Bioinformatics: samifier
Fixing the gff file
The top file in this picture (in pink box) is the short file that seems to work. The bottom file is an example of a gff file that needs to be edited to mimic the top.
https://www.evernote.com/shard/s242/sh/21fe6054-eb77-4754-b332-a42f086f41ba/5788c3de4f88a72eaa261006690e3017
List of things that need to be changed in bottom gff file (v9_p.gff):
1. gene line needs to be first entry for each scaffold
2. gene needs ID=geneXXXXX
3. gene format should be gene [2 numbers that are the same for cds and mRNA] . - . ID=....
4. mRNA and cds need ID=mRNAXXXXX or ID=cdsXXXXX
March 20, 2014
Bioinformatics: samifier
Running through all of the gff files that Steven made to see if any work.
java -jar samifier.jar -r /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/F003797.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/cnidarian/ets_v9_g.gff -c /Volumes/web/cnidarian/v9_multi -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out2014320 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_2014320 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_2014320.bed
List of files tried:
ets_v9_p.gff
ets_v9_o.gff
ets_v9_i.gff
ets_v9_h.gff
ets_v9_g.gff
March 19, 2014
Bioinformatics: samifier
Jimmy ran a mascot search on one of my files and it took 2 days to run. I exported the search results to use in samifier and the file export parameters can be found here - https://www.evernote.com/shard/s242/sh/a676bdf5-b33d-4484-a9f3-a1122d17c03b/5602e7de2eb5d31ae28c08aae42173a9
java -jar samifier.jar -r /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/F003797.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Users/emmatimminsschiffman/documents/winter_2014/Bioinformatics/ets_v9_r.giles.gff -c /Volumes/web/cnidarian/v9_multi -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out2014319 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_2014319 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_2014319.bed
March 13, 2014
Bioinformatics: samifier
Giles found a webpage (http://gmod.org/wiki/GFF3) that says that the #gff-version3 must be in the header of a gff3 file. I added this to one of the files that Steven had made (ets_v9_r.gff) and ran the following in samifier
java -jar samifier.jar -r /Volumes/web/oyster/proteomics/interact-20120821_103B_251_QE_02.prot.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/cnidarian/ets_v9_r.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20141313 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20141313 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20141313.bed
The code ran, but all I got was an empty log file. I think the error was "Run exception thrown", but I'm not sure what this means
Abbreviated gff file so that it only has a few entries and made sure that the "gene" line came before mRNA and exon lines. Ran the same code as above. Same error.
##gff-version3
C16582 GLEAN gene 35 385 0.555898 - . Name=CGI_10000001;
C16582 GLEAN mRNA 35 385 0.555898 - . Parent=CGI_10000001;
C16582 GLEAN exon 35 385 . - 0 Parent=CGI_10000001;
C17212 GLEAN gene 31 363 0.999572 + . Name=CGI_10000002;
C17212 GLEAN mRNA 31 363 0.999572 + . Parent=CGI_10000002;
C17212 GLEAN exon 31 363 . + 0 Parent=CGI_10000002;
C17316 GLEAN gene 30 257 0.555898 + . Name=CGI_10000003;
C17316 GLEAN mRNA 30 257 0.555898 + . Parent=CGI_10000003;
C17316 GLEAN exon 30 257 . + 0 Parent=CGI_10000003;
The above file did not actually look like the example the samifier developers had given me, so re-edited the file and reran code. Still got the same error.
##gff-version3
C16582 GLEAN gene 35 385 . - 0 Name=CGI_10000001;
C16582 GLEAN mRNA 35 385 0.555898 - . Parent=CGI_10000001;
C16582 GLEAN exon 35 385 . - 0 Parent=CGI_10000001;
C17212 GLEAN gene 31 363 . + 0 Name=CGI_10000002;
C17212 GLEAN mRNA 31 363 0.999572 + . Parent=CGI_10000002;
C17212 GLEAN exon 31 363 . + 0 Parent=CGI_10000002;
C17316 GLEAN gene 30 257 . + 0 Name=CGI_10000003;
C17316 GLEAN mRNA 30 257 0.555898 + . Parent=CGI_10000003;
C17316 GLEAN exon 30 257 . + 0 Parent=CGI_10000003;
Giles and I made another version of the gff file, but still got the same error. He looked around in the java code and figured out that the problem actually seems to be with the mzid file. I've emailed the samifier developers.
##gff-version 3
C16582 GLEAN gene 35 385 . - . ID=gene00001;Name=CGI_10000001;
C16582 GLEAN mRNA 35 385 0.555898 - . ID=mRNA00001;Parent=CGI_10000001;
C16582 GLEAN CDS 35 385 . - 0 ID=cds00001;Parent=CGI_10000001;
C17212 GLEAN gene 31 363 . + . ID=gene00002;Name=CGI_10000002;
C17212 GLEAN mRNA 31 363 0.999572 + . ID=mRNA00002;Parent=CGI_10000002;
C17212 GLEAN CDS 31 363 . + 0 ID=cds00002;Parent=CGI_10000002;
C17316 GLEAN gene 30 257 . + . ID=gene00003;Name=CGI_10000003;
C17316 GLEAN mRNA 30 257 0.555898 + . ID=mRNA00003;Parent=CGI_10000003;
C17316 GLEAN CDS 30 257 . + 0 ID=cds00003;Parent=CGI_10000003;
March 12, 2014
Bioinformatics: samifier
Validating gff as gff3 format (http://modencode.oicr.on.ca/cgi-bin/validate_gff3_online). File is in cnidarian ets_v9_f.gff
File is not correct format. Part of error report is below.
<span style="background-color: #f1f3ff;"># GFF3 File Validation Report # ontology_file(s): http://song.cvs.sourceforge.net/*checkout*/song/ontology/so.obo # generated: 12-Mar-14 11:55:25 ############################################################################### # THIS FILE HAS NOT BEEN VALIDATED, IT CONTAINS ERRORS, PLEASE REVIEW REPORT! # # (NO WARNINGS HAVE BEEN ISSUED FOR THIS FILE) # ############################################################################### ############################################################################### # THIS FILE HAS BEEN PROCESSED ENTIRELY AND ALL ERRORS/WARNINGS ARE REPORTED! # ############################################################################### # First 10 lines of the analyzed GFF3 file follows: # [line 1]> C16582 GLEAN CDS 35 385 0.555898 - . [line 1]> Parent=CGI_10000001; [line 2]> C16582 GLEAN exon 35 385 . - 0 [line 2]> ID=CGI_10000001; [line 3]> C17212 GLEAN CDS 31 363 0.999572 + . [line 3]> Parent=CGI_10000002; [line 4]> C17212 GLEAN exon 31 363 . + 0 [line 4]> ID=CGI_10000002; [line 5]> C17316 GLEAN CDS 30 257 0.555898 + . [line 5]> Parent=CGI_10000003; [line 6]> C17316 GLEAN exon 30 257 . + 0 [line 6]> ID=CGI_10000003; [line 7]> C17998 GLEAN CDS 196 387 1 - . [line 7]> Parent=CGI_10000005; [line 8]> C17998 GLEAN exon 196 387 . - 0 [line 8]> ID=CGI_10000005; [line 9]> C18346 GLEAN CDS 174 551 1 + . [line 9]> Parent=CGI_10000009; [line 10]> C18346 GLEAN exon 174 551 . + 0 [line 10]> ID=CGI_10000009; # ... Line Number Error/Warning ----------- ------------- 1 [ERROR] CDS does not have a phase (phase: .) 1 [ERROR] empty tag/value information (Parent=CGI_10000001;) 1 [ERROR] first line must be ##gff-version 3 (line: GLEAN) 1 [ERROR] invalid type pair - check all parents (at line 2; CDS to exon) 2 [ERROR] empty tag/value information (ID=CGI_10000001;) 3 [ERROR] CDS does not have a phase (phase: .) 3 [ERROR] empty tag/value information (Parent=CGI_10000002;) 3 [ERROR] invalid type pair - check all parents (at line 4; CDS to exon) </span>
samifier developers told me that each mRNA and CDS entry must have a gene entry so that the file should look like this:
<span style="background-color: #f8f8f8; color: #333333; font-family: Consolas,'Liberation Mono',Courier,monospace; font-size: 13px;">**C16582 GLEAN gene 35 385 . - 0 Name=CGI_10000001;** C16582 GLEAN mRNA 35 385 0.555898 - . Parent=CGI_10000001; C16582 GLEAN CDS 35 385 . - 0 **Parent=CGI_10000001;** </span>cnidarian: ets v9 f gff
-c should be individual fasta files in a directory (v9 multi)
February 21, 2014
Bioinformatics: samifier
new gff file (added "fixed" to name) where trying to troubleshoot samifier's problem with CGI_10000004. First, changed order of 2 CDSs, but got same error (except stop of sequence overflows gene). Then switched identifiers so that CDS = mRNA and vice versa, but this didn't work either.
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/xml_files/interact-20120821_103B_251_QE_02.pep.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.fixed.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20140220 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20140220 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20140220.bed
Try using Steven's new ensembl version of gff file. Edit first line so that it is Parent=CGI...get error for subsequent line that Parent attribute not found.
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/xml_files/interact-20120821_103B_251_QE_02.pep.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/whale/ensembl/ftp.ensemblgenomes.org/pub/release-21/metazoa/gtf/crassostrea_gigas/Cgtest.gtf -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20140220 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20140220 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20140220.bed
February 20, 2014
Bioinformatics: samifier
In all files, everything should be in terms of CGIs
for gff, make sure that only has gene info (i.e. no info for non-CGI elements) - checked and all are CGIs
created mapping file that is 3 columns of the CGI IDs identified in 103B_251_02
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/emma/interact-20120821_103B_251_QE_02.pep.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20140220 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20140220 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20140220.bed
mzIdentml files are from the wrong searches (against old database, not against oyster genome v9) so accession numbers are wrong.
Looking for software that will convert pep or protxml to mzIdentml. I tried to download proteowizard, but the download didn't work.
error:
Start of sequence in gene CGI_10000004 overflows gene
at au.org.intersect.samifier.parser.GenomeParserImpl.throwParsingException(GenomeParserImpl.java:98)
at au.org.intersect.samifier.parser.GenomeParserImpl.processSequence(GenomeParserImpl.java:178)
at au.org.intersect.samifier.parser.GenomeParserImpl.doParsing(GenomeParserImpl.java:84)
at au.org.intersect.samifier.parser.GenomeParserImpl.parseGenomeFile(GenomeParserImpl.java:46)
at au.org.intersect.samifier.runner.SamifierRunner.run(SamifierRunner.java:84)
at au.org.intersect.samifier.Samifier.main(Samifier.java:125)
Jimmy reran proteowizard on the correct files (saved in folder xml files).
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/xml_files/interact-20120821_103B_251_QE_02.prot.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20140220 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20140220 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20140220.bed
same error as above....try with pep file instead of prot
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/xml_files/interact-20120821_103B_251_QE_02.pep.mzid -m /Volumes/web/oyster/bioinformatics/103B_251_02_mappingfile.txt -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out20140220 -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/log_20140220 -b /Users/emmatimminsschiffman/Documents/winter_2014/Cg_20140220.bed
same error
February 13, 2014
Bioinformatics: Samifier
Downloaded samifier https://github.com/IntersectAustralia/ap11_samifier
Navigated to application file in terminal and typed "ant dist" to build application.
downloaded gff genome file from crassostreome (gene features)
Emma-Timmins-Schiffmans-MacBook-Pro:ap11_samifier-master emmatimminsschiffman$ samifier -r /Volumes/web/oyster/bioinformatics/emma/interact-20120821_103B_251_QE_02.pep.mzid -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_00297895.1.21.gtf -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.gff -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out021314.sam -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_log021314 -b /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier021314.bed
Can't get this to run. Tried following commands: ./samifier, -jar samifier.jar, samifier.jar
Found samifier.jar in dist folder. Ran above command: java -jar samifier.jar -r ...
This won't run without a mapping file (-m). Must make a mapping file...
Maybe gtf file can be used as mapping file and chromosome directory can be the genome file? Downloaded genome file (fasta) from ensembl site.
java -jar samifier.jar -r /Volumes/web/oyster/bioinformatics/emma/interact-20120821_103B_251_QE_02.pep.mzid -m /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_00297895.1.21.gtf -g /Volumes/web/oyster/bioinformatics/oyster.v9.glean.final.rename.gff -c /Volumes/web/oyster/bioinformatics/Crassostrea_gigas.GCA_000297895.1.21.dna.genome.fa.fa -o /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier_out021314.sam -l /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_log021314 -b /Users/emmatimminsschiffman/Documents/winter_2014/Bioinformatics/Cg_samifier021314.bed
error with gff file: start of sequence in gene CGI_10000004 overflows gene usage
issue is specifically with following entry: C17476 GLEAN CDS 34 74 . - 2 Parent=CGI_10000004
February 12, 2014
Bioinformatics: iPiG
Trying to find uniprot ID mapping file that I can use with the purple urchin data. I think this file is a list of uniprot IDs and corresponding IDs from other databases. From the uniprot ftp website, I'm checking out the file idmapping_selected.tab.gz. Explanation of this file is here: ftp://ftp.ebi.ac.uk/pub/databases/uniprot/current_release/knowledgebase/idmapping/README. Other file options are already divided up by taxonomic group and so would probably not work.
This and other files are saved in eagle in my bioinformatics file.
Downloaded oyster proteome (FASTA) and GTF file to use in ipig from http://metazoa.ensembl.org/info/data/ftp/index.html
Running ipig with oyster files
peptide spectrum matches: 103B_251_QE_02.pep.mzid
ensembl genes table: Crassostrea_gigas.GCA_000297895.1.21.gtf
ensembl amino acid sequences: Crassostrea_gigas.GCA_000297895.1.21.pep.all.fa
uniprot ID-mapping: idmapping_selected.tab
proteome fasta: same as amino acid sequences
error -
The content of element 'DatabaseName' is not complete
Tried running the same as above but peptide spectrum matches = 103B_251_QE_02.prot.mzid
error -
Duplicate unique value [] declared for identity constraint "PK_SCDBSEQ" of element "MzIdentML".
February 11, 2014
Proteomics: focus on immune
Joined all files together based on GO term
SELECT * FROM [emmats@washington.edu].[unique_immune_GO_terms.txt]
LEFT JOIN [emmats@washington.edu].[OA_immune_by_GO.csv]
ON [emmats@washington.edu].[unique_immune_GO_terms.txt].GO=[emmats@washington.edu].[OA_immune_by_GO.csv].[OA GO terms]
LEFT JOIN [emmats@washington.edu].[400MechS_immune_by_GO.csv]
ON [emmats@washington.edu].[unique_immune_GO_terms.txt].GO=[emmats@washington.edu].[400MechS_immune_by_GO.csv].[400MechS GO]
LEFT JOIN [emmats@washington.edu].[2800MechS_immune_by_GO.csv]
ON [emmats@washington.edu].[unique_immune_GO_terms.txt].GO=[emmats@washington.edu].[2800MechS_immune_by_GO.csv].[2800Mech GO terms]
February 10, 2014
Proteomics: focus on immune
Working with dataset of proteins that were originally subset based on GO terms related to the immune response. For each stress response (OA, Mech stress, OA + mech stress) protein sets were edited so that only proteins with a non-zero expression across all 8 oysters and at least a 2-fold change are included. In R, calculated number of proteins in each GO term for each stress response and average fold change for each GO term (by averaging fold change for all proteins included in that GO group).
February 6, 2014
Secondary stress: Glycogen
Recalculated glycogen content as µg glyc/mg tissue. For glycogen calculated as µg/µl multiplied by (200 µl/[mg glycogen used in extraction]. 200 µl is the volume in which the glycogen pellets were reconstituted. This correction made the means among the 3 treatments even more similar and an anova with pCO2 as a fixed factor yielded a p-value of 0.4.
Bioinformatics: iPiG
jimmy converted some of my files to mzIdentML. First file I tried was 103B_251_QE_02.pep.mzid (peptide spectrum matches file). ensembl genes table file = S. purpuratus from USCS (screenshot of download saved) - other option could be sea hare. Amino acid sequences = same entries for download as genes table except table = RefSeq genes. No uniprot ID mapping file is available for S. purpuratus so uploaded a blank txt file because I couldn't delete the file path that was already there. For FASTA file downloaded S. purpuratus peptides:
ftp://ftp.ensemblgenomes.org/pub/metazoa/release-21/fasta/strongylocentrotus_purpuratus/pep/
Tried running iPiG with all the files Jimmy converted but always got an error about DatabaseName not being complete or Duplicate unique value [] declared for identity constraint... I think I need to get rid of the uniprot ID-mapping file but I'm not sure how. If I don't change the file path, then it still doesn't work.
January 31, 2014
Bioinformatics: Module 3
Heard back from the iPiG developer and he pointed me in the direction of ProCon, which converts SEQUEST output into mxIdentML.
http://www.medizinisches-proteom-center.de/index.php/de/software-top/137-proteomics-conversion-tool-procon
I think I need to configure it first in command line (both generally and for sequest file conversion). Navigated to config file and ran:
./ProCon.properties MassSpecContactName=Emma MassSpecInstitution=UniversityofWashington MassSpecEmailPhoneFax=emmats@uw.edu DataSetContactName=Emma DataSetInstitution=UniversityofWashington DataSetEmailPhoneFax=emmats@uw.edu
got following error:
./ProCon.properties: line 1: E.: command not found
./ProCon.properties: line 2: Proteom-Center,: command not found
: No such file or directory: +49/234/32-22427
: command not found: line 4: Eisenacher
./ProCon.properties: line 5: Proteom-Center,: command not found
Following workflow for conversion of sequest outfiles to mzIdentML. For select folder with Sequest...selected a prot.xls file. Clicked parse SEQUEST out folder. Left default file (procon_mzIdentML.mzid) for output file and clicked export. Error: no Sequest import, export of mzIdentML only possible for Sequest out folder, but none imported. Hmmmm.....
Sam said to configure files manually. Opened Procon.properties in textwrangler and entered my contact info. Then opened log4j.properties and replaced \\ with
I've contacted Jimmy about the specific SEQUEST massvalues file. I also need to ask him about the sequest url and server name properties file.
January 29, 2014
Bioinformatics: Module 3
navigated to ipig folder in applications and ran graphical user interface: ./ipiggui
Jimmy sent me a sample mzIdentML from a mascot search (F003766.mzid)
Defaults for all other settings: genes table = knownGeneHuman.txt, amino acid sequences table = knownGenePep.txt, uniprot ID-mapping = HUMAN_9606_..., proteome fasta = HUMAN
Files downloaded following iPiG wiki instructions: http://sourceforge.net/p/ipig/wiki/Input%20Formats/.
January 28, 2014
Secondary stress: Glycogen
Redid samples from 1/25 that were too concentrated (diluted them 1:60 this time). There was not enough hydrolysis enzyme mix for the last replicate of 24, so it was only done in duplicate (the last being a sample blank control). Redid stats (ANOVA) and there is no difference among treatments. Below are means with 95% CI.
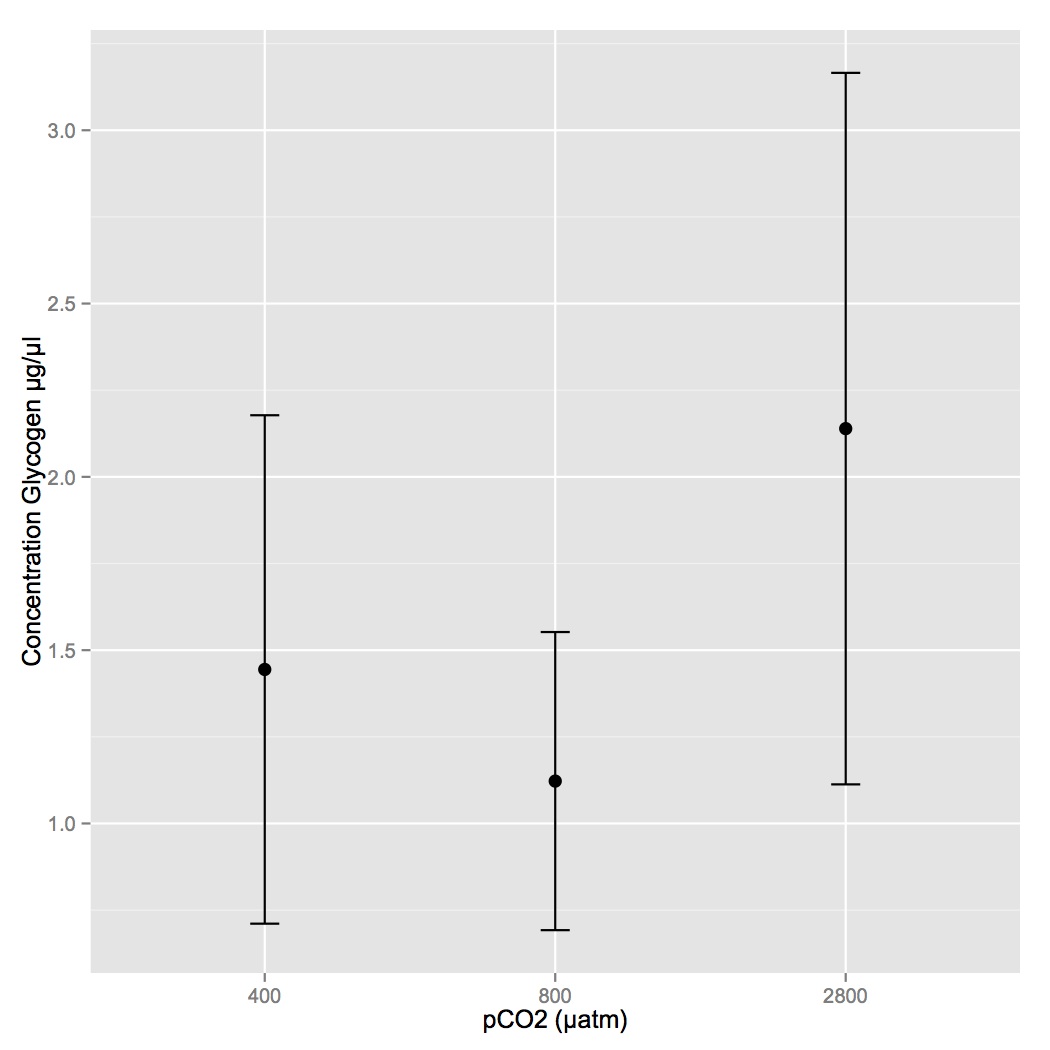
January 25, 2014
Secondary stress: Glycogen
Followed manufacturer's protocol for calculation of glycogen concentration (µg/µl) in oyster tissues. If the reaction turned brown for any of the oysters, the results were not included in the analysis (the concentration of the glycogen exceeded the limits of the reaction). The background was subtracted from each absorbance value. Coefficient of variation was <20% for all samples so all 3 replicates were included in averages.
For both plates, the standard curve was completely linear and the equation of the trendline was used to calculate glycogen concentration for each unknown sample. Samples concentrations were corrected for the 1:30 dilution and for the reaction volume.
There was no different in glycogen content among the 3 pCO2 treatment levels (400, 800, 2800 µatm). However, 4 of the samples that were too concentrated to measure at a 1:30 dilution were from the 400 µatm treatment and this may indicate that there was more glycogen content in the control treatment.
The following samples were excluded from analysis and will have to be rerun at a lower concentration: 3, 12, 15, 234, 24
January 24, 2014
Secondary stress: Glycogen
Did glycogen assay (sigma kit) on n = 8 samples from each of 3 pCO2 treatments (previously extracted by Sam) - 400, 800, and 2800 µatm. Samples were run in triplicate except for 0 standard and sample blanks. Sample blanks were a mixture of multiple samples to which no hydrolysis enzyme was added. All samples were diluted 1:30. I will have to redo a few samples at lower dilution because they maxed out the reaction (samples turned brown).
Bioinformatics: Module 3
making a .bed file from mass spec data
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0050246
January 23, 2014
Secondary stress: proteomics
SR did a blastp of oyster proteins against the mouse proteome to get a single species annotation (file is qdod_proteome_blastp in cnidarian). Make | into delimiters.
tr "|" "\t" </Volumes/web/cnidarian/qdod_proteome_blastp.txt /Volumes/web/oyster/proteomics/oyster_blastp_mouse
Uploaded dataset, kept only columns that are useful and renamed them:
SELECT [Column1] AS [CGI ID],
[Column3] AS [SPID],
[Column4] AS [Mouse Protein],
[Column13] AS [e-value]
FROM [emmats@washington.edu].[table_oyster_blastp_mouse]
Uploaded lists of differentially expressed proteins for each treatment. Joined to blastp output:
SELECT DISTINCT * FROM [emmats@washington.edu].[distinct oyster blastp mouse]
LEFT JOIN [emmats@washington.edu].[OA_CGIDs.txt]
ON [emmats@washington.edu].[distinct oyster blastp mouse].[CGI ID]=[emmats@washington.edu].[OA_CGIDs.txt].OA
LEFT JOIN [emmats@washington.edu].[400MechS_CGIDs.txt]
ON [emmats@washington.edu].[distinct oyster blastp mouse].[CGI ID]=[emmats@washington.edu].[400MechS_CGIDs.txt].[400MechS]
LEFT JOIN [emmats@washington.edu].[2800MechS_CGIDs.txt]
ON [emmats@washington.edu].[distinct oyster blastp mouse].[CGI ID]=[emmats@washington.edu].[2800MechS_CGIDs.txt].[2800MechS]
In Cytoscape, followed same steps as Jan 21 but did not use expression data as node attributes because this doesn't affect the layout (organic yfiles).
OA

mechanical stress at 400 µatm
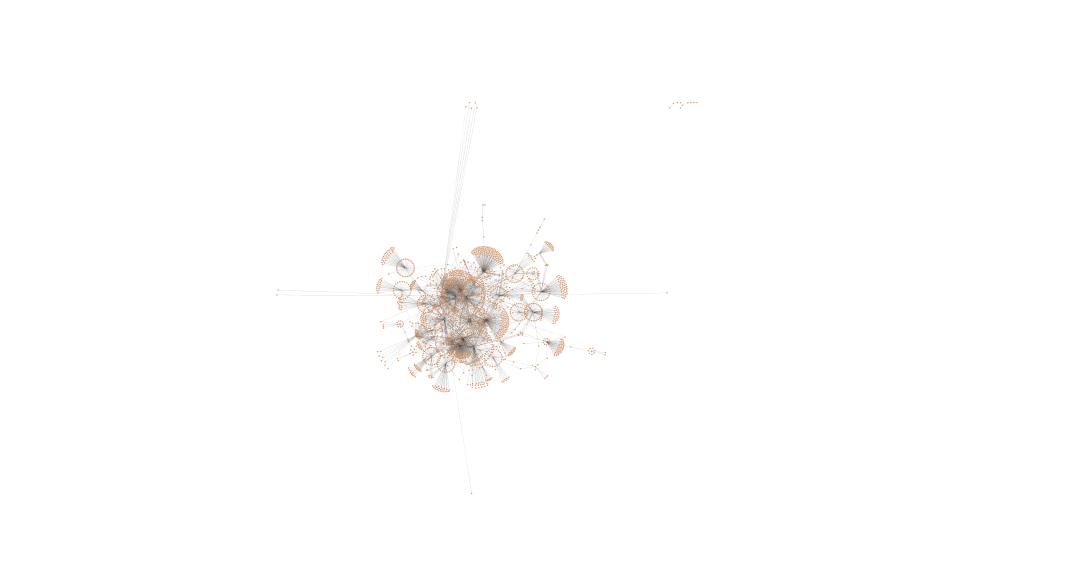
mechanical stress at 2800 µatm
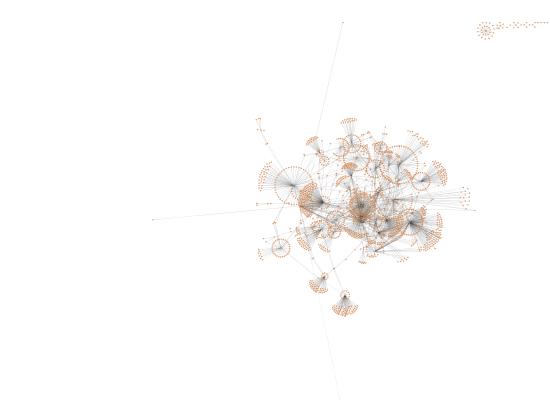
January 22, 2014
Secondary stress: Glycogen
Glycogen content assay using Sigma's MAK016 kit. Followed manufacturer's protocol for absorbance assay. Samples were run in duplicate. The 3 samples I extracted (3, 219, and 366) were also run at full concentration and diluted 1:2, 1:10, and 1:20 in water. After the master reaction mix was added, wells were mixed by pipetting up and down. I think this created too many bubbles and affected the replication for my plate read later.
Some of the samples maxed out the assay and it turned brown (instead of fuschia): All samples at full concentration and 1:2, sample 3 at 1:10. It also seems that for the first row (the standards) within each duplicate every other sample is lower than its partner. Mac votes "plate effect" and for the next plate I will avoid the external columns and rows.
I also think I will need to dilute the samples 1:30 in order to be within the range for the curve. I might add an extra standard on the high end of the curve to make sure.
January 21, 2014
Secondary stress: proteomics
Further exploration of possible protein-protein interaction network software. Navigator is a no-go due to limitations on annotations from multiple species. I've installed APID2NET v. 1.52 plugin in cytoscape, but it is only approved to work with an older version of cytoscape. APID seems perfect because it provides an option to find interactions between proteins from different species.
APID retrieval -> search list from file -> selected file OA for string (list of swissprot IDs for differentially expressed proteins in response to elevated pCO2)
in search filter dialogue box, checked "search interspecies protein..." and "search hypothetical protein...", connexion levels = 1, experimental methods = 1
in search list, selected find all. After results loaded, clicked Paint. APID Session -> save session -> OA APID
This seems to have worked! More to come....
In the NODE GO I can get a list of the frequencies of all GO terms represented in the network. I'm having trouble figuring out how to manipulate the network and actually zoom in to specific parts. Could be a versioning issue?
Imported list of differentially expressed swissprot IDs for response to mechanical stress and response to mechanical stress at elevated pCO2 and followed same steps as above.
Networks are here: https://www.evernote.com/shard/s242/sh/dec36fe0-46c0-4dad-815c-653ceed3aac4/7797ffb673aa690ea0f35afc1b765fd5
Downloaded cytoscape 3.0.2 and chose new network. Then import network from public databases.
data source: interaction database universal client
enter search conditions: pasted list of swissprot IDs for differentially expressed proteins in response to OA
search mode: search by ID (gene/protein/compound ID)
click "search"
selected string database
Made a node attributes file of proteins (swissprot IDs) and fold change between pCO2 levels. #DIV/0 were replaced with 100 (i.e. if a protein was expressed only at high pCO2 it is considered expressed 100-fold more than the 0 expression at low pCO2).
import -> table -> file -> OA node attributes
key column for network: shared name
import data as: node table columns
under show text file import options select that first row is column names
in show mapping options make sure column with node identifiers (SPIDs) is selected
layout -> yfiles layout -> organic (from the manual: The organic layout algorithm is a kind of spring-embedded algorithm that combines elements of the other algorithms to show the clustered structure of a graph)
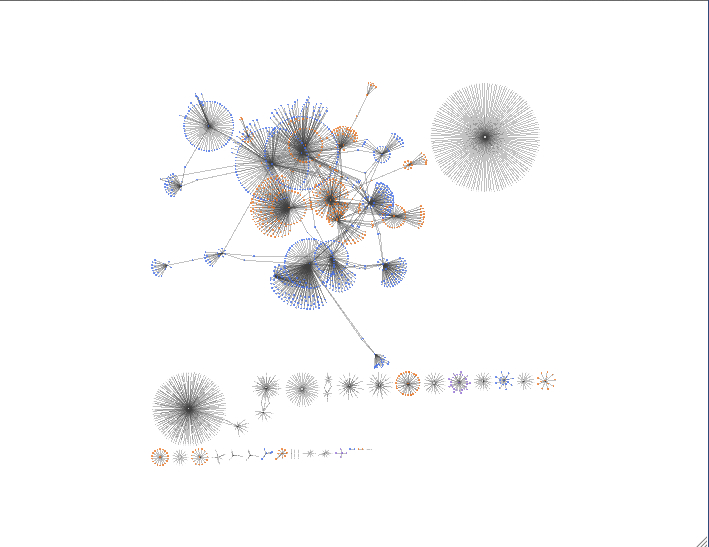
layouts can also be organized so that a shared attribute will be in its own circle. i did this for taxonomy of the annotation
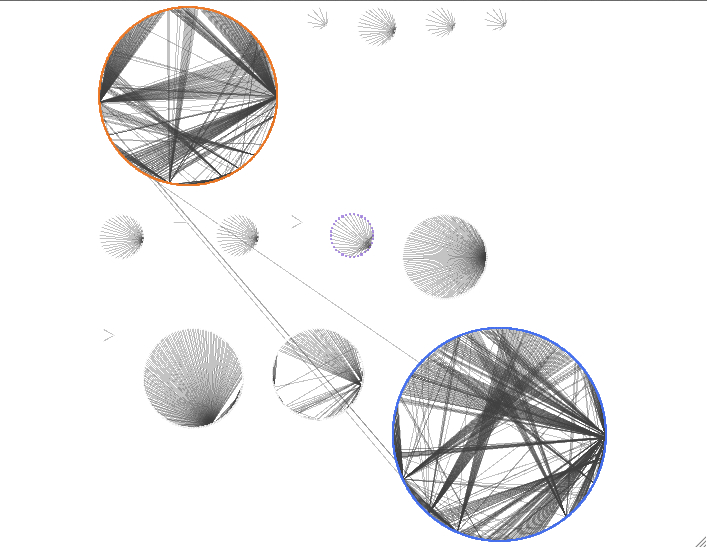
and for fold-change.
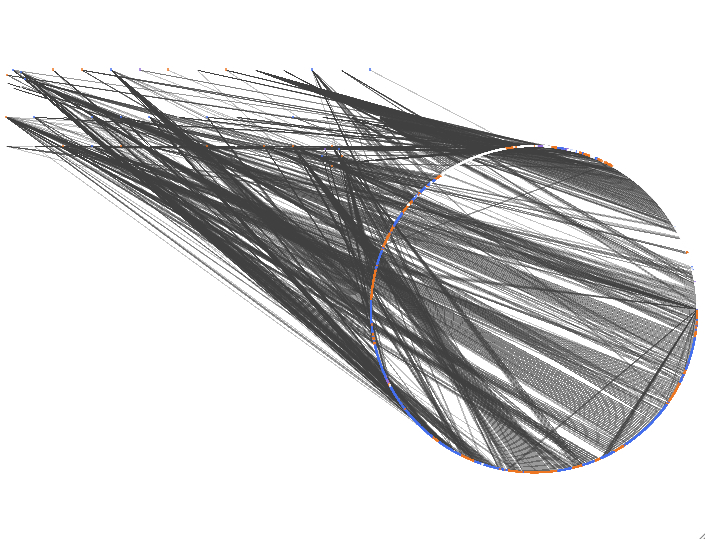
Adding gene ontology information: import -> ontology and annotation -> data type = node, annotation = gene association file for uniprot, ontology = gene ontology full -> import
After 37 minutes this still wasn't done and my computer was on the brink of crashing, so I canceled the import.
January 20, 2014
Bioinformatics: Module 2
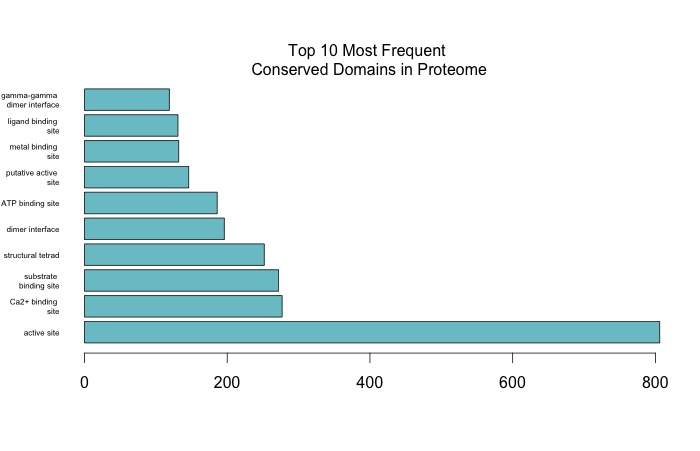
In RStudio made horizontal bar plots of top 10 and top 20 CDDs represented in proteome.
In SQL, subsetted annotated dataset and selected rows that only correspond to GO biological processes.
SELECT [CGI Number],[CDD annotation],[PSSM-ID],[feature description], [Gene Name], [term],[GOSlim_bin] FROM [emmats@washington.edu].[proteome CDD annotations, SPIDs, and GO slim]
WHERE [aspect]='P'

Make new column with numbers replacing GO Slim terms
SELECT [feature description], [GOSlim_bin],
CASE WHEN [GOSlim_bin]='cell adhesion' THEN 1
WHEN [GOSlim_bin]='cell cycle and proliferation' THEN 2
WHEN [GOSlim_bin]='cell organization and biogenesis' THEN 2
WHEN [GOSlim_bin]='cell-cell signaling' THEN 4
WHEN [GOSlim_bin]='death' THEN 5
WHEN [GOSlim_bin]='developmental processes' THEN 6
WHEN [GOSlim_bin]='DNA metabolism' THEN 7
WHEN [GOSlim_bin]='other biological processes' THEN 8
WHEN [GOSlim_bin]='other metabolic processes' THEN 9
WHEN [GOSlim_bin]='protein metabolism' THEN 10
WHEN [GOSlim_bin]='RNA metabolism' THEN 11
WHEN [GOSlim_bin]='signal transduction' THEN 12
WHEN [GOSlim_bin]='stress response' THEN 13
WHEN [GOSlim_bin]='transport' THEN 14
END
FROM [emmats@washington.edu].[proteome CDD bio processes]
Secondary Stress: proteomics
Exploring making protein-protein interaction networks.
On website for Mint (http://mint.bio.uniroma2.it/mint/Welcome.do) entered list of differentially expressed proteins for response to ocean acidification in search box (for connect proteins). Selected "only consider proteins in this list". This needs to be run in Safari due to non-compatibility between the new version of java and chrome. Everything seemed to work find except the visualization of the interaction wouldn't load because my security settings wouldn't let it (?). I tried to change the Java security settings but couldn't get it to work.
Navigator might also be interesting, but I have a feeling that it is very model-species centric (i.e will not accept lists of mixed species) - http://ophid.utoronto.ca/navigator/
APID is also worth exploring - http://bioinfow.dep.usal.es/apid/index.htm
January 18, 2014
Secondary stress: Glycogen
Samples from yesterday were spun at 4000xg for 30 minutes (4°C). Supernatant was removed and sample tubes were inverted for about 20 minutes to dry. 200 µl of nanopure water was added and samples were vortexed to dissolve glycogen pellets. Tubes were stored at -20°C.
January 17, 2014
Secondary stress: Glycogen
Extraction of 3 glycogen samples (same protocol that Sam used for all samples): 3, 219, and 366 from experiment 2. Samples were previously lyophilized and homogenized. Added 20-40 mg of oyster powder to 3 mL 15% trichloroacetic acid (15 g TCA powder + 100 mL Nanopure water). Vortexed well. Let incubate at 4°C for 1 hour.
| Sample |
Mass (mg) |
| 3 |
33.1 |
| 219 |
22.7 |
| 366 |
28 |
Bioinformatics: Module 2
still trying to remove gnl|CDD| from the file. I am running the command in the terminal (tr '|' "\t" </Volumes/web/oyster/bioinformatics/proteome_cdd_010813), but this just prints the correctly edited file in the terminal window. I would like to save a new file that I can then upload to SQL.
tr '|' "\tr" </Volumes/web/oyster/bioinformatics/proteome_cdd_010813> /Volumes/web/oyster/bioinformatics/proteome_cdd_sepnumb
uploaded to SQL and decreased file to just 3 columns, with new column names:
SELECT
Column1 AS [CGI number],
Column4 AS [CDD annotation],
Column13 AS [e-value]
FROM [emmats@washington.edu].[proteome_cdd_sepnumb]
Joined file with CDD annotations:
SELECT * FROM [emmats@washington.edu].[proteome CDD annot small file]
LEFT JOIN [emmats@washington.edu].[table_cddannot.txt]
ON [emmats@washington.edu].[proteome CDD annot small file].[CDD annotation]=[emmats@washington.edu].[table_cddannot.txt].[PSSM-ID]
Annotated with SPIDs and then with GO and GO Slim terms:
SELECT * FROM [emmats@washington.edu].[proteome CDD annot small file]
LEFT JOIN [emmats@washington.edu].[table_cddannot.txt]
ON [emmats@washington.edu].[proteome CDD annot small file].[CDD annotation]=[emmats@washington.edu].[table_cddannot.txt].[PSSM-ID]
LEFT JOIN [emmats@washington.edu].[table_TJGR_Gene_SPID_evalue_Description.txt]
ON [emmats@washington.edu].[proteome CDD annot small file].[CGI number]=[emmats@washington.edu].[table_TJGR_Gene_SPID_evalue_Description.txt].[CGI Protein]
SELECT * FROM [emmats@washington.edu].[proteome CDD annotations and SPIDs]
LEFT JOIN [dhalperi@washington.edu].[SPID_GOnumber.txt]
ON [emmats@washington.edu].[proteome CDD annotations and SPIDs].SPID=[dhalperi@washington.edu].[SPID_GOnumber.txt].A0A000
SELECT * FROM [emmats@washington.edu].[proteome CDD annotations, SPIDs, and GO]
LEFT JOIN [sr320@washington.edu].[GO_to_GOslim]
ON [emmats@washington.edu].[proteome CDD annotations, SPIDs, and GO].[GO:0003824]=[sr320@washington.edu].[GO_to_GOslim].GO_id
January 16, 2014
Secondary stress: Proteomics
Installed ClueGO v. 1.8 plugin in cytoscape to visualize differentially expressed protein data.
Imported list of differentially expressed proteins (in response to elevated pCO2) - this is just a list of uniprot IDs. The settings used for the analysis are here:
https://www.evernote.com/shard/s242/sh/16c1fb22-0ceb-4af8-8933-2d71ff7f65f6/23ff7c2a2ccea8527ed3da5cca32afa0
It apears that cluego ran, but I don't see a summary where I can click OK to view results. I wonder if this is because I picked Homo sapiens when I picked the gene cluster list. It seems that ClueGo only works with a single model species at a time (listed in dropdown menu). This is a bit limiting for my uses.
trying to remove gnl|CDD| from column 2 in blast output from 1/15/14 (in SQL)
UPDATE [emmats@washington.edu].[table_proteome_cdd_010813] SET [Column2] = REPLACE([Column2], 'gnl|CDD|', '')
January 15, 2014
Bioinformatics: Module 1
Reran deltablast with max target seqs = 5 to get multiple conserved domains per protein query. note: max_hsps_per_subject argument does not work with deltablast.
./deltablast -num_threads 8 -out /Users/Emma/Documents/cddblast/proteome_cdd_011513 -db /Users/Shared/Apps/ncbi-blast-2.2.29+/bin/cdd_delta -outfmt 6 -evalue 1E-10 -max_target_seqs 5 -query /Users/Emma/Documents/oyster.v9_90.fa.txt
error = Segmentation fault: 11
I'm not sure what this means but the output file is empty. I guess I won't get to see multiple conserved domains for my proteins :(
downloaded CDD annotations from here: http://www.ncbi.nlm.nih.gov/Ftp/
information on column names found here: http://www.biowebdb.org/cdd/README
Uploaded cddannot to sqlshare
January 14, 2014
Secondary stress: Proteomics
Using String v 9.1 to create a protein interaction network.
Uploaded file of differentially expressed (swissprot IDs, at least 2-fold) proteins in the OA response to string under the "multiple names" tab. Chose auto-detect for organism and for interactors chose proteins. This forced me to choose a single organism for the interaction network.
Repeated same steps as above except chose eukaryota as organism, however this still forces me to choose an organism on the next page. Tried again asking for COGs as interactor, this seemed to work.
Where I am now: I've downloaded the tab delim txt file from String and uploaded it as a protein interaction network into Cytoscape with column 1 as the source and column 2 as the target (based on this comment from a discussion board: If you download the "Text Summary" .txt file from STRING (instead of trying the "Graph Layout" .dat file), you can import it into Cytoscape using the table import function (File->Import->Network from Table (Test/MS Excel)...). The first two columns contain the interactions and the rest contain the weights of different interaction types from STRING. Unfortunately, the specific layout of the string network is not easy to import into Cytoscape right now, but the interactions are.). I would like to upload protein expression data as node attributes, but my network file is based on COGs and my protein expression is SPIDs. I'm having trouble finding a way to link COGs with SPIDs because it seem that NCBI doesn't maintain these files (here's the list of files I found: http://www.ncbi.nlm.nih.gov/COG/).
I've also uploaded just a list of proteins (swiss prot IDs) but since there are no interactions between the proteins nothing happens when I upload expression information and try to do a directed layout.
January 9, 2014
Bioinformatics: Module 1
Moved CDD database from Eagle to bin folder on local computer and reran code. It seems to be working this time.
./deltablast -num_threads 8 -out /Users/Emma/Documents/cddblast/proteome_cdd_010813 -db /Users/Shared/Apps/ncbi-blast-2.2.29+/bin/cdd_delta -outfmt 6 -evalue 1E-10 -max_target_seqs 1 -query /Volumes/web-1/oyster/oyster_v9_aa_format1.fasta
January 8, 2014
Bioinformatics: Module 1
blastp of oyster proteome against conserved domains database.
./blastp -num_threads 8 -out /Users/Emma/Documents/cddblast/proteome_cdd_010813 -db /Volumes/web-1/whale/blast/db/cdd_delta -outfmt 6 -evalue 1E-10 -max_target_seqs 1 -query /Users/Emma/Documents/oyster.v9_90.fa.txt
oops, wrong blast and wrong query file. Here is new code:
./deltablast -num_threads 8 -out /Users/Emma/Documents/cddblast/proteome_cdd_010813 -db /Volumes/web-1/whale/blast/db/cdd_delta -outfmt 6 -evalue 1E-10 -max_target_seqs 1 -query /Volumes/web-1/oyster/oyster_v9_aa_format1.fasta
but got following error:
BLAST Database error: No alias or index file found for protein database [cdd_delta] in search path [/Users/Shared/Apps/ncbi-blast-2.2.29+/bin::]