned all the tanks as described 1/1/12.
January 2, 2012
Secondary stress
Cooler 101 B was below its set point and the heater was never going on and 101A was above its set point and the heater wasn't going off (only a couple of degrees in each direction). The autotune function was not on for either heater so I turned it on.
Checked ammonia levels in one container from 101B, 101A, and 102A and from one of the sea tables. Ammonia was very low in all. Also took pH from the source water and one container from 102B and 104A and from the sea table (see table below). I have not done the dye correction for the new m-cresol purple yet, so the pH have not been properly adjusted. Additionally, the cuvettes were probably not very clean because many small bubbles formed inside and could not be dislodged so the pH values are probably not completely accurate. M-cresol purple was made by dissolving 0.016 g of MCP in 20 mL of milliQ water (2 mmol/L) and adding dilute NaOH until the pH = 7.9. pH values are calculated in CO2sys assuming 2100 µmol/kg TA and adjusted temperature of 13°C except for the sea table where the temperature is assumed to be 10°C. Salinity was measured in the source water in each container and used to correct pH.
| Sample |
Salinity |
MeasuredpH |
CorrectedpH |
| 102Bsource |
29.7 |
7.51 |
7.67 |
| 102Bcontainer |
29.7 |
7.43 |
7.58 |
| 104Asource |
29.9 |
7.50 |
7.67 |
| 104Acontainer |
29.9 |
7.40 |
7.55 |
| sea table |
29.5 |
7.44 |
7.63 |
Fed the oysters in the containers and sea tables by turning off flow and giving them ~120,000 cells per mL of shellfish diet. Put 19 mL of shellfish diet in 941 mL of seawater and gave 9 mL to the container oysters and 255 mL to the sea table oysters (estimated sea table dimensions used to calculate volume: 3' x 2' x 0.5').
January 1, 2012
Secondary stress
Oysters were fed as described 12/31/11 except they were fed ~120,000 cells per mL
Emptied all coolers and cleaned out by removing debris, wiping down with Vortex, rinsing and wiping down with freshwater and then salt water. Coolers were then refilled with salt water. Only 6 oysters were replaced in each of the containers (which had been cleaned by rinsing with fresh and then salt water) and only 6 coolers were used (8 reps of 6 per cooler). These will be the oysters used in the 1 week OA exposure. The remaining oysters were divided between 2 sea tables.
December 31, 2011
Secondary stress
Fed oysters in their containers about 60,000 cells of algae (Shellfish Diet 1800) per mL by diluting the SD in seawater and turning on flow in containers for ~30 minutes. Checked ammonia levels of a few containers and all were low.
December 30, 2011
Secondary stress
Brought oysters up to Friday Harbor. Changed filters with Michelle (put in new bag filters and clean 0.2 µm filters). Turned on water to coolers and let run through to rinse out lines for > 1 hour. Put ~110 oysters in each of 6 coolers and set temperature to 13°C. Moose later told me that having them in the coolers would contaminate the water for the remainder of the experiment, so put oysters in microcosm containers (3.5 L). 10 oysters were placed in each containers and 8 containers were put in each of 8 coolers. Green drippers (57.5 mL/ min) were used for flow of water into containers.
December 29, 2011
Secondary Stress
2 am: collected ~650 oysters (same group as used for most recent heat stress experiment) in Oyster Bay at low tide. Put oysters in mesh bags in a cooler, covered with towels and put a few ice packs on top of the towels. The oysters remained in the cooler (ice packs were replaced when thawed) until the afternoon of 12/30.










