6/2/2014
Reverse Transcription (see 1.17.2014 for detailed method) of 2012 & 2013, July & Sept Field samples| Count |
Sample |
ng/ul |
ug/ul |
V (ul) of RNA = 1ug |
DEPC 0.1% (ul) |
Total Rxn |
| 1 |
2013_61 |
120.31 |
0.12031 |
8.31 |
9.4 |
17.75 |
| 2 |
2013_62 |
178.53 |
0.17853 |
5.60 |
12.1 |
17.75 |
| 3 |
2013_63 |
164.64 |
0.16464 |
6.07 |
11.7 |
17.75 |
| 4 |
2013_64 |
169.92 |
0.16992 |
5.89 |
11.9 |
17.75 |
| 5 |
2013_65 |
117.92 |
0.11792 |
8.48 |
9.3 |
17.75 |
| 6 |
2013_66 |
158.87 |
0.15887 |
6.29 |
11.5 |
17.75 |
| 7 |
2013_67 |
127.7 |
0.1277 |
7.83 |
9.9 |
17.75 |
| 8 |
2013_68 |
165.15 |
0.16515 |
6.06 |
11.7 |
17.75 |
| 9 |
2013_69 |
77.51 |
0.07751 |
12.90 |
4.8 |
17.75 |
| 10 |
2013_70 |
70.51 |
0.07051 |
14.18 |
3.6 |
17.75 |
| 11 |
2013_71 |
76.28 |
0.07628 |
13.11 |
4.6 |
17.75 |
| 12 |
2013_72 |
179.23 |
0.17923 |
5.58 |
12.2 |
17.75 |
| 13 |
2013_73 |
169.95 |
0.16995 |
5.88 |
11.9 |
17.75 |
| 14 |
2013_74 |
135.52 |
0.13552 |
7.38 |
10.4 |
17.75 |
| 15 |
2013_75 |
93.44 |
0.09344 |
10.70 |
7.0 |
17.75 |
| 16 |
2013_21 |
155.64 |
0.15564 |
6.43 |
11.3 |
17.75 |
| 17 |
2013_22 |
159.02 |
0.15902 |
6.29 |
11.5 |
17.75 |
| 18 |
2013_24 |
168.19 |
0.16819 |
5.95 |
11.8 |
17.75 |
| 19 |
2013_25 |
172.36 |
0.17236 |
5.80 |
11.9 |
17.75 |
| 20 |
2013_26 |
148.32 |
0.14832 |
6.74 |
11.0 |
17.75 |
| 21 |
2013_27 |
148.01 |
0.14801 |
6.76 |
11.0 |
17.75 |
| 22 |
2013_28 |
95.41 |
0.09541 |
10.48 |
7.3 |
17.75 |
| 23 |
2013_29 |
136.3 |
0.1363 |
7.34 |
10.4 |
17.75 |
| 24 |
2013_30 |
182.7 |
0.1827 |
5.47 |
12.3 |
17.75 |
| 25 |
2012_21 |
143.88 |
0.14388 |
6.95 |
10.8 |
17.75 |
| 26 |
2012_22 |
153.18 |
0.15318 |
6.53 |
11.2 |
17.75 |
| 27 |
2012_23 |
154.13 |
0.15413 |
6.49 |
11.3 |
17.75 |
| 28 |
2012_24 |
159.87 |
0.15987 |
6.26 |
11.5 |
17.75 |
| 29 |
2012_25 |
159.46 |
0.15946 |
6.27 |
11.5 |
17.75 |
| 30 |
2012_26 |
175.49 |
0.17549 |
5.70 |
12.1 |
17.75 |
| 31 |
2012_27 |
63.83 |
0.06383 |
15.67 |
2.1 |
17.75 |
| 32 |
2012_28 |
143.29 |
0.14329 |
6.98 |
10.8 |
17.75 |
| 33 |
2012_29 |
133.67 |
0.13367 |
7.48 |
10.3 |
17.75 |
| 34 |
2012_30 |
87.77 |
0.08777 |
11.39 |
6.4 |
17.75 |
| 35 |
2012_31 |
172.68 |
0.17268 |
5.79 |
12.0 |
17.75 |
| 36 |
2012_32 |
152.56 |
0.15256 |
6.55 |
11.2 |
17.75 |
| 37 |
2012_33 |
155.19 |
0.15519 |
6.44 |
11.3 |
17.75 |
| 38 |
2012_34 |
107.45 |
0.10745 |
9.31 |
8.4 |
17.75 |
| 39 |
2012_35 |
151.32 |
0.15132 |
6.61 |
11.1 |
17.75 |
| 40 |
2012_61 |
165.44 |
0.16544 |
6.04 |
11.7 |
17.75 |
| 41 |
2012_62 |
130.12 |
0.13012 |
7.69 |
10.1 |
17.75 |
| 42 |
2012_63 |
171.02 |
0.17102 |
5.85 |
11.9 |
17.75 |
| 43 |
2012_64 |
74.99 |
0.07499 |
13.34 |
4.4 |
17.75 |
| 44 |
2012_65 |
163.03 |
0.16303 |
6.13 |
11.6 |
17.75 |
| 45 |
2012_66 |
159.56 |
0.15956 |
6.27 |
11.5 |
17.75 |
| 46 |
2012_67 |
137.52 |
0.13752 |
7.27 |
10.5 |
17.75 |
| 47 |
2012_68 |
169.18 |
0.16918 |
5.91 |
11.8 |
17.75 |
| 48 |
2012_69 |
118.47 |
0.11847 |
8.44 |
9.3 |
17.75 |
| 49 |
2012_70 |
168.58 |
0.16858 |
5.93 |
11.8 |
17.75 |
| 50 |
2012_71 |
92.8 |
0.0928 |
10.78 |
7.0 |
17.75 |
| 51 |
2012_72 |
170.31 |
0.17031 |
5.87 |
11.9 |
17.75 |
| 52 |
2012_73 |
158.76 |
0.15876 |
6.30 |
11.5 |
17.75 |
| 53 |
2012_74 |
200.8 |
0.2008 |
4.98 |
12.8 |
17.75 |
| 54 |
2012_75 |
146.04 |
0.14604 |
6.85 |
10.9 |
17.75 |
5/29/2014
qPCR on purified RNA (20ul rnx) 2012 & 2013, July & Sept Field samples54 samples
1 NTC
1 gDNA
2 error
= 59 total rxns
gDNA template = 0.5ul
pureRNA template = 0.5ul
Master Mix EF-1a P5:
2x Sso Fast Evogreen 10 x 59 = 590ul
Forward primer 2-1 0.5 x 59 = 29.5ul
Reverse primer 2-1 0.5 x 59 = 29.5ul
H20 DEPC 0.1% 8.5 x 59 = 501.5ul
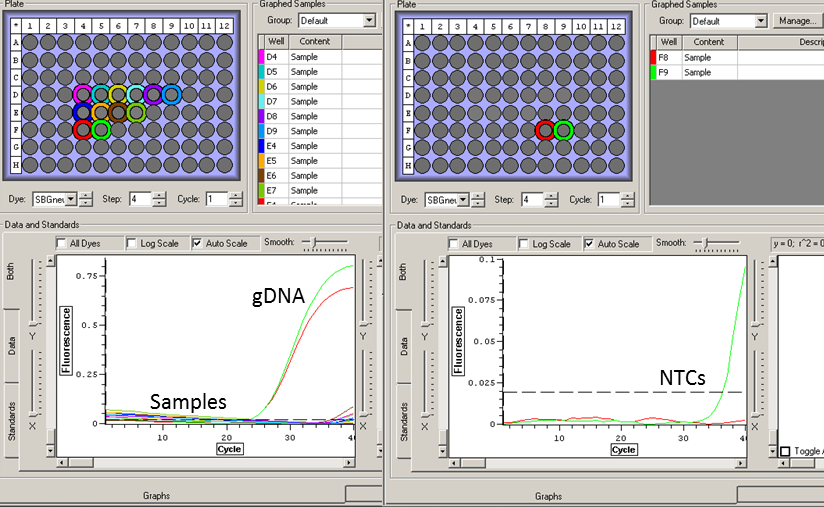
Results: Clean RNA
 |
| Amplification only in gDNA well (yellow) |
5/28/2014
Standard DNA-free protocol (see 1.16.2014 for details) on 2012 & 2013 July and September Field SamplesNanospec results:
| Sample ID |
ng/ul |
260/280 |
260/230 |
| 2013_61 |
120.31 |
1.92 |
0.94 |
| 2013_62 |
178.53 |
1.95 |
1.58 |
| 2013_63 |
164.64 |
1.94 |
1.22 |
| 2013_64 |
169.92 |
1.95 |
0.84 |
| 2013_65 |
117.92 |
1.92 |
1.04 |
| 2013_66 |
158.87 |
1.92 |
1.31 |
| 2013_67 |
127.7 |
1.92 |
1.39 |
| 2013_68 |
165.15 |
1.92 |
1.32 |
| 2013_69 |
77.51 |
1.85 |
0.83 |
| 2013_70 |
70.51 |
1.85 |
1.14 |
| 2013_71 |
76.28 |
1.86 |
1.09 |
| 2013_72 |
179.23 |
1.92 |
0.92 |
| 2013_73 |
169.95 |
1.95 |
1.67 |
| 2013_74 |
135.52 |
1.92 |
1.24 |
| 2013_75 |
93.44 |
1.89 |
1.00 |
| 2013_21 |
155.64 |
1.94 |
1.42 |
| 2013_22 |
159.02 |
1.89 |
0.99 |
| 2013_24 |
168.19 |
1.94 |
1.31 |
| 2013_25 |
172.36 |
1.94 |
1.26 |
| 2013_26 |
148.32 |
1.96 |
1.53 |
| 2013_27 |
148.01 |
1.93 |
1.52 |
| 2013_28 |
95.41 |
1.94 |
1.35 |
| 2013_29 |
136.3 |
2.00 |
1.64 |
| 2013_30 |
182.7 |
2.00 |
1.36 |
| 2012_21 |
143.88 |
1.92 |
1.45 |
| 2012_22 |
153.18 |
1.92 |
1.38 |
| 2012_23 |
154.13 |
1.93 |
1.23 |
| 2012_24 |
159.87 |
1.91 |
1.32 |
| 2012_25 |
159.46 |
1.92 |
1.41 |
| 2012_26 |
175.49 |
1.94 |
1.36 |
| 2012_27 |
63.83 |
1.79 |
0.53 |
| 2012_28 |
143.29 |
1.95 |
1.48 |
| 2012_29 |
133.67 |
1.94 |
1.53 |
| 2012_30 |
87.77 |
1.86 |
1.02 |
| 2012_31 |
172.68 |
1.91 |
0.92 |
| 2012_32 |
152.56 |
1.91 |
1.32 |
| 2012_33 |
155.19 |
1.93 |
1.28 |
| 2012_34 |
107.45 |
1.94 |
1.43 |
| 2012_35 |
151.32 |
1.93 |
1.29 |
| 2012_61 |
165.44 |
1.93 |
1.41 |
| 2012_62 |
130.12 |
1.9 |
1.32 |
| 2012_63 |
171.02 |
1.94 |
1.51 |
| 2012_64 |
74.99 |
1.86 |
0.99 |
| 2012_65 |
163.03 |
1.89 |
1.3 |
| 2012_66 |
159.56 |
1.91 |
1.34 |
| 2012_67 |
137.52 |
1.93 |
1.31 |
| 2012_68 |
169.18 |
1.93 |
1.52 |
| 2012_69 |
118.47 |
1.91 |
1.13 |
| 2012_70 |
168.58 |
1.93 |
1.39 |
| 2012_71 |
92.8 |
1.9 |
1.24 |
| 2012_72 |
170.31 |
1.94 |
1.48 |
| 2012_73 |
158.76 |
1.9 |
1.09 |
| 2012_74 |
200.8 |
1.94 |
1.52 |
| 2012_75 |
146.04 |
1.92 |
1.21 |
4/16-4/17/2014
RNA Extraction 2012 & 2013 Field samples (July & September) (see 1.15.2014 for detailed methods)
Nanodrop:| Sample |
ng/ul |
260/280 |
260/230 |
| 2013_61 |
168.79 |
1.84 |
1.36 |
| 2013_62 |
446.01 |
1.85 |
1.98 |
| 2013_63 |
402.04 |
1.91 |
1.57 |
| 2013_64 |
254.12 |
1.92 |
0.84 |
| 2013_65 |
167.86 |
1.81 |
1.69 |
| 2013_66 |
237.47 |
1.84 |
1.83 |
| 2013_67 |
178.7 |
1.8 |
1.85 |
| 2013_68 |
313.62 |
1.89 |
1.67 |
| 2013_69 |
109.11 |
1.73 |
1.55 |
| 2013_70 |
99.51 |
1.72 |
1.73 |
| 2013_71 |
106.83 |
1.71 |
1.74 |
| 2013_72 |
247.48 |
1.9 |
1.07 |
| 2013_73 |
1944.77 |
1.97 |
2.13 |
| 2013_74 |
312.59 |
1.88 |
1.85 |
| 2013_75 |
231.14 |
1.86 |
1.68 |
| 2013_21 |
366.7 |
1.86 |
1.87 |
| 2013_22 |
287.7 |
1.9 |
1.24 |
| 2013_24 |
315.46 |
1.93 |
1.13 |
| 2013_25 |
408.68 |
1.89 |
1.75 |
| 2013_26 |
756.4 |
1.97 |
2.03 |
| 2013_27 |
625.07 |
1.95 |
2.04 |
| 2013_28 |
827.09 |
1.97 |
2.02 |
| 2013_29 |
1359.92 |
2 |
2.19 |
| 2013_30 |
456.71 |
1.86 |
1.67 |
| 2012_21 |
444.88 |
1.85 |
1.95 |
| 2012_22 |
251.48 |
1.86 |
1.79 |
| 2012_23 |
293.23 |
1.9 |
1.11 |
| 2012_24 |
259.93 |
1.87 |
1.69 |
| 2012_25 |
298.81 |
1.87 |
1.82 |
| 2012_26 |
339.59 |
1.89 |
1.66 |
| 2012_27 |
113.63 |
1.77 |
0.79 |
| 2012_28 |
552 |
1.94 |
1.91 |
| 2012_29 |
563.57 |
1.95 |
2.06 |
| 2012_30 |
181.11 |
1.84 |
1.77 |
| 2012_31 |
245.64 |
1.93 |
0.57 |
| 2012_32 |
588.03 |
1.98 |
1.32 |
| 2012_33 |
204.57 |
1.9 |
0.78 |
| 2012_34 |
1329.69 |
2.01 |
2.14 |
| 2012_35 |
409.82 |
1.9 |
1.59 |
| 2012_61 |
311.98 |
1.86 |
1.9 |
| 2012_62 |
162.11 |
1.78 |
1.79 |
| 2012_63 |
289.12 |
1.89 |
1.53 |
| 2012_64 |
103.39 |
1.73 |
1.15 |
| 2012_65 |
263.36 |
1.86 |
1.8 |
| 2012_66 |
276.73 |
1.87 |
1.7 |
| 2012_67 |
181.5 |
1.82 |
1.74 |
| 2012_68 |
366.91 |
1.87 |
2.05 |
| 2012_69 |
158.96 |
1.78 |
1.5 |
| 2012_70 |
387.15 |
1.89 |
1.81 |
| 2012_71 |
132.72 |
1.75 |
1.8 |
| 2012_72 |
388.71 |
1.9 |
1.69 |
| 2012_73 |
312.09 |
1.9 |
1.54 |
| 2012_74 |
278.13 |
1.87 |
1.84 |
| 2012_74 |
203.24 |
1.83 |
1.55 |
| 2012_75 |
199.53 |
1.84 |
1.42 |
3/29/2014
qPCR - cDNA 2013 Field samples 98-99 & 2013 Field samples 99-100 (20ul rxn) HIF-1a & EF-1a
5 samples
5 duplicates
2 NTC
1 error
= 13 total rxns
cDNA template = 1ul
Master Mix HIF-1a P2-1:
2x Sso Fast Evogreen 10 x 13 = 130ul
Forward primer 2-1 0.5 x 13 = 6.5ul
Reverse primer 2-1 0.5 x 13 = 6.5ul
H20 DEPC 0.1% 8 x 13 = 104ul
Master Mix EF-1a P2:
2x Sso Fast Evogreen 10 x 13 = 130ul
Forward primer 2 0.5 x 13 = 6.5ul
Reverse primer 2 0.5 x 13 = 6.5ul
H20 DEPC 0.1% 8 x 13 = 104ul
qPCR - cDNA 2013 Field samples 1-43 & 44-97 (20ul rxn) HIF-1a & EF-1a
23 samples
23 duplicates
2 NTC
2 error
= 50 total rxns
cDNA template = 1ul
Master Mix HIF-1a P2-1:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2-1 0.5 x 50 = 25ul
Reverse primer 2-1 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
Master Mix EF-1a P2:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2 0.5 x 50 = 25ul
Reverse primer 2 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
3/31/2014
qPCR - cDNA 2012 Field samples 1-49 & 50-98(20ul rxn) HIF-1a & EF-1a23 samples
23 duplicates
2 NTC
2 error
= 50 total rxns
cDNA template = 1ul
Master Mix HIF-1a P2-1:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2-1 0.5 x 50 = 25ul
Reverse primer 2-1 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
Master Mix EF-1a P2:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2 0.5 x 50 = 25ul
Reverse primer 2 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
3/29/2014
Reverse Transcription
2013 Field samples3/28/2014
Reverse Transcription - see 1/17/2014 for method
2012 Field samples3/27/2014
qPCR DNased RNA - clean3/27/2014
Standard DNA-free treatment
2012 Field samples3/26/2014
Standard DNA-free treatment - see 1/16/2014 for method
2013 Field samples3/25/2014
RNA extraction (part 2)
2012 Field samples3/24/2014
RNA extraction (part 2) - see 1/15/2014 for method
2013 Field samples3/17/2014
RNA extraction (part 1)
Homogenized 2013 Field P. herring liver samples (n=48), stored -80C-Samples from June, August, and October
3/14/2014
RNA extraction (part 1)
Homogenized 2012 Field P. herring liver samples (n=48), stored -80C-Samples from June, August, and October
2/27/2014
qPCR - cDNA 'Time-course' samples 24-46 (20ul rxn) HIF-1a & EF-1a23 samples
23 duplicates
2 NTC
2 error
= 50 total rxns
cDNA template = 1ul
Master Mix HIF-1a P2-1:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2-1 0.5 x 50 = 25ul
Reverse primer 2-1 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
Master Mix EF-1a P2:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2 0.5 x 50 = 25ul
Reverse primer 2 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
qPCR - cDNA 'Time-course' samples 1-23 (20ul rxn) HIF-1a & EF-1a
23 samples
23 duplicates
2 NTC
2 error
= 50 total rxns
cDNA template = 1ul
Master Mix HIF-1a P2-1:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2-1 0.5 x 50 = 25ul
Reverse primer 2-1 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
Master Mix EF-1a P2:
2x Sso Fast Evogreen 10 x 50 = 500ul
Forward primer 2 0.5 x 50 = 25ul
Reverse primer 2 0.5 x 50 = 25ul
H20 DEPC 0.1% 8 x 50 = 400ul
2/26/2014
Reverse Transcription - TCL 101-144
44 samples
3 error
= 47 rxns
Mean amount = 1194.46ug/ul
Mean 260/280 = 1.71
Mean 260/230 = 1.65
2/25/2014
Reverse Transcription - TCL 51-100
50 samples
4 error
= 54 rxns
Mean amount = 1334.42ug/ul
Mean 260/280 = 1.69
Mean 260/230 = 1.74
2/19/2014
Reverse Transcription - TCL 1-50
50 samples
4 NTC
= 54 rxns
Mean amount = 1433ug/ul
Mean 260/280 = 1.69
Mean 260/230 = 1.69
2/18/2014
qPCR Time-course DNased RNA 51-144
94 samples
1 gDNA
1 NTC
= 96 rxns
Results: Clean (i.e., no amplification)
2/17/2014
qPCR Time-course DNased RNA 1-50
50 samples
1 gDNA
2 NTC
= 53 rxns
Results: Clean (i.e., no amplification)
2/13/2014
DNased RNA Time-course Sample 101-144
2/12/2014
DNased RNATime-course Sample 51-100
2/10/2014
Nandrop Time-course (Sample 1-50) DNased RNA:
| Sample ID |
ng/ul |
260/280 |
260/230 |
| TCL-1 |
286.71 |
1.97 |
1.43 |
| TCL-2 |
155.6 |
1.95 |
1.38 |
| TCL-3 |
153.5 |
1.96 |
1.01 |
| TCL-4 |
165.52 |
1.92 |
1.38 |
| TCL-5 |
163.01 |
1.97 |
1.42 |
| TCL-6 |
141.96 |
1.96 |
1.37 |
| TCL-7 |
90.09 |
1.89 |
1.13 |
| TCL-8 |
151.42 |
1.96 |
1.53 |
| TCL-9 |
158.92 |
1.95 |
1.37 |
| TCL-10 |
100.03 |
1.9 |
1.31 |
| TCL-11-2 |
181.95 |
1.83 |
1.03 |
| TCL-12 |
168.29 |
1.95 |
1.43 |
| TCL-13 |
103.82 |
1.92 |
1.37 |
| TCL-14 |
121.29 |
1.95 |
1.1 |
| TCL-15 |
150 |
1.94 |
0.62 |
| TCL-16 |
152.08 |
1.95 |
1.36 |
| TCL-17 |
106.98 |
1.9 |
1.31 |
| TCL-18 |
110.82 |
1.93 |
1.53 |
| TCL-19 |
165.86 |
1.95 |
1.43 |
| TCL-20 |
160.11 |
1.94 |
1.36 |
| TCL-21 |
190.23 |
1.96 |
1.65 |
| TCL-22 |
195.42 |
1.95 |
1.6 |
| TCL-23 |
129.87 |
1.93 |
1.17 |
| TCL-24 |
151.68 |
1.95 |
1.34 |
| TCL-25 |
153.89 |
1.91 |
1.54 |
| TCL-26 |
147.35 |
1.91 |
0.81 |
| TCL-27 |
160.08 |
1.96 |
1.41 |
| TCL-28 |
157.32 |
1.97 |
0.81 |
| TCL-29 |
90.66 |
1.93 |
1.43 |
| TCL-30 |
153.7 |
1.93 |
1.38 |
| TCL-31 |
117.77 |
1.91 |
0.97 |
| TCL-32 |
167.99 |
1.96 |
1.63 |
| TCL-33 |
134.84 |
1.9 |
1 |
| TCL-34-2 |
151.77 |
1.98 |
1.33 |
| TCL-35 |
109.33 |
1.91 |
1.31 |
| TCL-36 |
133.69 |
1.91 |
1.13 |
| TCL-37 |
88.72 |
1.91 |
1.36 |
| TCL-38 |
135.12 |
1.91 |
1.53 |
| TCL-39 |
106.91 |
1.94 |
1.36 |
| TCL-40 |
136.95 |
1.94 |
1.48 |
| TCL-41 |
161.95 |
1.96 |
1.58 |
| TCL-42 |
82.24 |
1.89 |
1.27 |
| TCL-43 |
121.64 |
1.96 |
1.55 |
| TCL-44 |
86.75 |
1.9 |
1.22 |
| TCL-45 |
111.54 |
1.93 |
1.23 |
| TCL-46 |
158.21 |
1.96 |
1.52 |
| TCL-47 |
107.82 |
1.9 |
1.23 |
| TCL-48 |
153.1 |
1.96 |
1.57 |
| TCL-49 |
226.39 |
1.97 |
1.23 |
| TCL-50 |
120.6 |
1.94 |
1.54 |
1/31/2014
Nandrop Time-course (Sample 51-100) RNA (pre-DNase):
| Sample |
260/280 |
260/230 |
ng/ul |
| TCL 51 |
1.96 |
1.94 |
707.7 |
| TCL 52 |
2.01 |
1.79 |
1022.5 |
| TCL 53 |
1.97 |
2.01 |
788.1 |
| TCL 54 |
1.95 |
1.82 |
636.2 |
| TCL 55 |
1.95 |
1.98 |
616.9 |
| TCL 56 |
1.84 |
2.06 |
435.5 |
| TCL 57 |
1.97 |
2.06 |
716.4 |
| TCL 58 |
1.86 |
1.71 |
285.9 |
| TCL 59 |
1.77 |
1.36 |
150.3 |
| TCL 60 |
1.98 |
1.95 |
763.7 |
| TCL 61 |
1.98 |
1.75 |
700.9 |
| TCL 62 |
2 |
2.07 |
1035.6 |
| TCL 63 |
1.86 |
2.02 |
305 |
| TCL 64 |
1.87 |
1.83 |
394.9 |
| TCL 65 |
1.98 |
1.95 |
788.2 |
| TCL 66 |
1.88 |
1.92 |
319.2 |
| TCL 67 |
1.71 |
1.79 |
115.6 |
| TCL 68 |
1.96 |
1.99 |
678.3 |
| TCL 69 |
1.99 |
1.93 |
1217 |
| TCL 70 |
2 |
2..16 |
1102.6 |
| TCL 71 |
1.97 |
2.02 |
773 |
| TCL 72 |
1.93 |
2.07 |
644.5 |
| TCL 73 |
2.01 |
2.11 |
1204.7 |
| TCL 74 |
1.85 |
1.97 |
442.6 |
| TCL 75 |
1.96 |
1.75 |
634.1 |
| TCL 76 |
1.95 |
2 |
752.7 |
| TCL 77 |
2 |
2.12 |
1381 |
| TCL 78 |
1.98 |
1.98 |
866.3 |
| TCL 79 |
1.78 |
1.88 |
419.6 |
| TCL 80 |
2 |
1.8 |
855.6 |
| TCL 81 |
1.87 |
1.62 |
436.1 |
| TCL 82 |
2 |
1.93 |
1236.8 |
| TCL 83 |
1.94 |
2.06 |
735.2 |
| TCL 84 |
2 |
1.89 |
118.2 |
| TCL 85 |
1.88 |
1.66 |
243 |
| TCL 86 |
2 |
1.96 |
1026.6 |
| TCL 87 |
2 |
2.12 |
1640.9 |
| TCL 88 |
1.88 |
1.78 |
413.6 |
| TCL 89 |
1.89 |
1.8 |
383.2 |
| TCL 90 |
1.77 |
1.36 |
139.3 |
| TCL 91 |
1.94 |
2.04 |
651.6 |
| TCL 92 |
2 |
2.19 |
1963.9 |
| TCL 93 |
1.97 |
2.17 |
833.7 |
| TCL 94 |
1.97 |
2.11 |
791.8 |
| TCL 95 |
2 |
2.11 |
1101.9 |
| TCL 96 |
1.99 |
1.93 |
916.6 |
| TCL 97 |
2 |
2.14 |
1104.8 |
| TCL 98 |
1.86 |
1.76 |
257.6 |
| TCL 99 |
1.98 |
2.17 |
850.2 |
| TCL 100 |
2 |
2.22 |
1758.3 |
1/30/2014
Nandrop Time-course (Sample 1-50) RNA (pre-DNase):
| Sample |
260/280 |
260/230 |
ng/ul |
| TCL 1 |
2.06 |
1.61 |
179.3 |
| TCL 2 |
1.87 |
1.84 |
258.3 |
| TCL 3 |
1.87 |
1.9 |
261.1 |
| TCL 4 |
1.85 |
1.9 |
253.3 |
| TCL 5 |
1.89 |
1.71 |
421.9 |
| TCL 6 |
1.98 |
1.63 |
612.8 |
| TCL 7 |
2 |
1.57 |
574.3 |
| TCL 8 |
1.86 |
1.99 |
460 |
| TCL 9 |
1.9 |
1.68 |
332.8 |
| TCL 10 |
2 |
1.97 |
846.9 |
| TCL 11 |
1.63 |
0.65 |
50.9 |
| TCL 12 |
1.89 |
1.8 |
353.9 |
| TCL 13 |
1.97 |
2.07 |
677.4 |
| TCL 14 |
2 |
1.48 |
688.3 |
| TCL 15 |
1.91 |
0.78 |
242.4 |
| TCL 16 |
1.9 |
1.88 |
357.9 |
| TCL 17 |
1.99 |
1.99 |
654.3 |
| TCL 18 |
1.88 |
2.26 |
922.9 |
| TCL 19 |
1.73 |
1.83 |
343.1 |
| TCL 20 |
1.7 |
1.92 |
234.1 |
| TCL 21 |
176 |
2.06 |
393.3 |
| TCL 22 |
1.72 |
1.99 |
343.2 |
| TCL 23 |
1.7 |
1.8 |
175.2 |
| TCL 24 |
1.71 |
1.81 |
264.7 |
| TCL 25 |
1.71 |
1.91 |
337.1 |
| TCL 26 |
1.68 |
1.83 |
197.5 |
| TCL 27 |
1.72 |
2.11 |
373.1 |
| TCL 28 |
1.72 |
1.93 |
406.2 |
| TCL 29 |
1.79 |
2.07 |
666.3 |
| TCL 30 |
1.69 |
2.06 |
228.5 |
| TCL 31 |
1.79 |
1.81 |
686.2 |
| TCL 32 |
1.83 |
2.13 |
859 |
| TCL 33 |
1.66 |
1.56 |
175.3 |
| TCL 34 |
1.61 |
8 |
88.4 |
| TCL 35 |
1.79 |
1.97 |
685.8 |
| TCL 36 |
1.7 |
1.91 |
262.1 |
| TCL 37 |
1.81 |
2.21 |
798.2 |
| TCL 38 |
1.68 |
2.1 |
458 |
| TCL 39 |
1.8 |
2.12 |
604.5 |
| TCL 40 |
1.69 |
2.11 |
263.8 |
| TCL 41 |
1.72 |
2.19 |
291.9 |
| TCL 42 |
1.82 |
2.04 |
755.8 |
| TCL 43 |
2 |
2.16 |
1141.5 |
| TCL 44 |
2 |
2.08 |
912.9 |
| TCL 45 |
1.78 |
1.72 |
145.7 |
| TCL 46 |
1.98 |
1.93 |
636.1 |
| TCL 47 |
1.99 |
1.95 |
796.2 |
| TCL 48 |
1.97 |
2.01 |
607 |
| TCL 49 |
1.89 |
1.27 |
456.6 |
| TCL 50 |
2 |
2.25 |
997.3 |
1/27 - 1/29/2014
RNA Extraction (Part 1) - Time-course P. herring liver tissue samples (TCL 1-144)
homogenized tissue samples stored -80C
1/22/2014
qPCR - cDNA 'Threshold' Reps 2 & 3 (20ul rxn) HIF-1a & EF-1a
Note: two rounds of qPCR were conducted separately for the HIF and EF genes
36 samples
36 duplicates
2 NTC
6 error
= 80 total rxns
cDNA template = 1ul
Master Mix:
2x Sso Fast Evogreen 10 x 80 = 800ul
Forward primer 2 0.5 x 80 = 40ul Used HIF-1a P2-1 & EF-1a P2
Reverse primer 2 0.5 x 80 = 40ul
H20 DEPC 0.1% 8 x 82 = 656ul
1/17/2014 & 1/21/2014
Reverse Transcription (Promega M-MLV Protocol)
RT the 'Threshold' lab samples (Replicates 2 & 3) to create cDNA
A single reaction volume = 25uL**. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA.
1) Calculate volume of RNA = 1ug of RNA:
| Sample |
ng/ul |
ug/ul |
V (ul) of 1ug of RNA |
DEPC 0.1% |
Total V (ul) |
| 31-H |
222.5 |
0.2225 |
4.5 |
13.3 |
17.75 |
| 32-H |
133.6 |
0.1336 |
7.5 |
10.3 |
17.75 |
| 33-H |
157.2 |
0.1572 |
6.4 |
11.4 |
17.75 |
| 34-H |
150.8 |
0.1508 |
6.6 |
11.1 |
17.75 |
| 35-H |
194.2 |
0.1942 |
5.1 |
12.6 |
17.75 |
| 36-H |
178.2 |
0.1782 |
5.6 |
12.1 |
17.75 |
| 31-M |
188.8 |
0.1888 |
5.3 |
12.5 |
17.75 |
| 32-M |
146.8 |
0.1468 |
6.8 |
10.9 |
17.75 |
| 33-M |
111.1 |
0.1111 |
9.0 |
8.7 |
17.75 |
| 34-M |
155 |
0.155 |
6.5 |
11.3 |
17.75 |
| 35-M |
137.9 |
0.1379 |
7.3 |
10.5 |
17.75 |
| 36-M |
87.1 |
0.0871 |
11.5 |
6.3 |
17.75 |
| 31-C |
136.7 |
0.1367 |
7.3 |
10.4 |
17.75 |
| 32-C |
157 |
0.157 |
6.4 |
11.4 |
17.75 |
| 33-C |
143.2 |
0.1432 |
7.0 |
10.8 |
17.75 |
| 34-C |
154.1 |
0.1541 |
6.5 |
11.3 |
17.75 |
| 35-C |
177.2 |
0.1772 |
5.6 |
12.1 |
17.75 |
| 36-C |
119.9 |
0.1199 |
8.3 |
9.4 |
17.75 |
| 49-H |
164.5 |
0.1645 |
6.1 |
11.7 |
17.75 |
| 50-H |
142.5 |
0.1425 |
7.0 |
10.7 |
17.75 |
| 51-H |
91.8 |
0.0918 |
10.9 |
6.9 |
17.75 |
| 52-H |
179.1 |
0.1791 |
5.6 |
12.2 |
17.75 |
| 53-H |
170 |
0.17 |
5.9 |
11.9 |
17.75 |
| 54-H |
134.8 |
0.1348 |
7.4 |
10.3 |
17.75 |
| 49-M |
167.6 |
0.1676 |
6.0 |
11.8 |
17.75 |
| 50-M |
134.1 |
0.1341 |
7.5 |
10.3 |
17.75 |
| 51-M |
154.1 |
0.1541 |
6.5 |
11.3 |
17.75 |
| 52-M |
186.1 |
0.1861 |
5.4 |
12.4 |
17.75 |
| 53-M |
181.3 |
0.1813 |
5.5 |
12.2 |
17.75 |
| 54-M |
165.3 |
0.1653 |
6.0 |
11.7 |
17.75 |
| 49-C |
156.9 |
0.1569 |
6.4 |
11.4 |
17.75 |
| 50-C |
240.6 |
0.2406 |
4.2 |
13.6 |
17.75 |
| 51-C |
134.2 |
0.1342 |
7.5 |
10.3 |
17.75 |
| 52-C |
166.5 |
0.1665 |
6.0 |
11.7 |
17.75 |
| 53-C |
247.6 |
0.2476 |
4.0 |
13.7 |
17.75 |
| 54-C |
153.5 |
0.1535 |
6.5 |
11.2 |
17.75 |
17.75ulRNA + 0.5ul Promega oligo dT = 18.25ul
4) Heat samples at 70C for 5 min in thermocycler.
5) Placed samples on ice IMMEDIATELY.
6) Made Master Mix (40 RXNs)
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer) x 40 = 200ul
1.25 uL 2.5mM dNTPs x 40 = 50ul
0.5 uL M-MLV RT per ug of RNA x 40 = 20ul
7) Mix well.
8) Added 6.75uL of master mix to each reaction.
9) Mix well, flicked lightly
10) Spot spun
11) Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12) Heat inactivate @ 95C for 3 min.
13) Spot spun.
1/16/2014
Standard DNA-free treatment:
1. Each RNA sample was diluted to equal 10ug of RNA for 50ul rxn in a 0.5mL tube:
2. 5ul of the TURBO DNase buffer to each sample
3. 1ul of TURBO DNase was added to each sample
4. Sample were incubated for 30min @ 37C
6. DNase Inactivation Reagent was resuspended (flicked)
7. 5ul of the Inactivation Reagent was added to each sample
8. Samples incubated for ~2min @ RT, each mixed/flicked an additional time to resuspend the reagent
9. Samples centrifuge at 10,000 x g for 1.5min
10. Supernatent was carefully pipetted to new (labeled) 0.5mL tubes.
| Sample |
ng/ul |
ug/ul |
Dilute = 10ug RNA = V ul of RNA |
DEPC 0.1% |
Total V (ul) |
Turbo Dnase Buffer (0.1 V ul) |
Turbo Dnase (ul) |
DNase Inactivetion Reagent (ul) |
| 31-H |
342 |
0.342 |
29.24 |
20.76 |
50.00 |
5.00 |
1.00 |
5 |
| 32-H |
308 |
0.308 |
32.47 |
17.53 |
50.00 |
5.00 |
1.00 |
5 |
| 33-H |
328.4 |
0.3284 |
30.45 |
19.55 |
50.00 |
5.00 |
1.00 |
5 |
| 34-H |
621.9 |
0.6219 |
16.08 |
33.92 |
50.00 |
5.00 |
1.00 |
5 |
| 35-H |
422.8 |
0.4228 |
23.65 |
26.35 |
50.00 |
5.00 |
1.00 |
5 |
| 36-H |
329.2 |
0.3292 |
30.38 |
19.62 |
50.00 |
5.00 |
1.00 |
5 |
| 31-M |
216.3 |
0.2163 |
46.23 |
3.77 |
50.00 |
5.00 |
1.00 |
5 |
| 32-M |
1104.3 |
1.1043 |
9.06 |
40.94 |
50.00 |
5.00 |
1.00 |
5 |
| 33-M |
145.5 |
0.1455 |
50.00 |
0.00 |
50.00 |
5.00 |
1.00 |
5 |
| 34-M |
113.6 |
0.1136 |
50.00 |
0.00 |
50.00 |
5.00 |
1.00 |
5 |
| 35-M |
792.8 |
0.7928 |
12.61 |
37.39 |
50.00 |
5.00 |
1.00 |
5 |
| 36-M |
137.6 |
0.1376 |
50.00 |
0.00 |
50.00 |
5.00 |
1.00 |
5 |
| 31-C |
1033.6 |
1.0336 |
9.67 |
40.33 |
50.00 |
5.00 |
1.00 |
5 |
| 32-C |
306.1 |
0.3061 |
32.67 |
17.33 |
50.00 |
5.00 |
1.00 |
5 |
| 33-C |
303.4 |
0.3034 |
32.96 |
17.04 |
50.00 |
5.00 |
1.00 |
5 |
| 34-C |
651.7 |
0.6517 |
15.34 |
34.66 |
50.00 |
5.00 |
1.00 |
5 |
| 35-C |
304.8 |
0.3048 |
32.81 |
17.19 |
50.00 |
5.00 |
1.00 |
5 |
| 36-C |
644.2 |
0.6442 |
15.52 |
34.48 |
50.00 |
5.00 |
1.00 |
5 |
| 49-H |
166.4 |
0.1664 |
50.00 |
0.00 |
50.00 |
5.00 |
1.00 |
5 |
| 50-H |
397.4 |
0.3974 |
25.16 |
24.84 |
50.00 |
5.00 |
1.00 |
5 |
| 51-H |
405.7 |
0.4057 |
24.65 |
25.35 |
50.00 |
5.00 |
1.00 |
5 |
| 52-H |
427.9 |
0.4279 |
23.37 |
26.63 |
50.00 |
5.00 |
1.00 |
5 |
| 53-H |
244.3 |
0.2443 |
40.93 |
9.07 |
50.00 |
5.00 |
1.00 |
5 |
| 54-H |
388.9 |
0.3889 |
25.71 |
24.29 |
50.00 |
5.00 |
1.00 |
5 |
| 49-M |
316.4 |
0.3164 |
31.61 |
18.39 |
50.00 |
5.00 |
1.00 |
5 |
| 50-M |
641.5 |
0.6415 |
15.59 |
34.41 |
50.00 |
5.00 |
1.00 |
5 |
| 51-M |
360.3 |
0.3603 |
27.75 |
22.25 |
50.00 |
5.00 |
1.00 |
5 |
| 52-M |
444.9 |
0.4449 |
22.48 |
27.52 |
50.00 |
5.00 |
1.00 |
5 |
| 53-M |
332.3 |
0.3323 |
30.09 |
19.91 |
50.00 |
5.00 |
1.00 |
5 |
| 54-M |
283.7 |
0.2837 |
35.25 |
14.75 |
50.00 |
5.00 |
1.00 |
5 |
| 49-C |
384.9 |
0.3849 |
25.98 |
24.02 |
50.00 |
5.00 |
1.00 |
5 |
| 50-C |
1020.7 |
1.0207 |
9.80 |
40.20 |
50.00 |
5.00 |
1.00 |
5 |
| 51-C |
649.6 |
0.6496 |
15.39 |
34.61 |
50.00 |
5.00 |
1.00 |
5 |
| 52-C |
282.8 |
0.2828 |
35.36 |
14.64 |
50.00 |
5.00 |
1.00 |
5 |
| 53-C |
217.1 |
0.2171 |
46.06 |
3.94 |
50.00 |
5.00 |
1.00 |
5 |
| 54-C |
374.1 |
0.3741 |
26.73 |
23.27 |
50.00 |
5.00 |
1.00 |
5 |
qPCR 'clean' RNA 'Threshold' Reps 1 & 4 (20ul rxn)
36 samples
3 NTC
1 gDNA
4 error
= 43 total rxns
RNA template = 0.5ul
gDNA template = 0.5ul
Master Mix: *Note: used EF-1a primer pair 1 that works but is not being used for normalization
2x Sso Fast Evogreen 10 x 43 = 430ul
Forward EF-1a p1 0.5 x 43 = 21.5ul
Reverse EF-1a p1 0.5 x 43 = 21.5ul
H20 DEPC 0.1% 8.5 x 43 = 365.5ul
RESULTS: 20140117_105648_Halley_RNA_1.17.2014.tad
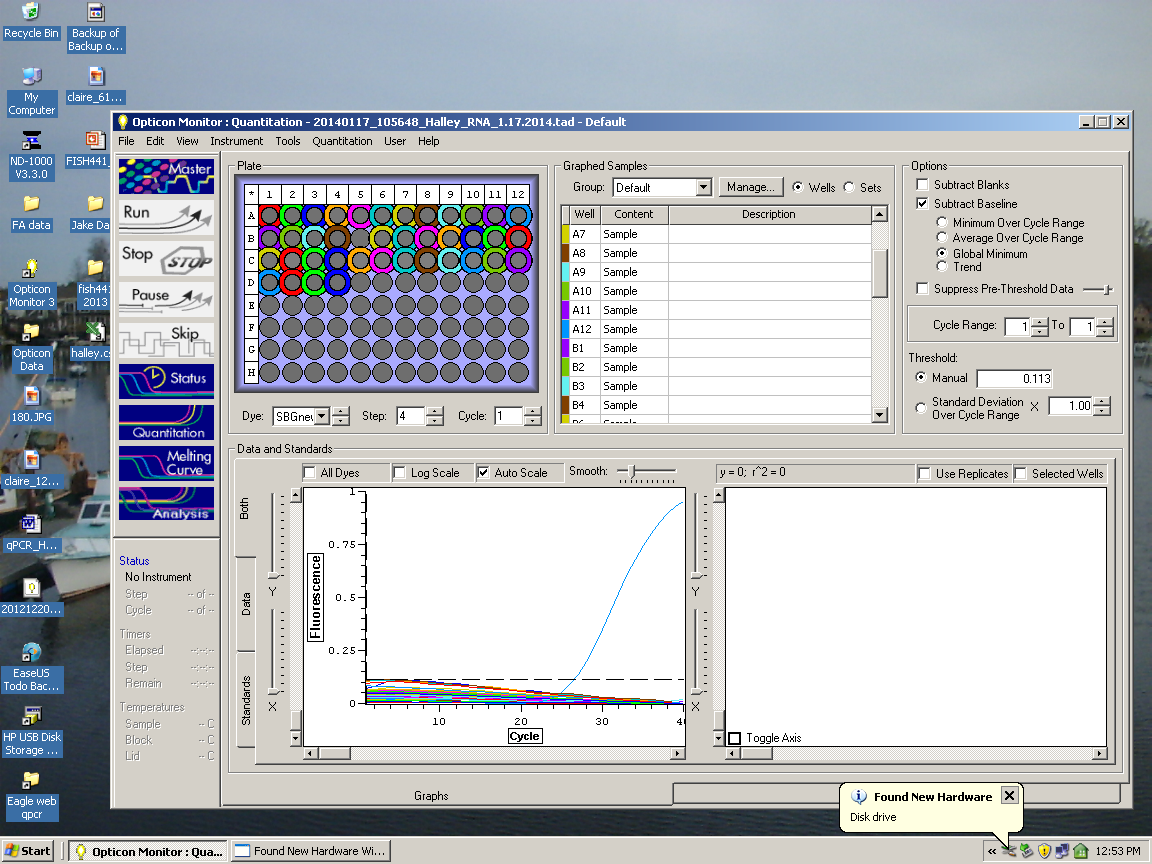
1/15/2014
RNA extraction continued (see 1.14.2014):
1) Tubes incubated at RT for 5min
2) Under fume hood 200ul of chloroform was added to each sample
3) Each sample vortexed vigorously for 30sec for 'milky' emulsion to occur
4) Incubated at RT for 5min
5) Tubes spun down for 15min, max speed, 4C
6) Tubes gently removed
7) Aqueous phase (top layer) carefully transferred to new 1.5ml tubes
8) 500ul of isopropanol added to new tube containing the RNA
9) Invert several time to mix
10) Incubated 10min at RT
11) Spun down max speed, 8min, 4C; tube hinge point up
12) Small white pellet present (RNA)
13) Supernatent removed
14) 1ml of 75% EtOH added to tube with pellet.
15) Vortex briefly to dislodge pellet
16) Spun down at 7500g for 5min
17) Carefully remove supernatent
18) Briefly spun down again (~15s) to pool residual EtOH
19) EtOH removed with P10 pipette
20) Tubes left open for no more than 5min
21) Pellets re-suspended with 100ul of 0.1% DEPC-H2O
22) Tubes incubated at 55C for 5min (waterbath)
23) Tubes flicked several times
24) Stored at -80C
1/14/2014
qPCR Normalizing gene (EF-1a) - cDNA 'Threshold' Reps 1 & 4 (20ul rxn)
36 samples
36 duplicates
3 NTC
8 error
= 82 total rxns
cDNA template = 1ul
Master Mix:
2x Sso Fast Evogreen 10 x 82 = 820ul
Forward 2-1 0.5 x 82 = 41ul
Reverse 2-1 0.5 x 82 = 41ul
H20 DEPC 0.1% 8 x 82 = 656ul
RESULTS: (20140114_113921_Halley_EF-1a(p2).tad)
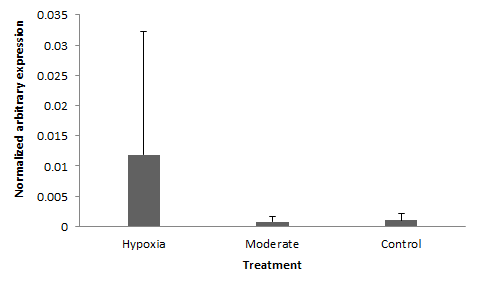 |
| Normalized (EF-1a) HIF-1a 'Threshold' Reps 1 & 4 |
RNA Extraction (TriReagent) of 'Threshold' experimental samples from MMFS: Replicates 2 & 3
Rep 2: 31-H to 36-H; 31-M to 36-M; 31-C to 36-C
Rep 3: 49-H to 54- H; 49-M to 54-M; 39-C to 54-C
1) 500ul of TriReagent added to each sample in a 1.5ml snap-cap tube
2) Each sample homogenized with sterile pestle
3) An additional 500ul added
4) Each sample vortexed vigorously for 15s
5) Stored at -80C
7/31/2013
cDNA nanodrop
| Sample |
ng/ul |
260/280 |
260/230 |
| 13-H |
222 |
1.76 |
1.95 |
| 14-H |
309.7 |
1.8 |
2 |
| 15-H |
221.4 |
1.78 |
2.03 |
| 16-H |
301.1 |
1.78 |
2.05 |
| 17-H |
415.7 |
1.78 |
2.02 |
| 18-H |
368.3 |
1.77 |
2.05 |
| 67-H |
346.3 |
1.39 |
1.49 |
| 68-H |
294.5 |
1.34 |
1.4 |
| 69-H |
283.9 |
1.4 |
1.51 |
| 70-H |
263.4 |
1.41 |
1.54 |
| 71-H |
332.3 |
1.33 |
1.3 |
| 72-H |
232.5 |
1.54 |
1.56 |
| 72.5-H |
308.2 |
1.35 |
1.45 |
| 13-M |
296 |
1.32 |
1.39 |
| 14-M |
290.8 |
1.32 |
1.41 |
| 15-M |
318.9 |
1.37 |
1.49 |
| 16-M |
309.1 |
1.33 |
1.35 |
| 17-M |
296.8 |
1.34 |
1.45 |
| 18-M |
286.9 |
1.33 |
1.44 |
| 67-M |
297.2 |
1.31 |
1.2 |
| 68-M |
294.3 |
1.31 |
1.44 |
| 69-M |
282.1 |
1.31 |
1.46 |
| 70-M |
310.1 |
1.3 |
1.28 |
| 71-M |
281.4 |
1.32 |
1.46 |
| 72-M |
326.1 |
1.33 |
1.25 |
| 13-C |
311 |
1.32 |
1.45 |
| 14-C |
311.5 |
1.31 |
1.44 |
| 15-C |
306.3 |
1.32 |
1.3 |
| 16-C |
291 |
1.3 |
1.47 |
| 17-C |
284.2 |
1.31 |
1.4 |
| 18-C |
286.3 |
1.31 |
1.46 |
| 67-C |
309.5 |
1.34 |
1.37 |
| 68-C |
316.6 |
1.32 |
1.27 |
| 69-C |
301.5 |
1.31 |
1.29 |
| 70-C |
309.8 |
1.35 |
1.45 |
| 71-C |
280.1 |
1.32 |
1.45 |
| 72-C |
254.8 |
1.3 |
2.1 |
qPCR - cDNA 'Threshold' Reps 1 & 4 (20ul rxn)
37 samples
37 duplicates
2 NTC
4 error
= 80 total rxns
cDNA template = 1ul
Master Mix:
2x Sso Fast Evogreen 10 x 80 = 800ul
Forward 2-1 0.5 x 80 = 40ul
Reverse 2-1 0.5 x 80 = 40ul
H20 DEPC 0.1% 8 x 80 = 640ul
Results: 20130730_151908_Halley_THRESHOLD_cDNA.tad
7/30/2013
Reverse Transcription (Promega M-MLV Protocol)
RT the 'Threshold' lab samples (Replicates 1 & 4) to create cDNA
A single reaction volume = 25uL. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA. You may need to make changes to accommodate your own conditions.
1) Calculate volume of RNA = 1ug of RNA:
| Sample |
ng/ul |
ug/ul |
V (ul) of RNA = 1ug |
DEPC 0.1% (ul) |
Total Rxn |
| 13-H |
133.4 |
0.1334 |
7.50 |
10.3 |
17.75 |
| 14-H |
113.4 |
0.1134 |
8.82 |
8.9 |
17.75 |
| 15-H |
147.4 |
0.1474 |
6.78 |
11.0 |
17.75 |
| 16-H |
157.4 |
0.1574 |
6.35 |
11.4 |
17.75 |
| 17-H |
133.1 |
0.1331 |
7.51 |
10.2 |
17.75 |
| 18-H |
145.5 |
0.1455 |
6.87 |
10.9 |
17.75 |
| 67-H |
103.9 |
0.1039 |
9.62 |
8.1 |
17.75 |
| 68-H |
140.2 |
0.1402 |
7.13 |
10.6 |
17.75 |
| 69-H |
108.3 |
0.1083 |
9.23 |
8.5 |
17.75 |
| 70-H |
89.7 |
0.0897 |
11.15 |
6.6 |
17.75 |
| 71-H |
136.4 |
0.1364 |
7.33 |
10.4 |
17.75 |
| 72-H |
187.2 |
0.1872 |
5.34 |
12.4 |
17.75 |
| 72.5-H |
132.2 |
0.1322 |
7.56 |
10.2 |
17.75 |
| 13-M |
132.3 |
0.1323 |
7.56 |
10.2 |
17.75 |
| 14-M |
142.1 |
0.1421 |
7.04 |
10.7 |
17.75 |
| 15-M |
148.7 |
0.1487 |
6.72 |
11.0 |
17.75 |
| 16-M |
144.1 |
0.1441 |
6.94 |
10.8 |
17.75 |
| 17-M |
167.7 |
0.1677 |
5.96 |
11.8 |
17.75 |
| 18-M |
174.4 |
0.1744 |
5.73 |
12.0 |
17.75 |
| 67-M |
155.7 |
0.1557 |
6.42 |
11.3 |
17.75 |
| 68-M |
97.9 |
0.0979 |
10.21 |
7.5 |
17.75 |
| 69-M |
175.4 |
0.1754 |
5.70 |
12.0 |
17.75 |
| 70-M |
201.5 |
0.2015 |
4.96 |
12.8 |
17.75 |
| 71-M |
157.1 |
0.1571 |
6.37 |
11.4 |
17.75 |
| 72-M |
152.4 |
0.1524 |
6.56 |
11.2 |
17.75 |
| 13-C |
139.3 |
0.1393 |
7.18 |
10.6 |
17.75 |
| 14-C |
135.1 |
0.1351 |
7.40 |
10.3 |
17.75 |
| 15-C |
140.4 |
0.1404 |
7.12 |
10.6 |
17.75 |
| 16-C |
152.5 |
0.1525 |
6.56 |
11.2 |
17.75 |
| 17-C |
169 |
0.169 |
5.92 |
11.8 |
17.75 |
| 18-C |
140.2 |
0.1402 |
7.13 |
10.6 |
17.75 |
| 67-C |
155.03 |
0.15503 |
6.45 |
11.3 |
17.75 |
| 68-C |
180 |
0.18 |
5.56 |
12.2 |
17.75 |
| 69-C |
146.8 |
0.1468 |
6.81 |
10.9 |
17.75 |
| 70-C |
150.5 |
0.1505 |
6.64 |
11.1 |
17.75 |
| 71-C |
141.7 |
0.1417 |
7.06 |
10.7 |
17.75 |
| 72-C |
163 |
0.163 |
6.13 |
11.6 |
17.75 |
3) Add appropriate amount of primer to sample. Use 0.25ug primer per 1ug of RNA in sample (= 0.5uL of Promega oligo dT in this example). Total volume (RNA + primers) should equal 18.25uL.
17.75ulRNA + 0.5ul Promega oligo dT = 18.25ul
4) Heat samples at 70C for 5 min in thermocycler.
5) Placed samples on ice IMMEDIATELY.
6) Made Master Mix (40 RXNs)
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer) x 40 = 200ul
1.25 uL 2.5mM dNTPs x 40 = 50ul
0.5 uL M-MLV RT per ug of RNA x 40 = 20ul
7) Mix well.
8) Added 6.75uL of master mix to each reaction.
9) Mix well, flicked lightly
10) Spot spun
11) Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12) Heat inactivate @ 95C for 3 min.
13) Spot spun.
14)Stored @ -20C.
7/26/2013
qPCR - DNased RNA 'Threshold' Reps 1 & 436 samples
1 gDNA
2 NTC
1 error
= 40 total rxns
Master Mix:
2x Sso Fast Evogreen 10 x 40 = 400ul
Forward 2-1 0.5 x 40 = 20ul
Reverse 2-1 0.5 x 40 = 20ul
H20 DEPC 0.1% 8.5 x 40 = 340ul
Results: RNA clean (no amplification)
7/25/2013
Nanodrop RNA:| Sample |
ng/ul |
260/280 |
260/230 |
| 13-H |
656.8 |
2.07 |
2.09 |
| 14-H |
787.3 |
2.02 |
2.33 |
| 15-H |
1021.7 |
2.05 |
2.07 |
| 16-H |
942.8 |
2.05 |
2.18 |
| 17-H |
1516.3 |
2.11 |
2.13 |
| 18-H |
660.8 |
2.05 |
1.87 |
| 67-H |
327.6 |
2 |
1.84 |
| 68-H |
367.7 |
2 |
1.82 |
| 69-H |
1526.9 |
2.1 |
2.23 |
| 70-H |
597 |
2.03 |
2.17 |
| 71-H |
565.2 |
2.08 |
1.47 |
| 72-H |
707.3 |
2.08 |
1.81 |
| 72.5-H |
576.7 |
2.05 |
1.79 |
| 13-M |
817.9 |
2.08 |
1.89 |
| 14-M |
588.4 |
2.06 |
1.68 |
| 15-M |
386.34 |
1.94 |
1.95 |
| 16-M |
210.2 |
1.9 |
2 |
| 17-M |
804.2 |
2.08 |
1.79 |
| 18-M |
381.3 |
1.98 |
1.92 |
| 67-M |
688.9 |
2.09 |
0.78 |
| 68-M |
948.9 |
2.08 |
2.14 |
| 69-M |
421.2 |
1.97 |
2.03 |
| 70-M |
926.3 |
2.07 |
2.13 |
| 71-M |
1042.2 |
2.07 |
2.02 |
| 72-M |
384 |
1.95 |
2.14 |
| 13-C |
730.4 |
2.05 |
1.95 |
| 14-C |
645.1 |
2.07 |
1.47 |
| 15-C |
841 |
2.04 |
2.02 |
| 16-C |
773.5 |
2.06 |
1.95 |
| 17-C |
455.8 |
1.96 |
1.91 |
| 18-C |
940.4 |
2.07 |
2.08 |
| 67-C |
1207 |
2.1 |
1.51 |
| 68-C |
459.7 |
1.97 |
1.9 |
| 69-C |
680.2 |
2.06 |
1.8 |
| 70-C |
809.2 |
2.07 |
1.71 |
| 71-C |
772.8 |
2.07 |
1.79 |
| 72-C |
293 |
197 |
1.96 |
1. Each RNA sample was diluted to equal 10ug of RNA for 50ul rxn in a 0.5mL tube:
2. 5ul of the TURBO DNase buffer to each sample
3. 1ul of TURBO DNase was added to each sample
4. Sample were incubated for 30min @ 37C
6. DNase Inactivation Reagent was resuspended (flicked)
7. 5ul of the Inactivation Reagent was added to each sample
8. Samples incubated for ~2min @ RT, each mixed/flicked an additional time to resuspend the reagent
9. Samples centrifuge at 10,000 x g for 1.5min
10. Supernatent was carefully pipetted to new (labeled) 0.5mL tubes.
| Sample |
ng/ul |
ug/ul |
Dilute = 10ug RNA = V ul of RNA |
DEPC 0.1% |
Total (ul) |
Turbo Dnase Buffer (ul) |
Turbo Dnase (ul) |
DNase Inactivetion Reagent (ul) |
| 13-H |
656.8 |
0.6568 |
15.23 |
34.77 |
50.00 |
5.00 |
1.00 |
5 |
| 14-H |
787.3 |
0.7873 |
12.70 |
37.30 |
50.00 |
5.00 |
1.00 |
5 |
| 15-H |
1021.7 |
1.0217 |
9.79 |
40.21 |
50.00 |
5.00 |
1.00 |
5 |
| 16-H |
942.8 |
0.9428 |
10.61 |
39.39 |
50.00 |
5.00 |
1.00 |
5 |
| 17-H |
1516.3 |
1.5163 |
6.60 |
43.40 |
50.00 |
5.00 |
1.00 |
5 |
| 18-H |
660.8 |
0.6608 |
15.13 |
34.87 |
50.00 |
5.00 |
1.00 |
5 |
| 67-H |
327.6 |
0.3276 |
30.53 |
19.47 |
50.00 |
5.00 |
1.00 |
5 |
| 68-H |
367.7 |
0.3677 |
27.20 |
22.80 |
50.00 |
5.00 |
1.00 |
5 |
| 69-H |
1526.9 |
1.5269 |
6.55 |
43.45 |
50.00 |
5.00 |
1.00 |
5 |
| 70-H |
597 |
0.597 |
16.75 |
33.25 |
50.00 |
5.00 |
1.00 |
5 |
| 71-H |
565.2 |
0.5652 |
17.69 |
32.31 |
50.00 |
5.00 |
1.00 |
5 |
| 72-H |
707.3 |
0.7073 |
14.14 |
35.86 |
50.00 |
5.00 |
1.00 |
5 |
| 72.5-H |
576.7 |
0.5767 |
17.34 |
32.66 |
50.00 |
5.00 |
1.00 |
5 |
| 13-M |
817.9 |
0.8179 |
12.23 |
37.77 |
50.00 |
5.00 |
1.00 |
5 |
| 14-M |
588.4 |
0.5884 |
17.00 |
33.00 |
50.00 |
5.00 |
1.00 |
5 |
| 15-M |
386.34 |
0.38634 |
25.88 |
24.12 |
50.00 |
5.00 |
1.00 |
5 |
| 16-M |
210.2 |
0.2102 |
47.57 |
2.43 |
50.00 |
5.00 |
1.00 |
5 |
| 17-M |
804.2 |
0.8042 |
12.43 |
37.57 |
50.00 |
5.00 |
1.00 |
5 |
| 18-M |
381.3 |
0.3813 |
26.23 |
23.77 |
50.00 |
5.00 |
1.00 |
5 |
| 67-M |
688.9 |
0.6889 |
14.52 |
35.48 |
50.00 |
5.00 |
1.00 |
5 |
| 68-M |
948.9 |
0.9489 |
10.54 |
39.46 |
50.00 |
5.00 |
1.00 |
5 |
| 69-M |
421.2 |
0.4212 |
23.74 |
26.26 |
50.00 |
5.00 |
1.00 |
5 |
| 70-M |
926.3 |
0.9263 |
10.80 |
39.20 |
50.00 |
5.00 |
1.00 |
5 |
| 71-M |
1042.2 |
1.0422 |
9.60 |
40.40 |
50.00 |
5.00 |
1.00 |
5 |
| 72-M |
384 |
0.384 |
26.04 |
23.96 |
50.00 |
5.00 |
1.00 |
5 |
| 13-C |
730.4 |
0.7304 |
13.69 |
36.31 |
50.00 |
5.00 |
1.00 |
5 |
| 14-C |
645.1 |
0.6451 |
15.50 |
34.50 |
50.00 |
5.00 |
1.00 |
5 |
| 15-C |
841 |
0.841 |
11.89 |
38.11 |
50.00 |
5.00 |
1.00 |
5 |
| 16-C |
773.5 |
0.7735 |
12.93 |
37.07 |
50.00 |
5.00 |
1.00 |
5 |
| 17-C |
455.8 |
0.4558 |
21.94 |
28.06 |
50.00 |
5.00 |
1.00 |
5 |
| 18-C |
940.4 |
0.9404 |
10.63 |
39.37 |
50.00 |
5.00 |
1.00 |
5 |
| 67-C |
1207 |
1.207 |
8.29 |
41.71 |
50.00 |
5.00 |
1.00 |
5 |
| 68-C |
459.7 |
0.4597 |
21.75 |
28.25 |
50.00 |
5.00 |
1.00 |
5 |
| 69-C |
680.2 |
0.6802 |
14.70 |
35.30 |
50.00 |
5.00 |
1.00 |
5 |
| 70-C |
809.2 |
0.8092 |
12.36 |
37.64 |
50.00 |
5.00 |
1.00 |
5 |
| 71-C |
772.8 |
0.7728 |
12.94 |
37.06 |
50.00 |
5.00 |
1.00 |
5 |
| 72-C |
293 |
0.293 |
34.13 |
15.87 |
50.00 |
5.00 |
1.00 |
5 |
"Clean" RNA was then spec'ed in the Nano-Drop:
| Sample |
ng/ul |
260/280 |
260/230 |
| 13-H |
133.4 |
2.01 |
1.53 |
| 14-H |
113.4 |
1.95 |
1.65 |
| 15-H |
147.4 |
1.93 |
1.67 |
| 16-H |
157.4 |
1.99 |
1.69 |
| 17-H |
133.1 |
1.97 |
1.66 |
| 18-H |
145.5 |
1.94 |
1.47 |
| 67-H |
103.9 |
1.96 |
1.31 |
| 68-H |
140.2 |
1.95 |
1.33 |
| 69-H |
108.3 |
2.01 |
1.61 |
| 70-H |
89.7 |
1.98 |
1.47 |
| 71-H |
136.4 |
1.94 |
1.35 |
| 72-H |
187.2 |
1.98 |
1.69 |
| 72.5-H |
132.2 |
1.97 |
1.44 |
| 13-M |
132.3 |
2 |
1.62 |
| 14-M |
142.1 |
1.97 |
1.45 |
| 15-M |
148.7 |
1.97 |
1.6 |
| 16-M |
144.1 |
1.95 |
1.54 |
| 17-M |
167.7 |
1.99 |
1.5 |
| 18-M |
174.4 |
1.96 |
1.49 |
| 67-M |
155.7 |
2.01 |
0.76 |
| 68-M |
97.9 |
1.98 |
1.46 |
| 69-M |
175.4 |
1.97 |
1.69 |
| 70-M |
201.5 |
1.98 |
1.66 |
| 71-M |
157.1 |
1.96 |
1.63 |
| 72-M |
152.4 |
1.94 |
1.6 |
| 13-C |
139.3 |
1.99 |
1.55 |
| 14-C |
135.1 |
1.99 |
1.44 |
| 15-C |
140.4 |
2 |
1.64 |
| 16-C |
152.5 |
1.98 |
1.54 |
| 17-C |
169 |
1.99 |
1.4 |
| 18-C |
140.2 |
1.98 |
1.63 |
| 67-C |
155.03 |
1.98 |
1.20 |
| 68-C |
180 |
1.99 |
1.64 |
| 69-C |
146.8 |
1.97 |
1.14 |
| 70-C |
150.5 |
1.98 |
1.62 |
| 71-C |
141.7 |
1.98 |
1.6 |
| 72-C |
163 |
1.99 |
1.56 |
RNA stored at -80C
7/24/2013
RNA extraction continued:
1) Tubes incubated at RT for 5min
2) Under fume hood 200ul of chloroform was added to each sample
3) Each sample vortexed vigorously for 30sec for 'milky' emulsion to occur
4) Incubated at RT for 5min
5) Tubes spun down for 15min, max speed, 4C
6) Tubes gently removed
7) Aqueous phase (top layer) carefully transferred to new 1.5ml tubes
8) 500ul of isopropanol added to new tube containing the RNA
9) Invert several time to mix
10) Incubated 10min at RT
11) Spun down max speed, 8min, 4C; tube hinge point up
12) Small white pellet present (RNA)
13) Supernatent removed
14) 1ml of 75% EtOH added to tube with pellet.
15) Vortex briefly to dislodge pellet
16) Spun down at 7500g for 5min
17) Carefully remove supernatent
18) Briefly spun down again (~15s) to pool residual EtOH
19) EtOH removed with P10 pipette
20) Tubes left open for no more than 5min
21) Pellets re-suspended with 100ul of 0.1% DEPC-H2O
22) Tubes incubated at 55C for 5min (waterbath)
23) Tubes flicked several times
24) Stored at -80C
7/23/2013
RNA Extraction (TriReagent) of experimental samples from MMFS: Replicates 1 & 4Rep 1: 13-H to 18-H; 13-M to 18-M; 13-C to 18-C
Rep 4: 67-H to 72.5- H; 67-M to 72-M; 67-C to 72-C
1) 500ul of TriReagent added to each sample in a 1.5ml snap-cap tube
2) Each sample homogenized with sterile pestle
3) An additional 500ul added
4) Each sample vortexed vigorously for 15s
5) Stored at -80C
6/14/2013
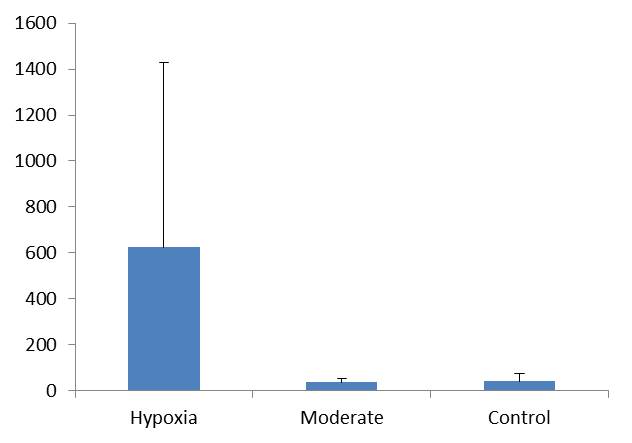
Preliminary graphs of HIF-1a gene expression from the first 20 samples: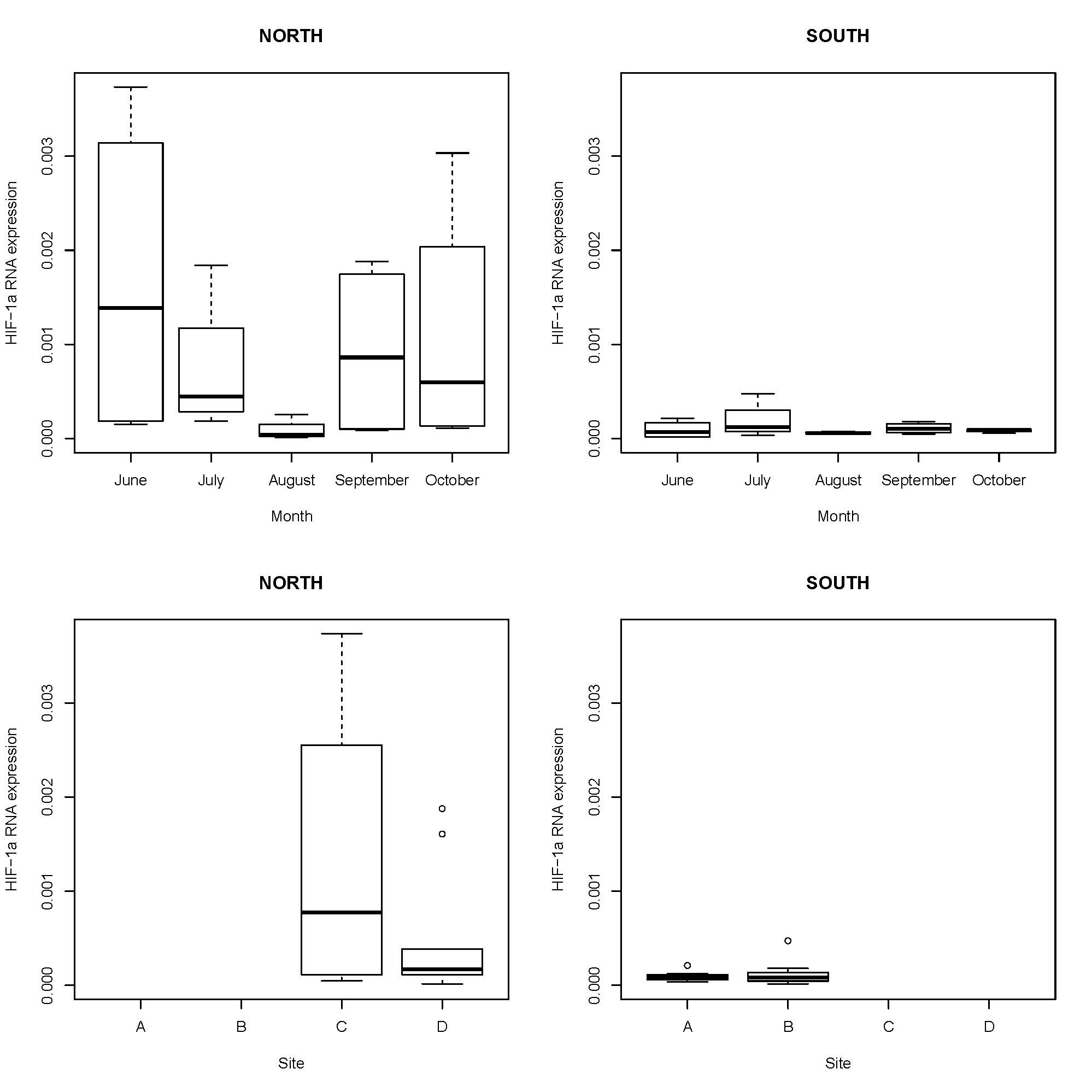 |
| HIF-1a mRNA expression range. Normalized with EF-1a. |
6/5/2013
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
qPCR on RT 20 cDNA field samples (6/7/2013):
RNA (20ul RXN)
Master Mix for HIF-1a:
Total # rxns = 20 samples+ 20 duplicates + 3 NTCs + 1 error = 44
2x Sso Fast EvaGreen 10 x 44 = 440ulHIF Forward Primer 2-1 0.5 x 44 = 22ul
HIF Reverse Primer 2-1 0.5 x 44 = 22ul
DEPC H20 8 x 44 =352ul
DEPC H20 =1ul
cDNA template = 1ul
Master Mix for EF-1a:
Total # rxns = 20 samples+ 20 duplicates + 3 NTCs + 1 error = 44
2x Sso Fast EvaGreen 10 x 44 = 440ulEF-1 Forward Primer 2 0.5 x 44 = 22ul
EF-1 Reverse Primer 2 0.5 x 44 = 22ul
DEPC H20 8 x 44 =352ul
DEPC H20 =1ul
cDNA template = 1ul
Plate:
Results: 20130611_111137_Halley_cDNA.tad
6/7/2013
Reverse Transcription (Promega M-MLV Protocol)
RT the 20 field samples to create cDNA
A single reaction volume = 25uL. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA. You may need to make changes to accommodate your own conditions.
1) Calculate volume of RNA = 1ug of RNA:
| Sample |
ng/ul |
ug/ul |
ul of RNA |
| 2 |
141.8 |
0.14 |
7.05 |
| 7 |
191.5 |
0.19 |
5.22 |
| 11 |
98.0 |
0.10 |
10.20 |
| 18 |
103.8 |
0.10 |
9.63 |
| 22 |
170.3 |
0.17 |
5.87 |
| 30 |
188.4 |
0.19 |
5.31 |
| 33 |
106.7 |
0.11 |
9.37 |
| 40 |
109.9 |
0.11 |
9.10 |
| 41 |
416.3 |
0.42 |
2.40 |
| 47 |
158.4 |
0.16 |
6.31 |
| 55 |
148.9 |
0.15 |
6.72 |
| 56 |
215.1 |
0.22 |
4.65 |
| 64 |
201.7 |
0.20 |
4.96 |
| 69 |
163.4 |
0.16 |
6.12 |
| 72 |
230.9 |
0.23 |
4.33 |
| 77 |
108.0 |
0.11 |
9.26 |
| 85 |
184.9 |
0.18 |
5.41 |
| 87 |
168.1 |
0.17 |
5.95 |
| 91 |
122.7 |
0.12 |
8.15 |
| 99 |
337.4 |
0.34 |
2.96 |
| Sample |
ul of RNA |
DEPC 0.1% |
| 2 |
7.05 |
10.7 |
| 7 |
5.22 |
12.5 |
| 11 |
10.20 |
7.5 |
| 18 |
9.63 |
8.1 |
| 22 |
5.87 |
11.9 |
| 30 |
5.31 |
12.4 |
| 33 |
9.37 |
8.4 |
| 40 |
9.10 |
8.7 |
| 41 |
2.40 |
15.3 |
| 47 |
6.31 |
11.4 |
| 55 |
6.72 |
11.0 |
| 56 |
4.65 |
13.1 |
| 64 |
4.96 |
12.8 |
| 69 |
6.12 |
11.6 |
| 72 |
4.33 |
13.4 |
| 77 |
9.26 |
8.5 |
| 85 |
5.41 |
12.3 |
| 87 |
5.95 |
11.8 |
| 91 |
8.15 |
9.6 |
| 99 |
2.96 |
14.8 |
17.75ulRNA + 0.5ul Promega oligo dT = 18.25ul
4) Heat samples at 70C for 5 min in thermocycler.
5) Placed samples on ice IMMEDIATELY.
6) Made Master Mix (21 RXNs)
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer) x 21 = 105ul
1.25 uL 2.5mM dNTPs x 21 = 26.3ul
0.5 uL M-MLV RT per ug of RNA x 21 = 10.5ul
7) Mix well.
8) Added 6.75uL of master mix to each reaction.
9) Mix well, but do not vortex (invert)
10) Spot spin.
11) Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12) Heat inactivate @ 95C for 3 min.
13) Spot spin.
14)Stored @ -20C.
6/5/2013
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)qPCR to get test 'clean' RNA samples compared to concentrated gDNA (4/10/2013):
RNA (20ul RXN)
Master Mix:
2x Sso Fast EvaGreen 10 x 38 = 380ul
Forward Primer 2 0.5 x 38 = 19ul
Reverse Primer 2 0.5 x 38 = 19ul
DEPC H20 8.5 x 38 =323ul
DEPC H20 = 0.5ul
gDNA template = 0.5ul
Total # rxns = 32sampls + 3 NTCs + 2 gDNA + 1 error = 38
Results: 20130605_113955_Halley_RNA.tad
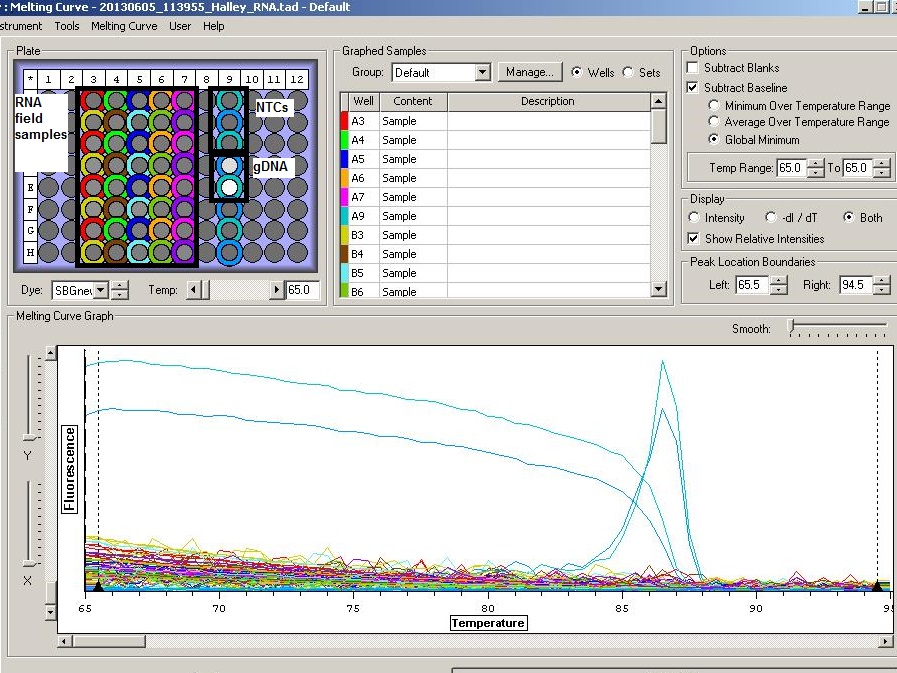
NTCs clean, no amplification of HIF for RNA samples. Proceed to reverse transcription of RNA.
5/29/2013
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
qPCR to get clean NTC compared to concentrated gDNA (4/10/2013):
RNA (20ul RXN)
Master Mix:
2x Sso Fast EvaGreen 10 x 7 = 70ul
Forward Primer 2 0.5 x 7 = 3.5ul
Reverse Primer 2 0.5 x 7 = 3.5ul
DEPC H20 8.5 x 7 =59.5ul
DEPC H20 = 0.5ul
gDNA template = 0.5ul
Total # rxns = 4 NTCs + 2 gDNA + 1 error = 7
Changed:
1) Bleached station
2) Used 1/3 fresh cut plate
3) Invert Sso Fast
4) Used same pipet tip to distribute master-mix
5) Used different pipet tip to distribute gDNA and NTC replicates
6) Wiped down top of plate (kim wipe) after placing in Bio-Rad
Results:
20130529_112511_Halley_NTC.tad
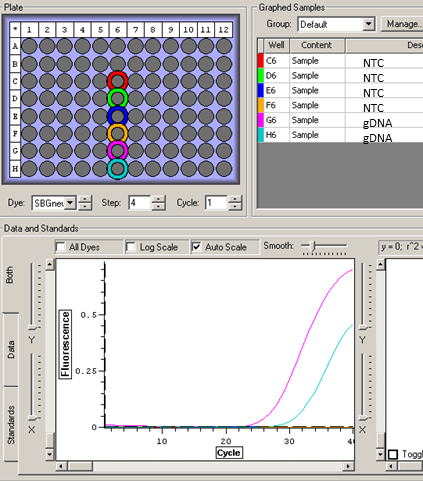
All NTCs clean!!!! - I will now run qPCR on a subsample of my RNA (5 out of 20 samples).
4/11/2013
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
qPCR on DNased RNA and newly concentrated gDNA (4/10/2013):
RNA (20ul RXN)
Master Mix:
2x Sso Fast EvaGreen 10 x 8 = 80ul
Forward Primer 2 0.5 x 8 = 4ul
Reverse Primer 2 0.5 x 8 = 4ul
H20 8.5 x 8 = 68ul
RNA template = 0.5ul
gDNA template = 0.5ul
Results: 20130411_HIF_NTC.tad (for full results)
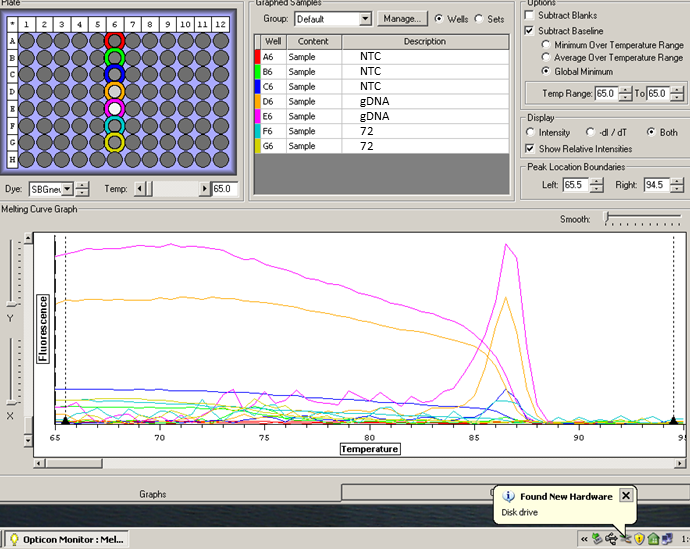
One replicate NTC (dark blue) and one replicate sample (#72; aqua) contaminated...otherwise, the remaining controls and samples look good.
4/10/2013
Ethanol precipitate of gDNA in order to concentrate
Made 25ml solution of 3M sodium acetateXg * 1mol/82.03g * 1/25ml * 1000ml/L = 3M
Xg = 6.15g of sodium acetate
1) 100ul of 'old' gDNA (see 12/5/2013 for DNA extraction)
2) Add 0.1 volumes (10ul) of 3M sodium acetate
3) incubate at -20C for at least 30min
4) pellet DNA at 16,000g for 15mins at 4C
5) wash pellet with 1ml of 70% ethanol
6) pellet DNA at 16,000g for 15 mins at 4C
7) discard supernatant
8) resuspend pellet (gently) in desired volume (50ul) of water/buffer
Nanospec
| gDNA ng/ul |
260/280 |
260/230 |
| 188.8 |
1.78 |
1.38 |
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
qPCR on DNased RNA and newly concentrated gDNA (above):
RNA (20ul RXN)
Master Mix:
2x Sso Fast EvaGreen 10 x 16 = 160ul
Forward Primer 2 0.5 x 16 = 8ul
Reverse Primer 2 0.5 x 16 = 8ul
H20 8.5 x 16 = 136ul
RNA template = 0.5ul
gDNA template = 0.5ul
Results: 20130410_HIF_RNA_sub.tad
Relative to the gDNA, the samples look relatively good...but, yet again, only one of my NTC controls is clean.
12/20/2012
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
qPCR on DNased RNA and new EF-1a primers:RNA (20ul RXN)
Master Mix:
2x Sso Fast EvaGreen 10 x 46 = 460ul
Forward Primer 2 0.5 x 46 = 23ul
Reverse Primer 2 0.5 x 46 = 23ul
H20 8.5 x 46 = 391ul
RNA template = 0.5ul
EF-1a Primer Pair 1 & Primer Pair 2
Two Seperate Master Mixes:
2x Sso Fast EvaGreen 10 x 7= 70ul
Forward Primer 2 0.5 x 7= 3.5ul
Reverse Primer 2 0.5 x 7= 3.5ul
H20 8 x 7= 56ul
cDNA-1 template = 1ul
RESULTS:
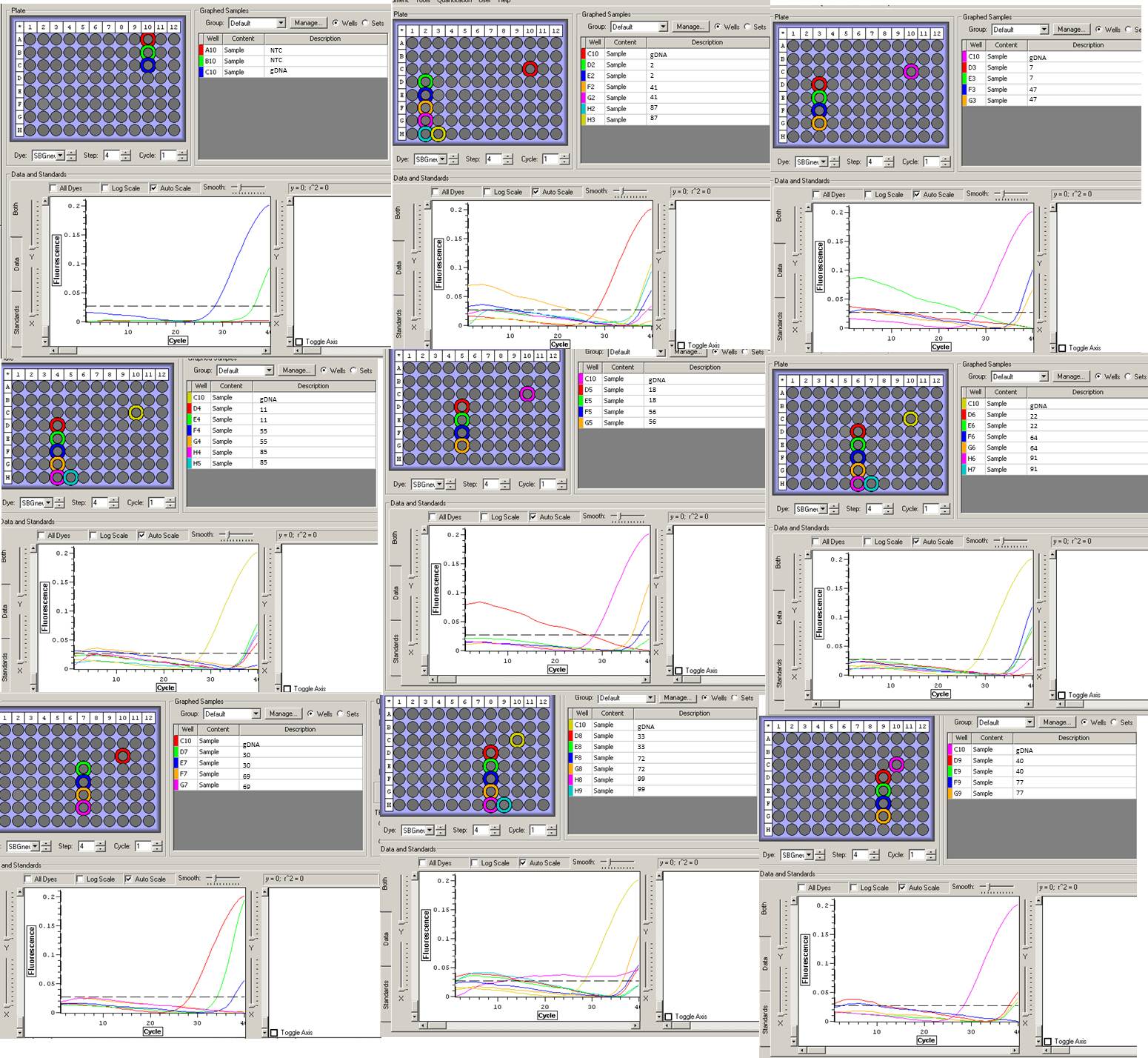 |
| RNA with HIF primer 2 |
One NTC was clean. Although most of the samples showed none or reduced amplification, some samples exhibited that early florescence...
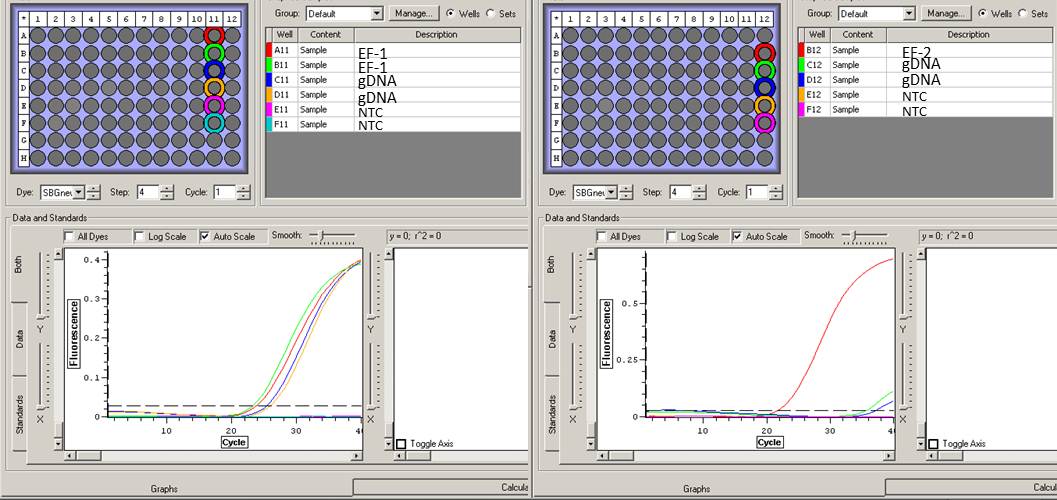 |
| EF-1a primers |
Both primers appear to work, however EF-2 had much more amplification.
12/19/2012
TURBO DNA-free Protocol
1. Each RNA sample was diluted to equal 10ug of RNA for 50ul rxn in a 0.5mL tube:| Sample |
ug/ul |
Dilution for 10ug of RNA |
0.1% DEPC |
| 2 |
1.18 |
8.50 |
41.5 |
| 7 |
1.14 |
8.78 |
41.2 |
| 11 |
1.27 |
7.89 |
42.1 |
| 18 |
0.94 |
10.68 |
39.3 |
| 22 |
0.78 |
12.77 |
37.2 |
| 30 |
0.78 |
12.78 |
37.2 |
| 33 |
1.19 |
8.41 |
41.6 |
| 40 |
0.74 |
13.53 |
36.5 |
| 41 |
1.26 |
7.94 |
42.1 |
| 47 |
1.37 |
7.31 |
42.7 |
| 55 |
0.85 |
11.81 |
38.2 |
| 56 |
1.39 |
7.21 |
42.8 |
| 64 |
0.26 |
38.25 |
11.8 |
| 69 |
0.41 |
24.39 |
25.6 |
| 72 |
1.47 |
6.80 |
43.2 |
| 77 |
2.44 |
4.09 |
45.9 |
| 87 |
1.07 |
9.37 |
40.6 |
| 85 |
1.75 |
5.72 |
44.3 |
| 91 |
1.53 |
6.54 |
43.5 |
| 99 |
1.49 |
6.70 |
43.3 |
2. 5ul of the TURBO DNase buffer to each sample
3. 0.5ul of TURBO DNase was added to each sample
4. Sample were incubated for 30min @ 37C
5. Remaining 0.5ul of TURBO DNase was added to each sample (for a total of 1ul of DNase enzyme)
6. DNase Inactivation Reagent was resuspended (flicked)
7. 5ul of the Inactivation Reagent was added to each sample
8. Samples incubated for ~2min @ RT, each mixed/flicked an additional time to resuspend the reagent
9. Samples centrifuge at 10,000 x g for 1.5min
10. Supernatent was carefully pipetted to new (labeled) 0.5mL tubes.
"Clean" RNA was then spec'ed in the Nano-Drop:
| Sample |
ng/ul |
260/280 |
260/230 |
| 2 |
141.8 |
1.96 |
1.09 |
| 7 |
191.5 |
1.97 |
1.00 |
| 11 |
98.0 |
1.95 |
1.29 |
| 18 |
103.8 |
1.97 |
0.77 |
| 22 |
170.3 |
1.97 |
1.29 |
| 30 |
188.4 |
1.95 |
0.78 |
| 33 |
106.7 |
1.93 |
1.29 |
| 40 |
109.9 |
1.98 |
1.10 |
| 41 |
416.3 |
1.95 |
1.56 |
| 47 |
158.4 |
1.97 |
1.51 |
| 55 |
148.9 |
1.97 |
1.31 |
| 56 |
215.1 |
1.99 |
1.45 |
| 64 |
201.7 |
1.98 |
1.52 |
| 69 |
163.4 |
2.00 |
1.46 |
| 72 |
230.9 |
1.99 |
1.54 |
| 77 |
108.0 |
1.90 |
1.11 |
| 85 |
184.9 |
1.98 |
1.28 |
| 87 |
168.1 |
1.97 |
1.18 |
| 91 |
122.7 |
1.94 |
1.29 |
| 99 |
337.4 |
1.97 |
1.49 |
12/11/2012
The ConsensusfromContig6914 (Q92005) Elongation factor 1-alpha for Pacific herring (Herring Hepatic Transcriptome) was BLASTed in NCBI to evaluate the most conserved region. Primer BLAST then was used on the contig sequence and two primers were selected and order from IDT: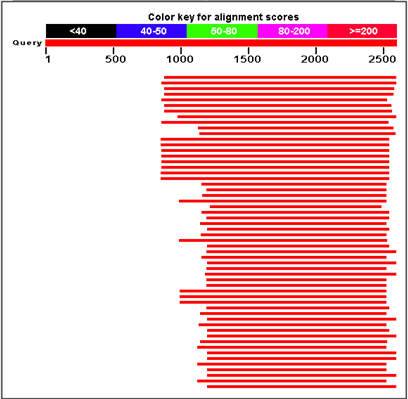 |
| EF-1a BLAST results |
Primer pair 1
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
CTCCGCATTTGTAGATGAGA |
Plus |
20 |
2414 |
2433 |
54.98 |
45.00 |
4.00 |
2.00 |
| Reverse primer |
CTTAAGCAATCATGGGCAAG |
Minus |
20 |
2521 |
2502 |
55.00 |
45.00 |
6.00 |
2.00 |
| Product length |
108 |
||||||||
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
AGAGCAATGTCAATGGTGAT |
Plus |
20 |
2281 |
2300 |
55.00 |
40.00 |
5.00 |
2.00 |
| Reverse primer |
TCTCATCTACAAATGCGGAG |
Minus |
20 |
2433 |
2414 |
54.98 |
45.00 |
4.00 |
2.00 |
| Product length |
153 |
||||||||
12/6/2012
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix)
"Test" qPCR for primers and corrected cDNA and gDNA:No. of RXNs
2 cDNA-1
2 cDNA-2
2 gDNA
2 NTCs
1 pipette error
=
9 RXNs
Master Mix:
2x Sso Fast EvaGreen 10 x 9 = 90ul
Forward Primer 2 0.5 x 9 = 4.5ul
Reverse Primer 2 0.5 x 9 = 4.5ul
H20 8 x 9 = 72ul
cDNA template = 1ul (**instead of 2ul)
Order (place in columns 6 & 7):
S
---
cDNA-1
cDNA-1
cDNA-2
cDNA-2
gDNA
gDNA
N
---
NTC-1
NTC-2
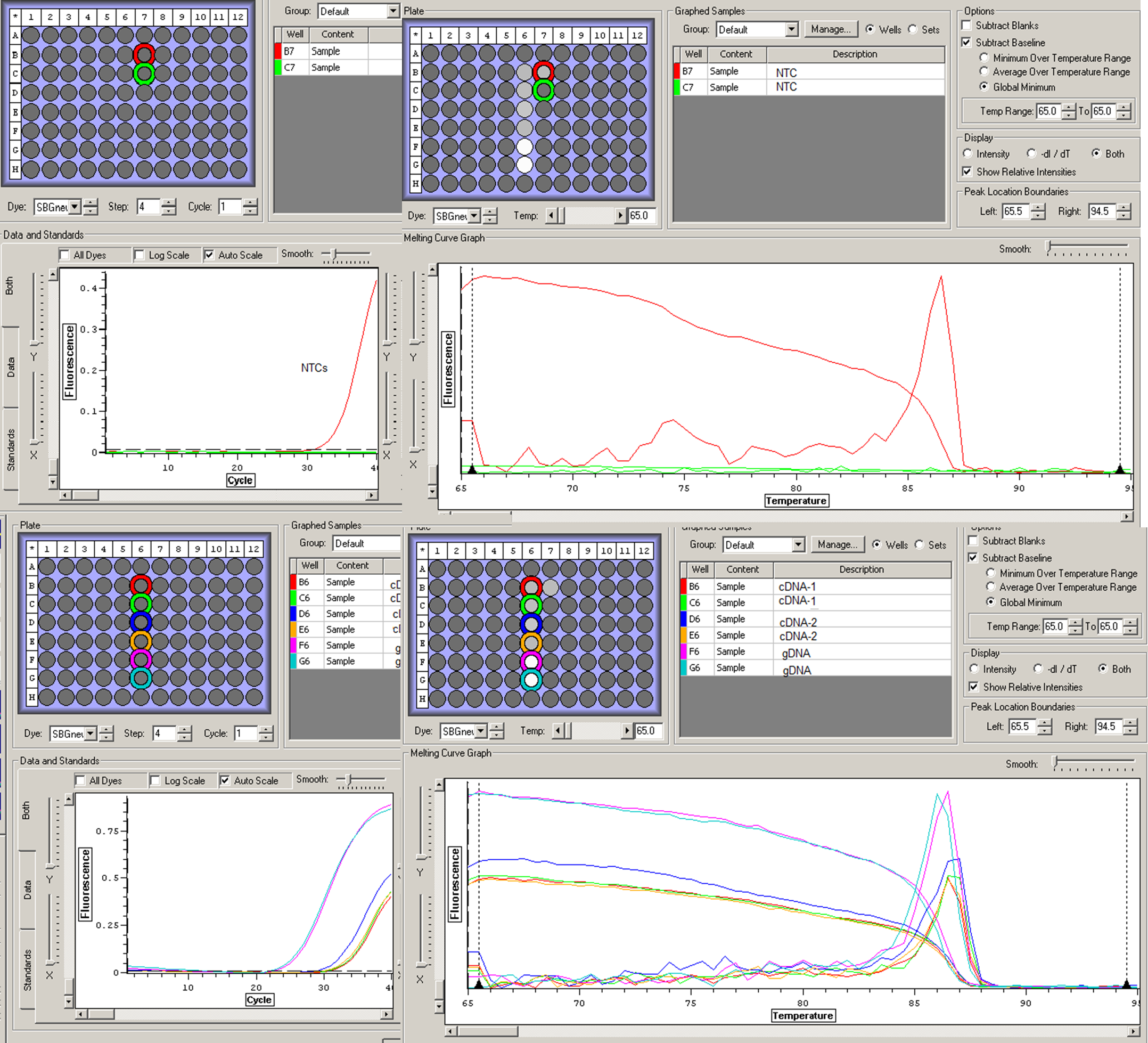
RESULTS: Clean amplification (i.e., no early florescence), duplicates almost identical, a singular peak for all samples. However, one of the two NTCs was contaminated. I will now proceed to DNasing my 'real' RNA samples and run a qPCR on that RNA to assure purity of samples.
12/5/2012
Nano-drop Spec. results on Robert's Lab Herring RNA:
| Sample |
ng/ul |
260/280 |
260/230 |
| RNA-1 |
771.3 |
2.07 |
1.66 |
| RNA-2 |
1786.8 |
2.00 |
2.02 |
Reverse Transcription (Promega M-MLV Protocol)
A single reaction volume = 25uL. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA. You may need to make changes to accommodate your own conditions.
1) Calculate volume of RNA = 1ug of RNA:
RNA-1 = 0.7713 ug/ul * Xul of RNA = 1ug of RNA
RNA-2 = 1.79 ug/ul * Xul of RNA = 1ug of RNA
2) Transfer calculated volume(s) of RNA to 0.5mL snap cap tubes or PCR plate. Adjust volumes of individual samples to 17.75uL with H2O.
V of RNA-1 = 1.3ul --> 1.3ul + 16.5ul DEPC 0.1% = 17.75ul
V of RNA-2 = 0.56ul --> 0.56ul + 17.19ul DEPC 0.1% = 17.75ul
3) Add appropriate amount of primer to sample. Use 0.25ug primer per 1ug of RNA in sample (= 0.5uL of Promega oligo dT in this example). Total volume (RNA + primers) should equal 18.25uL.
17.75ulRNA-1 + 0.5ul Promega oligo dT = 18.25ul
17.75ulRNA-2 + 0.5ul Promega oligo dT = 18.25ul
4) Heat samples at 70C for 5 min in thermocycler.
5) Placed samples on ice IMMEDIATELY.
6) Made Master Mix (3 RXNs)
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer) x 3 = 15ul
1.25 uL 10mM dNTPs x 3 = 3.75ul
0.5 uL M-MLV RT per ug of RNA =1.5ul
7) Mix well.
8) Added 6.75uL of master mix to each reaction.
9) Mix well, but do not vortex (invert)
10) Spot spin.
11) Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12) Heat inactivate @ 95C for 3 min.
13) Spot spin.
14)Stored @ -20C.
DNA Extraction with DNazol
1) With a sterile pestile, homogenized tissue sample (Herring liver: field sample #7) in 500ul of DNazol in a 1.5ml sterile micorfuge tube. After homogenized, an additional 500ul of DZanol was added and mixed well (flicked and slowly inverted)
2) Sampled incubated for 5min @ room temperature (RT)
3) Sample spun at 10,000 x g (RT) for 10min
4) Transfer supernatant to a new, labeled 1.5 micofuge tube
5) Added 500ul of 100% ethanol (EtOH) to supernatant
6) Mixed by inverting (slowly/gently) 5-8 times
7) Stored sample at RT for 1min
8) DNA was a cloudy precipitate. Degraded DNA & small quantities of DNA (<15ug) do not spool onto a pipette tip. So, the DNA precipitate was sedimentized by centrifugation @ 5,000g for 5min at 4C (NOTE: White DNA precipitate clung to side of tube after centrifuge).
9) The liquid was carefully discarded
10) The sample sat for 1min @ RT then the rest of the lysate (liquid that is not the DNA) was removed
11) DNA was washed with 1000ul of 75% EtOH: EtOH pipetted into the DNA tube, carefully inverted 6 times, and let sit for 1 min. Ethanol was then removed and this step was repeated.
12) Remaining EtOH at the bottom of the tube after the 2nd wash was removed with a small pipette
13) 300ul of 0.1% DEPC water was then added to the DNA and slowly pipetted up and down multiple time to dissolve DNA into solution
14) DNA sample was then quantified using Nanodrop (see result below):
| Sample |
ng/ul |
260/280 |
260/230 |
| DNA-7 |
103.4 |
1.64 |
0.79 |
11/27/2012
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix) [Cost per rxn ~$0.42]
Single reaction (20uL) set up is listed below with the FRESH primers:Master Mix:
2x Sso Fast EvaGreen 10 x 4 = 40ul
Forward Primer 2 0.5 x 4 = 2ul
Reverse Primer 2 0.5 x 4 = 2ul
H20 7 x 4 = 70ul
cDNA template = 2ul
Order:
A
---
P2
NTC
NTC
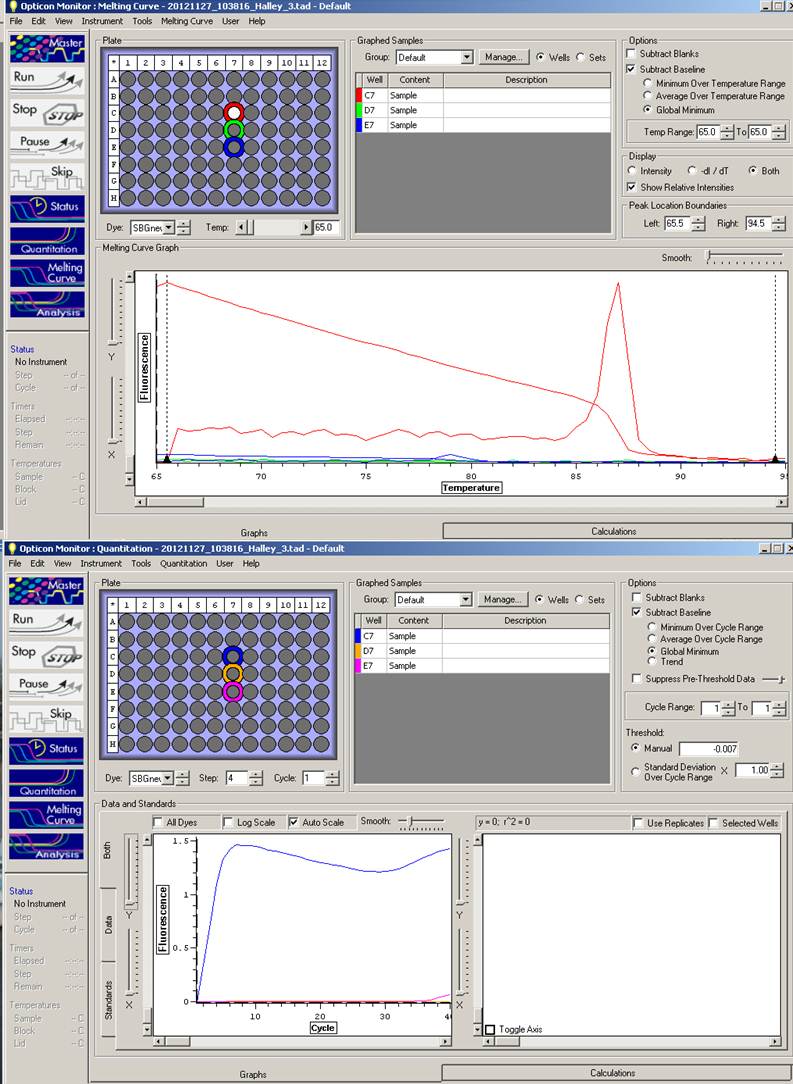
Amplification curve still fluorescing too soon. Sam is going to take a stab at it to pin point the problem - possibly 'wonky' cDNA?
Good news, the control samples are clean!
Conventional PCR was preformed on the Pacific herring (liver) cDNA:
MASTER MIX CALCULATIONS:
12.5ul 2xApex Red *24 = 300ul
0.5ul Forward primer *24= 12ul
0.5ul Reverse primer *24 = 12ul
9.5ul PCR H20 *24 = 228ul
Above calculations used for primer pair #2
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
62C - 15s
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Ran one medium agarose gel for primer pair 2:
100ml 1x TAE
1.90 Agarose
10 EtBr
Loaded 5ul of Hyperladder II
8ul of sample into each well (primer pair & NTCs)

The thermocycler malfunctioned in the beginning (over heated my samples) which may have degraded the samples and thus the results. The NTCs were clean (not shown).
11/16/2012
Nano-drop Results from RNA (20 samples). Clean RNA: 260/280 range should be 1.8-2.0 and 260/230 should range between 1.5-2.0.| Sample # |
ng/ul |
260/280 |
260/230 |
| 2 |
1176.51 |
2.01 |
1.43 |
| 7 |
1139.20 |
2.00 |
1.34 |
| 11 |
1267.65 |
1.99 |
1.96 |
| 18 |
936.53 |
1.98 |
1.86 |
| 22 |
782.78 |
1.99 |
1.74 |
| 30 |
782.51 |
1.99 |
0.97 |
| 33 |
1188.60 |
2.01 |
1.95 |
| 40 |
739.16 |
1.98 |
1.74 |
| 41 |
1259.86 |
2.01 |
1.04 |
| 47 |
1367.16 |
2.02 |
2.12 |
| 55 |
846.43 |
1.99 |
1.80 |
| 56 |
1386.20 |
2.02 |
1.19 |
| 64 |
261.45 |
1.84 |
2.01 |
| 69 |
409.99 |
1.86 |
1.99 |
| 72 |
1471.31 |
2.02 |
1.91 |
| 77 |
2442.94 |
1.96 |
1.69 |
| 87 |
1067.66 |
2.02 |
1.08 |
| 85 |
1749.34 |
2.02 |
1.58 |
| 91 |
1529.40 |
2.02 |
1.98 |
| 99 |
1493.62 |
2.01 |
1.57 |
| Mean |
1164.92 |
1.99 |
1.65 |
| SD |
483.58 |
0.05 |
0.36 |
Reverse Transcription (RT) of the 20 Pacific herring liver RNA samples (above):
1. Once our stock RNA thawed, inverted tube to mix sample.
2. Labeled 0.5ml PCR tube with "Sample # cDNA".
3. Pipetted 5µl of our stock RNA into the PCR tube
4. Added 1µl of oligo dT
5. Added 4µl of nuclease free H2O (i.e., DEPC)
6. In a thermocycler, we incubated the mix at 70C for 5min
7. After incubation, we place the mix on ice for 2min
8. Spun the sample down
9. Added 5µl of M-MLV 5x Reaction buffer
10. Added 5µl of dNTPs (2.5uM)
11. Added 1µl of M-MLV Reverse Transcriptase (RT)
12. Added 4µl more of M-MLV nuclease free H2O
13. Vortexed the mix for several seconds
14. Spun it down
15. Placed back in the thermocycler, the mix was incubated for 60min at 42C, then heat inactivate at 94C for 3min.
16. Mix spun down and kept on ice (ultimately stored @ -20C).
Waiting for fresh (uncontaminated) primers to proceed...
11/15/2012
Finished extracting the RNA from homogenized samples (10/31/2012)11/13/2012
*qPCR run on 11/9/2012 resulted in odd results. So, to ensure clean samples and NTCs a new batch of primer working stock (10mM) and PCR H2O were used. In addition, the plate was placed in the center of the thermocycler.*Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix) [Cost per rxn ~$0.42]
Single reaction (20uL) set up is listed below:Master Mix:
2x Sso Fast EvaGreen 10 x 5 = 50ul
Forward Primer 2 0.5 x 5 = 2.5ul
Reverse Primer 2 0.5 x 5 = 2.5ul
H20 7 x 5 = 35ul
2ul Template
Order:
A
---
P2
P2
NTC
NTC
RESULTS:
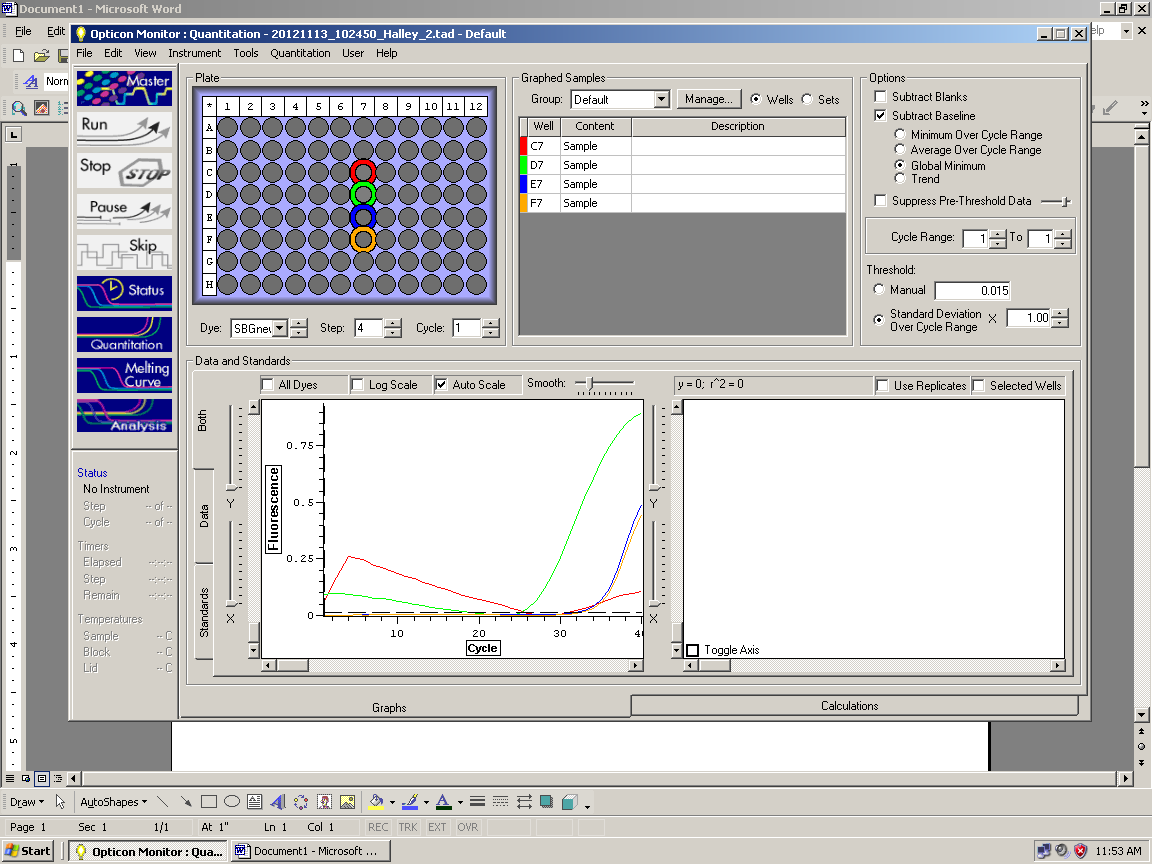
Problems:
1) Amplification of product in NTCs -- SOLUTION: Ordered new Primer Pair 2 from IDT
Primer pair 2
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
GTTGAGCAGCTTCCTCATGC |
Plus |
20 |
2498 |
2517 |
53.85 |
55.00 |
6.00 |
2.00 |
| Reverse primer |
GGAGTCGGAGGTGTTCTACG |
Minus |
20 |
2626 |
2607 |
53.85 |
60.00 |
3.00 |
2.00 |
| Product length |
129 |
||||||||
2) Irregular amplification for rxns with template -- Possible reasons:

Sigmoidal amplification curves
If your amplification curves look sigmoidal, it is likely you have one of these problems:
- The baseline setting in your instrument’s data analysis software may be too low
- You may have a high level of fluorescent noise during the early cycles of PCR
11/9/2012
Quantitative PCR (qPCR)/Real-time PCR (2x Sso Fast EvaGreen Supermix) [Cost per rxn ~$0.42]
Single reaction (20uL) set up is listed below. Be sure to make a master mix volume that will accommodate the following: all of your samples, two water (no template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 20uL) to white PCR plate. Add template. Cap with optical PCR caps. Spin plate for 1min @ 3000g. Put in thermalcycler.| Reaction_Components |
Volume |
Final Concentration |
| 2x Sso Fast EvaGreen |
10 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 20uL |
Master Mix:
2x Sso Fast EvaGreen 10 x 5 = 50ul
Forward Primer 2 0.5 x 5 = 2.5ul
Reverse Primer 2 0.5 x 5 = 2.5ul
H20 7 x 5 = 35ul
RESULTS: Funky....redo.
11/2/2012
Conventional PCR was preformed on the Pacific herring (liver) cDNA:MASTER MIX CALCULATIONS:
12.5ul 2xApex Red *4 = 50ul
0.5ul Forward primer *4= 2ul
0.5ul Reverse primer *4 = 2ul
9.5ul PCR H20 *4 = 38ul
12.5ul 2xApex Red *4 = 50ul 75% volume of primers to reduce primer-dimers
0.375ul Forward primer *4= 1.5ul
0.375ul Reverse primer *4 = 1.5ul
9.75ul PCR H20 *4 = 39ul
Above calculations used for each primer pair #2
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
62C - 15s
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Ran one small agarose gel for primer pair 2:
Loaded 5ul of Hyperladder II
12ul of sample into each well (primer pair & NTCs)
Results: Controls clean, primer-dimers reduced, and product maintained (although not as tight a band as I would like...but I'll take what I can get!) using full volume (0.5ul) of primer. Next step use qPCR to determine melt and amplification quantities.
10/31/2012
Conventional PCR was preformed on the Pacific herring (liver) cDNA:Conventional PCR (2x Apex Red)**
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes or PCR plate. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.25 |
0.2uM |
| Reverse Primer (10uM) |
0.25 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *4 = 50ul
0.25ul Forward primer *4= 1ul
0.25ul Reverse primer *4 = 1ul
10ul PCR H20 *4 = 40ul
Above calculations used for each primer pair #2
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
62C - 15s
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Ran one small agarose gel for primer pair 2 (see 5/30/2012).
TAE 75ml
Agarose 1g
EtBr 5ul
Loaded 5ul of Hyperladder II
12ul of sample into each well (primer pair & NTCs)

Results: The primer-dimers were reduced in the NTC, but now there is a "smear" for the sample rxn with no clear band.
On Herring Field Samples (see 10/23/2012 for samples processed)
RNA Extraction**
1) ~0.002-0.003g of liver tissue (n=20) was homogenized with a sterile pestle in 500ul of TriReagent in a 1.5ml microfuge tube
2) Shortly vortex
3) Additional 500ul of TriReagent was then added to the test tube
4) Vortexed for 15s
5) Homogenized tissues were incubated at RT for 5min
6) Stored at -80C
10/29/2012
Ran one medium agarose gel for primer pair 2 (see 5/30/2012).
TAE 125ml
Agarose 1.72g
EtBr 12ul
Loaded 5ul of Hyperladder II
12ul of sample into each well (primer pair & NTCs)
Results:
Temperatures greater than or equal to 63C should be avoided. However, the primer-dimers are still a nightmare.
10/25/2012
Conventional PCR was preformed on the Pacific herring (liver) cDNA:
Conventional PCR (2x Apex Red)**
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes or PCR plate. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *25 = 312.5ul
0.5ul Forward primer *25= 12.5ul
0.5ul Reverse primer *25 = 12.5ul
9.5ul PCR H20 *25 = 237.5ul
Above calculations used for each primer pair #2
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 30s
60-65C - 30s NOTE: PCR was conducted in the Real Time PCR Detector to perform a temp gradient (60C-65C) of annealing temperatures
72C - 90sec
1 cycle:
72C - 3min
4C - Inf.
Temp gradient:
| 60C |
60.2C |
60.6C |
61.2C |
62C |
63C |
64.2C |
65.2C |
|
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
| A |
P2-1 |
P2-2 |
P2-3 |
P2-4 |
P2-5 |
P2-6 |
P2-7 |
P2-8 |
| B |
N1-1 |
N1-2 |
N1-3 |
N1-4 |
N1-5 |
N1-6 |
N1-7 |
N1-8 |
| C |
N2-1 |
N2-2 |
N2-3 |
N2-4 |
N2-5 |
N2-6 |
N2-7 |
N2-8 |
10/24/2012
Reverse Transcription (RT) of Pacific herring liver RNA provided by the Robert's Lab:
1. Once our stock RNA thawed, inverted tube to mix sample.
2. Labeled 0.5ml PCR tube with "HIF cDNA".
3. Pipetted 5µl of our stock RNA into the PCR tube
4. Added 1µl of oligo dT
5. Added 4µl of nuclease free H2O (i.e., DEPC)
6. In a thermocycler, we incubated the mix at 70C for 5min
7. After incubation, we place the mix on ice for 2min
8. Spun the sample down
9. Added 5µl of M-MLV 5x Reaction buffer
10. Added 5µl of dNTPs (2.5uM)
11. Added 1µl of M-MLV Reverse Transcriptase (RT)
12. Added 4µl more of M-MLV nuclease free H2O
13. Vortexed the mix for several seconds
14. Spun it down
15. Placed back in the thermocycler, the mix was incubated for 60min at 42C, then heat inactivate at 94C for 3min.
16. Mix spun down and kept on ice (ultimately stored @ -20C).
10/23/2012
All 100 liver tissue samples have been collected from the field.
 |
| Hood Canal, WA. Each letter corresponds to a sampling station. Samples were collected June-October 2012. |
| Example of the liver collection process. |
20 samples will be processed at a time; the first 20 (in green) have been randomly selected to include five samples from each collection month and station.

8/23/2012
Ran one small agarose gel for primer pair 2 (see 5/30/2012).(gel from 8/21/12)
Loaded 5ul of Hyperladder II
20ul of samples (primer pair & NTCs)
Results:
Got rid of the primer-dimers, but at the expense on the PCR product. Need to test smaller annealing thermoregimes with the 60-65C range.
8/21/2012
Conventional PCR was preformed on the Pacific herring (liver) cDNA (see 6/20/12):Conventional PCR (2x Apex Red)
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *4 = 50ul
0.5ul Forward primer *4 = 2ul
0.5ul Reverse primer *4 = 2ul
9.5ul PCR H20 *4 = 38ul
Above calculations used for primer pair 2
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
65C - 15s NOTE: Annealing temperature was increased from 60C to 65C to remove primer-dimers
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Also prepared small agarose gel (16 wells)
75ml of 1XTAE
1g agarose
6ul of EtBr
8/15/12
Conventional PCR was preformed on the Pacific herring (liver) cDNA (see 6/20/12):Conventional PCR (2x Apex Red)
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *4 = 50ul
0.5ul Forward primer *4 = 2ul
0.5ul Reverse primer *4 = 2ul
9.5ul PCR H20 *4 = 38ul
Above calculations used for all three primer pairs
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
60C - 15s NOTE: Annealing temperature was increased from 55C to 60C to remove primer-dimers
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Ran one small agarose gel for all primer pairs (see 5/30/2012).
(gel from 8/13/12)
Placed gel in 1X TAE box (with comb)
Removed comb
Loaded 5ul of Hyperladder II
12ul of samples (primer pairs & NTCs)
Results:
Still have primer-dimers. Stumped. But primer 4 is totally out of the running!
8/13/12
Ran one small agarose gel for two of the primer pairs (see 5/30/2012).
75ml of 1x TAE
1g of agrarose
Microwaved ~3min, mix
6ul of Ethedium Bromide (EtBr)
Mix
Pour into container
Place well-comb
Let sit for ~30min
Placed gel in 1X TAE box (with comb)
Removed comb
Loaded 7ul of Hyperladder II
15ul of samples (primer pairs & NTCs)
Results:
Still have primer-dimers. Will try another round (8/15/2012) of cPCR with all three primers at an annealing temp of 60C (instead of 55C).
8/03/12
Conventional PCR was preformed on the Pacific herring (liver) cDNA (see 6/20/12):Conventional PCR (2x Apex Red)
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *4 = 50ul
0.5ul Forward primer *4 = 2ul
0.5ul Reverse primer *4 = 2ul
9.5ul PCR H20 *4 = 38ul
Above calculations used for each set of primer pairs (only using primer pair 1 and 2, no 4)
Cycling parameters:
1 cycle:
95C - 10mins
39 cyles of:
95C - 15s
55C - 15s NOTE: Annealing temperature was increased from 50C to 55C to remove primer-dimers
72C - 30sec
1 cycle:
72C - 8min
4C - Inf.
Stored at 20C for running gel on Monday 8/13/2012
6/27/2012*
Ran one medium agarose gel for all three primer pairs (see 5/30/2012).
150ml of 1x TAE
2g of agrarose
Microwaved ~3min, mix
12ul of Ethedium Bromide (EtBr)
Mix
Results:
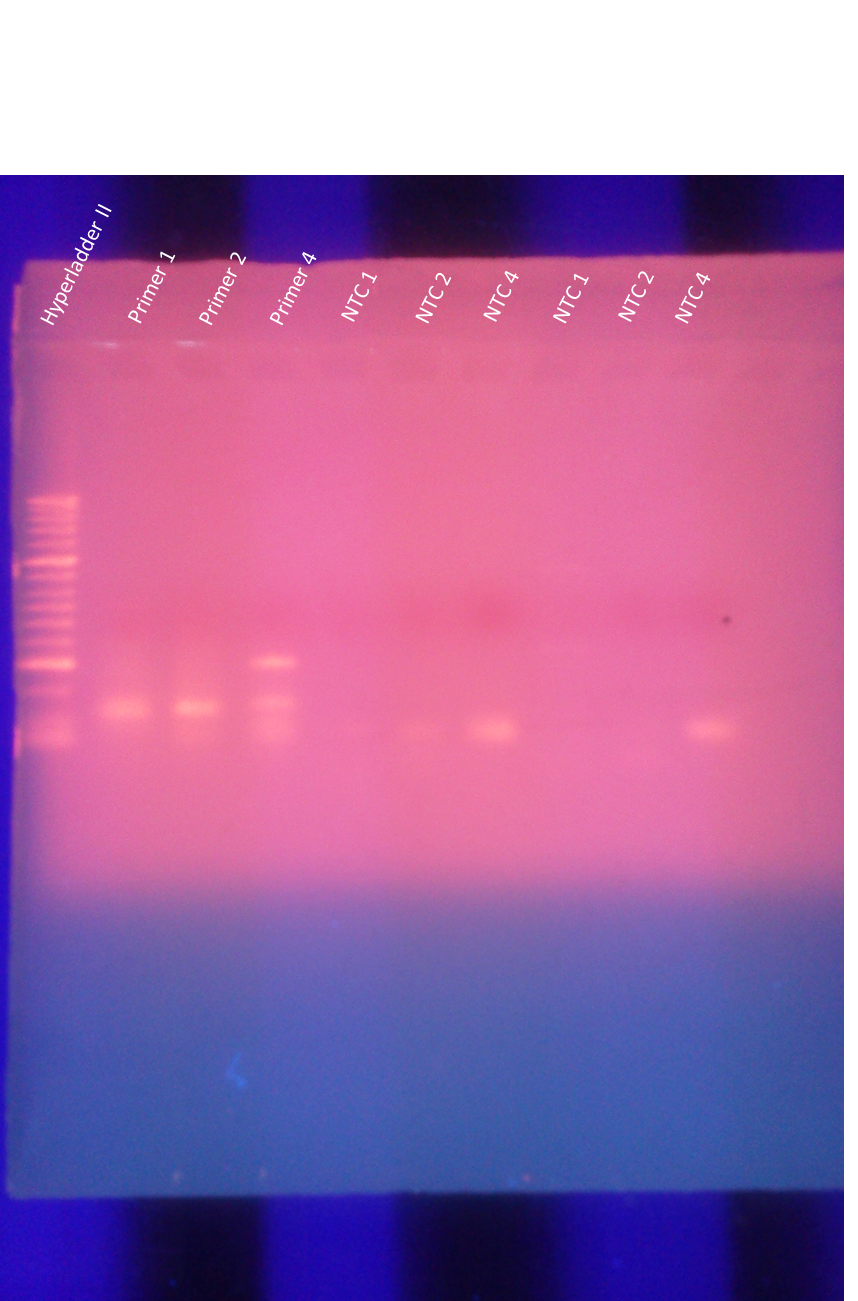
Hyperladder II in the 1st well, 3 samples and 2 non-template controls (NTCs) for each primer pair in the remaining wells.
Ran gel for 50 min at 100v
Results:
Primer 1: 116bp
Primer 2: 129bp
Primer 4: 138bp
Primer 1 & 2 products were the correct size (between 100-200bp, closer to 100bp) and the NTCs were clean (although the first NTC 2 did have a very slight band). Primer 4 appears contaminated.
6/20/2012*
Reverse Transcription (RT) of Pacific herring liver RNA provided by the Robert's Lab:
1. Once our stock RNA thawed, inverted tube to mix sample.
2. Labeled 0.5ml PCR tube with "HIF cDNA".
3. Pipetted 5µl of our stock RNA into the PCR tube
4. Added 1µl of oligo dT
5. Added 4µl of nuclease free H2O (i.e., DEPC)
6. In a thermocycler, we incubated the mix at 70C for 5min
7. After incubation, we place the mix on ice for 2min
8. Spun the sample down
9. Added 5µl of M-MLV 5x Reaction buffer
10. Added 5µl of dNTPs (2.5uM)
11. Added 1µl of M-MLV Reverse Transcriptase (RT)
12. Added 4µl more of M-MLV nuclease free H2O
13. Vortexed the mix for several seconds
14. Spun it down
15. Placed back in the thermocycler, the mix was incubated for 60min at 42C, then heat inactivate at 94C for 3min.
16. Mix spun down and kept on ice (ultimately stored @ -20C).
Conventional PCR was preformed on the Pacific herring (liver) cDNA (from above):
Conventional PCR (2x Apex Red)**
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (non-template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes or PCR plate. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red *4 = 50ul
0.5ul Forward primer *4 = 2ul
0.5ul Reverse primer *4 = 2ul
9.5ul PCR H20 *4 = 38ul
Above calculations used for each set of primer pairs (i.e., 3 x MM)
Cycling parameters:**
95C - 10mins
40 cyles of:
95C - 15s
50C - 15s
72C - 10s - 2mins (dependent on amplicon size; ~1000kb/min)
Stored at 20C for running gel on Friday 6/22/2012
6/11/ - 6/15/2012*
Collected liver tissue samples from four sites in Hood Canal, WA; 5 specimen from each site (20 total herring). On board, liver tissue samples (~1 mg) were collected and preserved in 5ml of RNAlater. Back at the lab, samples were stored at -20C.
5/30/2012*
Ordered three primer pairs (IDT) for Pacific herring (Clupea pallasii) HIF-1a mRNA analysis:
Primer pair 1
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
AGTGGAGCACCTTCCATGTG |
Plus |
20 |
2140 |
2159 |
54.09 |
55.00 |
4.00 |
3.00 |
| Reverse primer |
CAAGAAGGGCAAGGAGCAGA |
Minus |
20 |
2255 |
2236 |
54.09 |
55.00 |
2.00 |
0.00 |
| Product length |
116 |
||||||||
Primer pair 2
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
GTTGAGCAGCTTCCTCATGC |
Plus |
20 |
2498 |
2517 |
53.85 |
55.00 |
6.00 |
2.00 |
| Reverse primer |
GGAGTCGGAGGTGTTCTACG |
Minus |
20 |
2626 |
2607 |
53.85 |
60.00 |
3.00 |
2.00 |
| Product length |
129 |
||||||||
Primer pair 4
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
| Forward primer |
CACCTTCCATGTGGAGGACT |
Plus |
20 |
2147 |
2166 |
53.09 |
55.00 |
6.00 |
2.00 |
| Reverse primer |
AGGGAGATGTTGGTCCACAG |
Minus |
20 |
2284 |
2265 |
53.09 |
55.00 |
5.00 |
3.00 |
| Product length |
138 |
||||||||
The above primers fell within the most conserved region of HIF-1a (NCBI BLAST):
 |
| Result of BLAST HIF-1a herring contig sequence |
10/11/2011*
Collected 16 English sole liver and heart tissue samples (32 total samples) from Hood Canal, WA. Hypoxia is currently very shallow, so there is a high chance of these English sole being exposed to low DO conditions.
All samples were placed in 5ml of RNAlater and stored at -20oC.
7/28/2011***
Made new batch of 1x TAE
20ml of 50x TAE
980ml of NANO Pure H20
Made new small (75ml of 1x TAE) 1.3% gel (1g of Agarose, 6ul of EtBR)
Ran 7/22/2011 PCR products and NTCs for (100V 30min)
Once again got bands in all four NTCs.....
Next step? Run only (four) NTCs using these primers and perhaps run a known, single banded primer & sample of a different species for comparison. Will use all new PCR H2O AND Apex Red.
7/22/2011*
Diluted new primers to 100uM

Ran conventional PCR for the two new sets of primers (protocol 7/8/2011)
MASTER MIX CALCULATIONS (**X2):
12.5ul 2xApex Red * 7 = 87.5ul
0.5ul Forward primer * 7 = 3.5ul
0.5ul Reverse primer * 7 = 3.5ul
9.5ul PCR H20 * 7 = 66.5ul <- USED NEW BATCH OF H20 IN CASE PREVIOUS H20 WAS CONTAMINATED (i.e., NTC bands)
*Product stored at 4C for later gel run....
7/15/2011***
Ordered new primers (IDT) of known English sole sequences:
(GenBank EF119288) 16S ribosomal RNA gene, partial sequence; mitochondrial
| Primer pair 1 |
Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
| Forward |
CCCGCCTGCCCAGTGACAAC |
Plus |
20 |
34 |
53 |
60.25 |
70.00% |
| Reverse primer |
CCATGGTCGCCCCAACCGAA |
Minus |
20 |
327 |
308 |
59.34 |
65.00% |
| Product length |
294 |
(GenBank AF026744) GTPase K-rasB proto-oncogene mRNA, complete cds
| Primer pair 10 |
Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
| Forward |
GGTGGGAGCTGGTGGCGTTG |
Plus |
20 |
20 |
241 |
60.18 |
67.67% |
| Reverse primer |
CCACTGTCCGGAACGGGAGGT |
Minus |
21 |
593 |
573 |
60.18 |
66.67% |
| Product length |
353 |
link to Primer Search
7/13/2011***
Ran gel for re-do Primer 9 samples (same protocol as below: 7/11/2011):
No good!
7/11/2011***
Ran two small agarose gels for both primer pairs.
Used pre-existing gel for first run(Primer 3 samples)
Mixed up a new batch for small gel mold (Primer 9 samples):
75ml of 1x TAE
1g of agrarose
1ul of Ethedium Bromide (EtBr)
Hyperladder 1 in the 1st well, 4 samples and 2 non-template controls (NTCs) in the remaining wells.
Ran both gels (separately) for ~30min at 100v
Primer 3: Unsuccessful :'o(
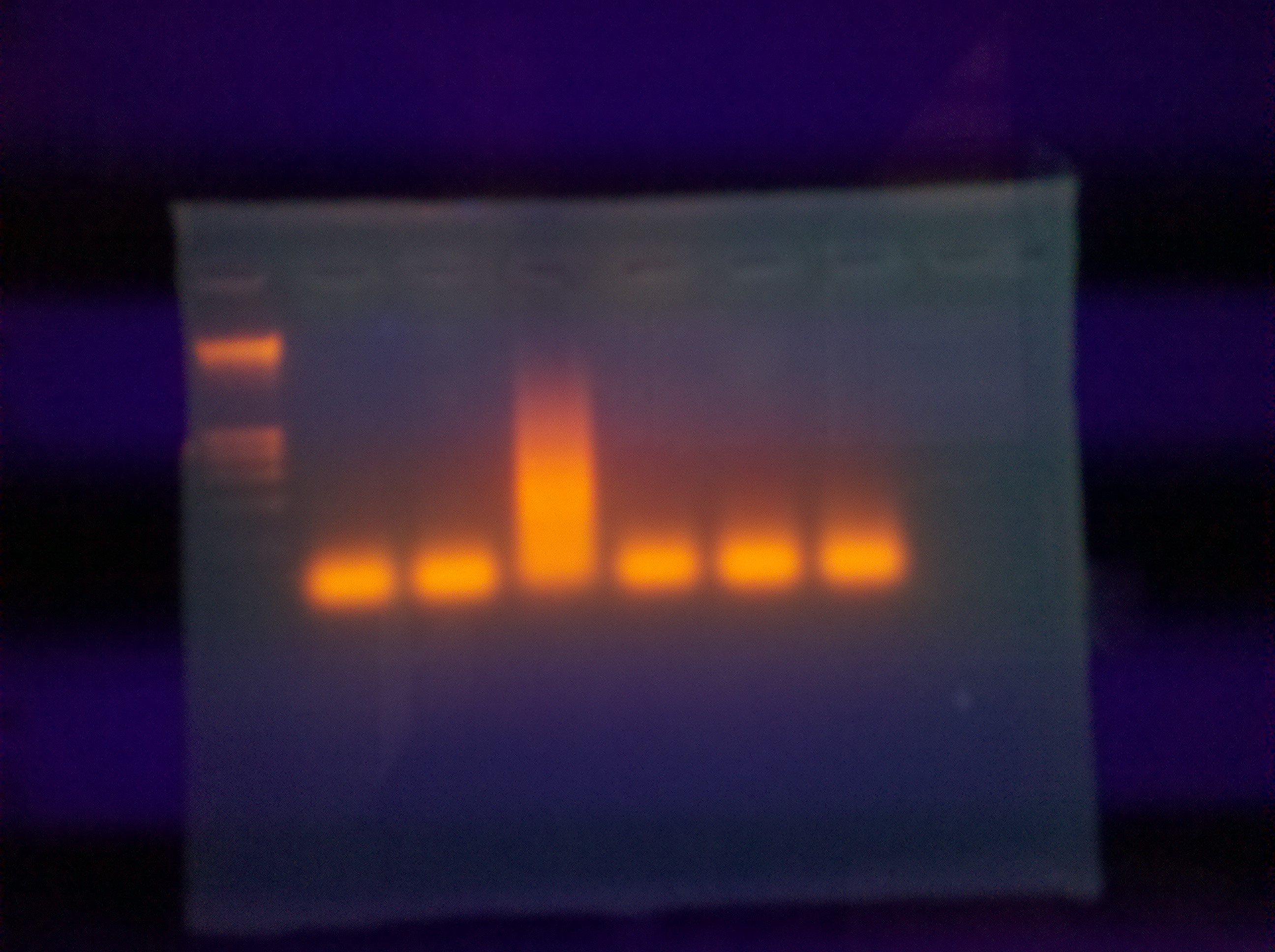
Primer 9: Contaminated NTCs :o/
No pic.
Proceeded by redoing Conventional PCR (2x Apex Red) for Primer pair 9.
NOTE: Same protocol and Master Mix proportions as below (7/8/2011)
7/8/2011*
Conventional PCR was preformed on the sole cDNA.
The following (Roberts Lab) protocol was followed:
Conventional PCR (2x Apex Red)**
Single reaction (25uL) set up is listed below. Be sure to make a master mix volume that will accommodate all of your samples, two water (no template controls; NTC) samples, plus an extra 10% to accommodate pipetting errors. Distribute appropriate amount of master mix (volume of master mix + template = 25uL) to PCR tubes or PCR plate. Make sure all tubes/caps are tightly closed. Put in thermalcycler.
| Reaction_Components |
Volume |
Final Concentration |
| 2x Apex Red |
12.5 |
1x |
| Forward Primer (10uM) |
0.5 |
0.2uM |
| Reverse Primer (10uM) |
0.5 |
0.2uM |
| Template |
Up to 5uL |
|
| H2O (PCR grade) |
variable |
Use to bring reaction volume up to 25uL |
12.5ul 2xApex Red * 7 = 87.5ul
0.5ul Forward primer * 7 = 3.5ul
0.5ul Reverse primer * 7 = 3.5ul
9.5ul PCR H20 * 7 = 66.5ul

NOTE: Did two separate batches of MM for each primer pair.
Total of 12 PCR tubes:
2 H1
2 L1
2 H3
2 L2
4 NTC
Typical cycling paramaters (ask for help on using the thermal cycler):
95C - 10mins
40 cyles of:
95C - 15s
50-60C - 15s
72C - 10s - 2mins (dependent on amplicon size; ~1000kb/min)
Stored at 4C for running gel next week....
7/6/2011***
Reverse Transcribed sole RNA (i.e., cDNA)
The following (Roberts Lab) protocol was followed:
Reverse Transcription (Promega M-MLV Protocol)
A single reaction volume = 25uL. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA. You may need to make changes to accommodate your own conditions.
- Use as much RNA as possible, max volume of RNA = 17.75uL. Generally, identify the RNA sample with the lowest concentration and multiply by 17.75uL. Use this quantity (ug) of RNA for each and every sample.
- Transfer calculated volume(s) of RNA to 0.5mL snap cap tubes or PCR plate. Adjust volumes of individual samples to 17.75uL with H2O.
- Add appropriate amount of primer to sample. Use 0.25ug primer per 1ug of RNA in sample (= 0.5uL of Promega oligo dT in this example). Total volume (RNA + primers) should equal 18.25uL.
- Heat samples at 70C for 5 min in thermocycler.
- Place samples on ice IMMEDIATELY.
- Make Master Mix:
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer)
1.25 uL 10mM dNTPs
0.5 uL M-MLV RT per ug of RNA
7. Mix well.
8. Add 6.75uL of master mix to each reaction.
9. Mix well, but do not vortex.
10.Spot spin.
11.Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12.Heat inactivate @ 95C for 3 min.
13.Spot spin.
14.Store @ -20C.
MASTER MIX CALCULATIONS:
5ul M-MLV RT Buffer * 4.5 = 22.5ul
1.25 dNTPs * 4.5 = 5.625ul --> rounded to 5.63ul
0.5 M-MLV RT (per ug of RNA) * 4.5 = 2.25ul

6/30/2011***
 |
| English sole (Parophrys vetulus) |
Successful RNA extraction of two heart and two liver samples of English Sole; apparent by the 260/280 ratios within 2 +/- 0.1.
The following (Roberts Lab) protocol was followed:
NOTE: no DNase treatment
RNA Extraction
Manufacturers' Protocol - MRC- Turn centrifuge on to cool to 4C
- Clean Homogenizer
- Get sample and thaw enough to get out of container
- Measure weight of sample
- Take sample out (screw out and use forceps) and chop up with sterile razor blade
- Put tri-reagent (stays on ice when not using & is light sensitive) into 50 ml falcon tube (or smaller tube depending on size of sample) – for a 0.7 gram sample I used 7 ml of tri-reagent. Note: in 50 ml falcon tube need at least 3 ml of tri-reagent to get it to work
- Add sample
- Keep on ice
- Blot homogenizer with paper towel to remove excess water
- Homogenize sample (don’t leave off ice for too long)
- Homogenize until sample is in solution
- Transfer all or part (I kept 6 ml) of mixture into a 13 ml tube (only add up to 7 ml)
- Let sit for 5 min at RT
- Rinse homogenizer in DEPC water (same tube used to clean in the beginning)
- Add 0.2 ml of chloroform (under hood, open only in hood, pour into glass beaker first) per 1 ml of tri-reagent
- Cover & shake
- Let sit for 15 min at room temp
- Change gloves
- Spin at 12,000 x g (11,500 rpm) for 15 min at 4C
- Transfer aqueous (top) phase to fresh tube (top layer has the RNA in it – bottom layer has DNA and proteins in it)
- Add 3 ml iso. (2 – Propanol, under hood) to precipitate out the RNA
- Cap and vortex
- Let sit at RT for 10 min
- Put waist with tri-reagent etc. in tri-reagent bottle in fume hood
- Clean up homogenizer and put away (put in 50 ml falcon tube with 5-7 ml 30% H2O2 and up to 40 ml with DEPC water
- Spin at 12,000 x g (11,500 rpm) for 15 min at 4C
- Remove supernatant (I want the pellet – RNA)
- Get glass beaker and paper towels (small stack)
- Pour off supernatant into beaker and place tube upside-down on paper towels
- Note: do not rock the tube back and fourth or will loosen pellet
- Add 1 ml 75% EtOH in DEPC water per 1 ml of tri-reagent added in beginning
- Cap & move – rock back and fourth to loosen pellet – vortex if necessary
- Spin 11,500 x rpm for 5 min at 4C
- Remove supernatant again & put on paper towel – be much more careful to make sure pellet does not slip out
- Spot Spin – turn on centrifuge, let go up to about 1000 rpm then shut off – note: place pellet facing upward
- Use filter pipette tips to remove excess EtOH
- Turn upside down on paper towels
- Wait 10 mins
- Depending on size of pellet add dnase free water to the tube – if taking to mRNA always use 500 ul (I used 500 ul for the ovary – large pellet, and 250 ul for the muscle)
- Dissolved into solution by pipetting
- Put in 1.5 ml tube
- Put on ice
- Spec to determine how much RNA you have
Also ordered (IDT) 2 pairs of the following Platichys flesus (European flatfish) HIF-1a primers (GenBank: EF100709) for PCR:
Primer Pair Three
Sequence (5'->3') Strand on template Length Start Stop Tm GC%
Forward primer GGCTCGGAGCGGAGGAAGGA Plus 20 37 56 60.04 70.00%
Reverse primer TCCCGCAGCTCCTCCTGGTC Minus 20 440 421 59.97 70.00%
Product length 404
Primer Pair Nine
Sequence (5'->3') Strand on template Length Start Stop Tm GC%
Forward primer CCACAGCGTCAGCTCCAGCC Plus 20 138 157 60.04 70.00%
Reverse primer TGACGTTGACAGTGCGGCCC Minus 20 553 534 59.91 65.00%
Product length 416