September 29, 2012,
DNA.txt
DNA version of the mature.fa file.
MUMmer 3.23 has not been very helpful. I will consult this link to find miRNA prediction software.
https://sites.google.com/site/mirnatools/mirna-targets
Much of the software is species specific. However, using
http://www.mirz.unibas.ch/cgi/pred_miRNA_genes.cgi
I was able to locate some potential miRNA regions in the sequence
https://mail-attachment.googleusercontent.com/attachment/?ui=2&ik=ee77591da2&view=att&th=13a15100c661a26f&attid=0.1&disp=inline&safe=1&zw&saduie=AG9B_P9fq4F2E0OBLpdOC8zD5Qt1&sadet=1348973347057&sads=588XldVcq7zhHx20R0ugumnT3Is
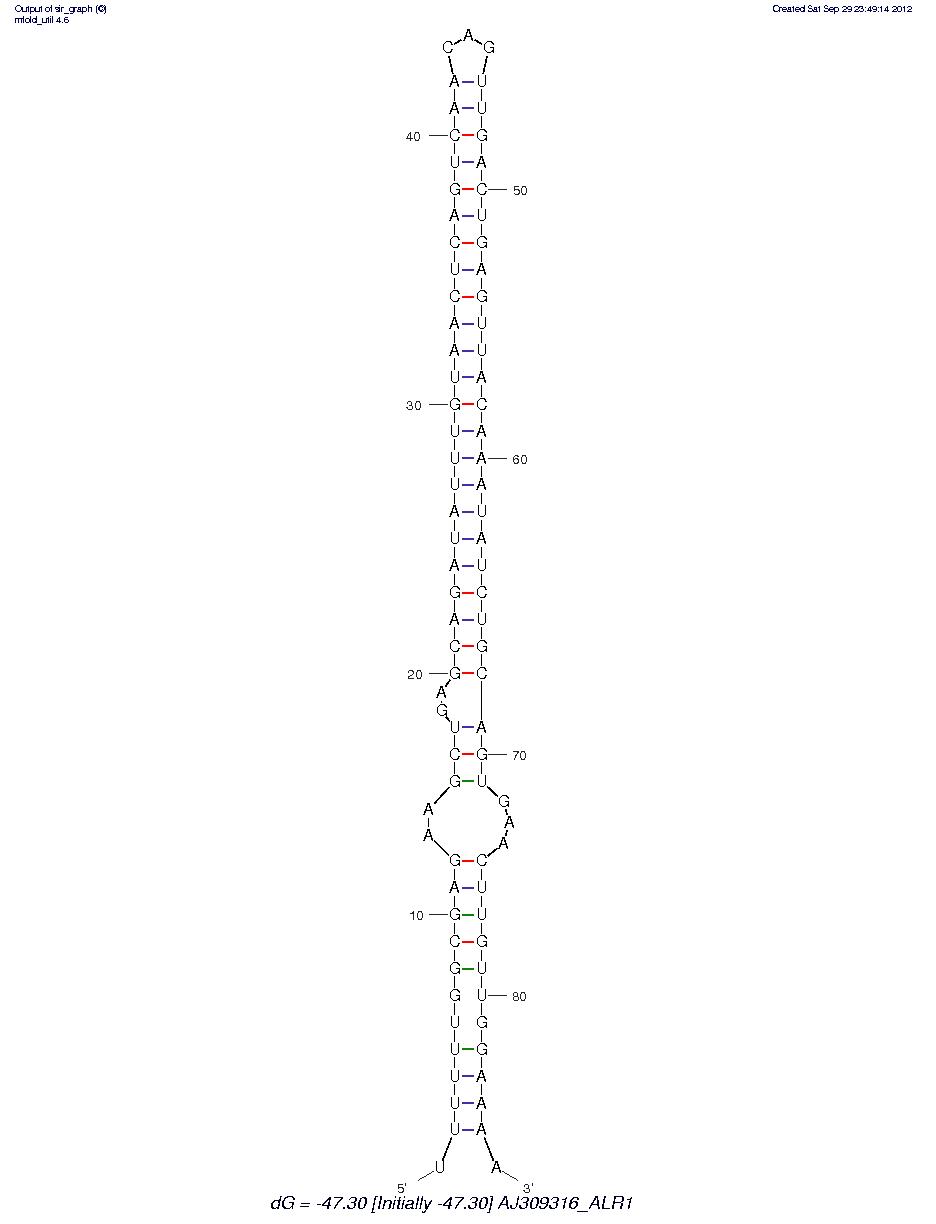
AJ309316_ALR1
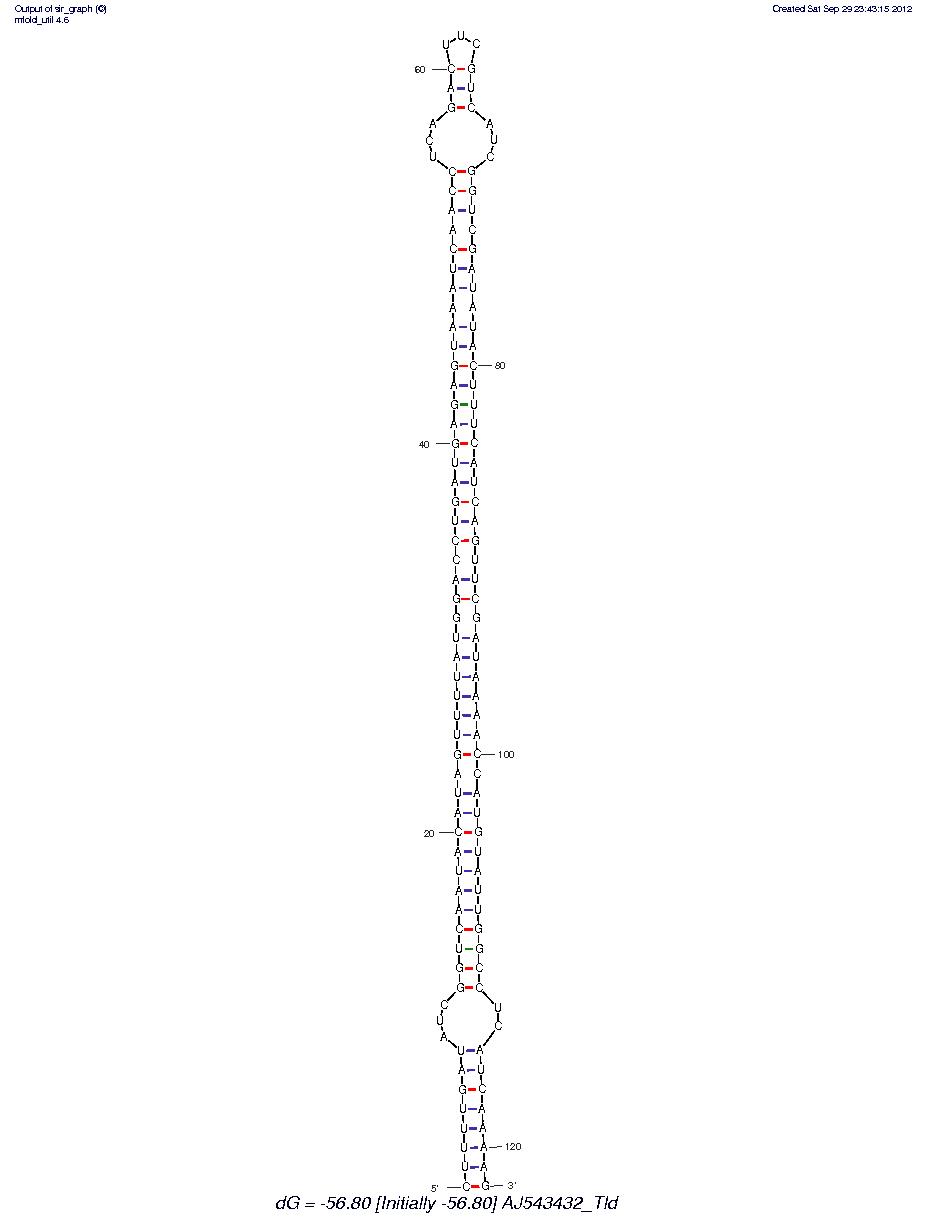
AJ543432_Tld
AM905317_gnrhr, EU342886_hexokinase are too large to analyze.
FJ347754_LAP
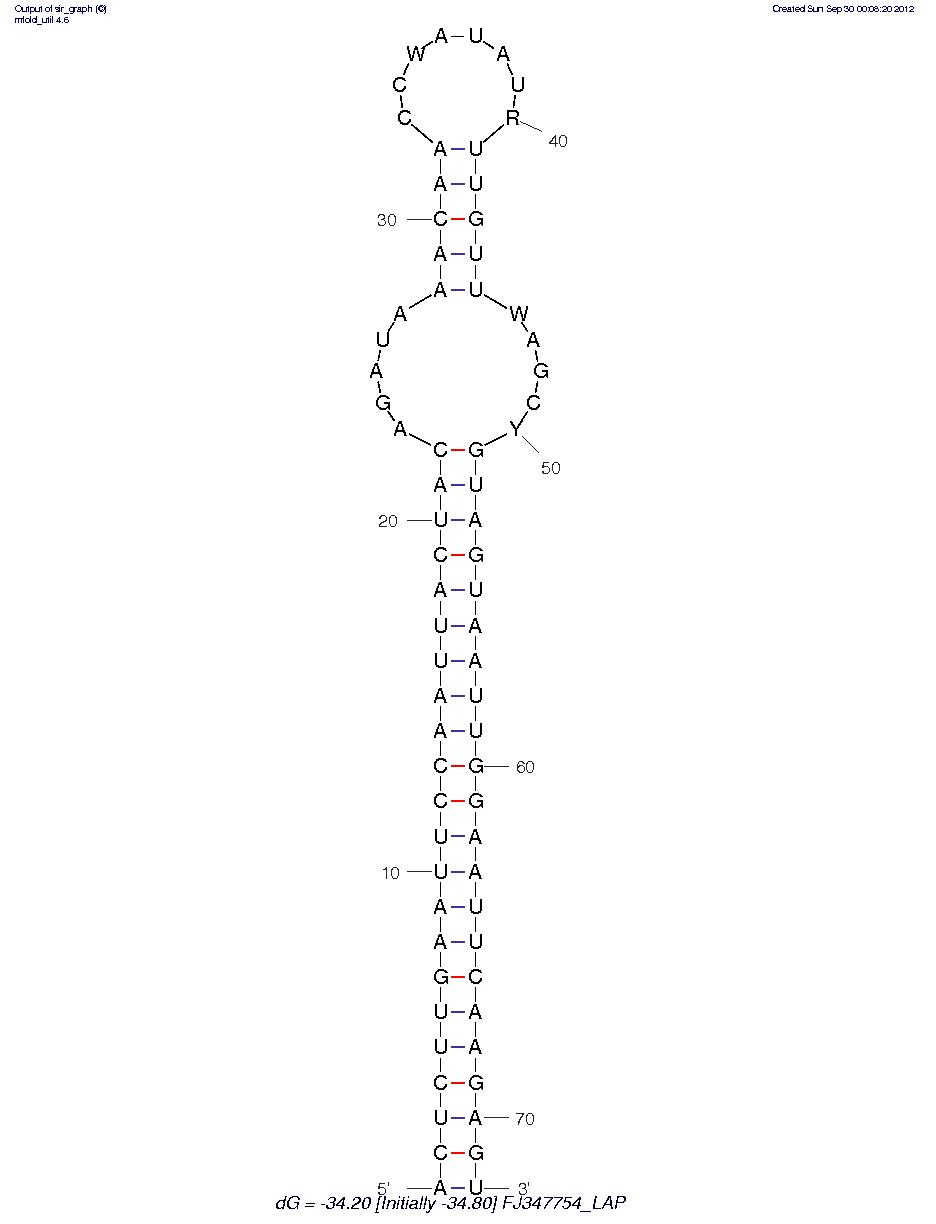
I see some goofy non-existent nucleotides, which needs to be considered.
Until I find another way of doing this I would like to simply go through all the sequences <10000bp with this. After I have done I can analyze the remaining sequences some other way.
Graphics are from this website,
http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form
These appear to be hairpin miRNA? I need to do some research to see if this is the right software to be using. However, that said this may be a valid direction of research as well. More to come.
September 28, 2012,
Just ran MUMmer 3.23 with the mature.fa file and cgigas_alpha_v032.fa file and it did not yield any hits whatsoever. To make sure I am doing it correctly, I will now run it against my repeat families file. If I did it correctly, this will surely yield some hits, otherwise I must be doing something goofy.
There appears to be some sort of bug with the nucmer command in MUMmer 3.23 >:O. I am trying some workarounds.
mature.fa doesn't appear to be a DNA fasta file perhaps this might be the problem?
We should replace all of the U's with T's and redo the analysis. But before I do that I would like to see if BLAST can handle the files.
September 25, 2012,
Getting back to research, moving away from TE's for a little bit. Lets begin looking for microRNA evidence in the Crassostrea gigas genome.
http://www.mirbase.org/ftp.shtml has mature miRNA fasta files.
BLAST is an option but I feel like MUMmer might be a better choice.
http://mummer.sourceforge.net/manual/ is the manual for MUMmer 3.23.
I will likely have to use the mummer command
I have to run mummer in my Ubuntu linux simulator. That will be the plan for next time.
August 24,2012,
This code here represents what represents the second half of the two part program that will
A.) Clean the data from MUMmer 3.23 to remove duplicate matches
B.) Group repeat regions into families and create 2 GFF files one of all the keys and one with all repeat regions including keys.
Anyways here is the java code,
find_repeat_families.java
and the R code,
MUMmer_data_cleaner.R
The next step will be to try and make both of these programs work together. Which apparently can be done (see Calling R from Java by Duncan Temple Lang)
However, for now I have improved on the previous algorithm and yielded even more repeat regions previously omitted in the first analysis.
If anyone would like to try out the first program it works with the halfsize_coordinates_file.csv file from August 4th. However, it will have to be saved as a text file for the program to read it.
I will post directions on using the first program soon. So that it may be used by others in the future.
My attempts to make this java program into an exe file have so far been fruitless,
Nonetheless I want to have my program be useful so I will help get a user guide for running my program in Command Prompt
Step 1: Download find_repeat_families.java. You should need jdk version 1.5 or higher.
Step 2: Download the accompanying text file that is created from the MUMmer_data_cleaner program. Admittedly, this program takes a prohibitively long time to run but until further data comes along you will only need this already cleaned text file (It is simply a text form of a csv file).
halfsize_coordinates_file.txt
Now you have the right program and the proper input for Crassostrea gigas repeat regions. Of course this program can be used for other genomes btw, just run MUMmer 3.23 with your genomic data, take that input run it in the MUMmer_data_cleaner R script. Take the csv output from that program and convert that into a text file (that may be unnecesarry, I am unsure).Then finally, use that text instead of the halfsize_coordinates_file.
Step 3: Make sure that you have the classpaths set correctly. You want to make sure your java and javac JAR files in your Java directory are linked to your command line. The following link explained the process in the most clear and concise way possible
http://introcs.cs.princeton.edu/java/15inout/windows-cmd.html
I am running on Windows 7 Java Version 1.7 and the path I used to link my java to command line was
C:\Program Files (x86)\Java\jdk1.7.0\bin
Step 4: Once you have your classpaths set correctly you need to open the directory that contains the find_repeat_families.java script in command line (Using the cd command).
Step 5: Compile the program by typing in the same directory " javac find_repeat_families.java" then as long as the path was set correctly and you have typed this exactly this should go to the next line and nothing should happen and you shouldn't get any errors. If you do you likely set your class path wrong so go back to step 3.
Step 6:Run the program by typing java find_repeat_families.
Step 7: At this time you are now actually in the program I wrote. Great. Type the path leading to and including the halfsize_coordinates_file.txt file. (ie: "C:\Users\Harry\Documents\halfsize_coordinates_file.txt"). Press enter.
Step 8: Type a name to identify the output of this program. Press enter.
Step 9: The rest of the program will guide you through cleaning the data the user will find relevant in her/his analysis. But the only other user input required will be the minimum and maximum allowable repeat family size (number of different repeat regions).
Step 10: The output will be in the same path as where the find_repeat_families program is. There will be two GFF3 files one contains the keys to the repeat families the other is the bona fide list of repeat families groups.
No actual transposable element analysis yet. It would be nice to automate the microsatellite identification process as when I first did this analysis all I had to do was remove microsatellite regions to significantly reduce the number of relevant families to look at in my analysis. More to come. I really encourage you to try this program and give me your input. It is beta/heuristically motivated and many steps may be taken to further optimize it, but it is still very helpful in reducing the large/redundant dataset created from MUMmer 3.23 self-similarity analysis for people interested in doing de novo TE analysis (which is challenging).
August 8, 2012,
Trying to make a very user friendly interface that will take the MUMmer 3.23 output and find repeat families. The code I used was already used to find repeat families for my first GFF file. As new data comes along
or if other people looking for this type of program need it, it would be available. I currently, have two different programs I would like to turn into an exe or JAR file for an open source program. The first is written in R, simply converts the MUMmer 3.23 output into a concise, clean and efficient format which will yield a csv file. This file would then be sent to another program that would be responsible for the grouping of families and here some features could be added that would allow you to define groups of interest. There could be many potential avenues but the ultimate goal would be to streamline the de novo discovery of repeat regions.
Currently I have the first two programs written but not compiled into a user friendly format. Also, I have not added the user defined features yet but that is where I am ending tonight. Once I am at the stage of combining the the programming languages into a Java-R interface I shouldn't need to modify the R code (or the Java code) very much. This is the first time I have tried to do anything like this but it does not seem to be anything beyond my ability. This would be a nice tool to leave the lab with so future TE discovery can be done very rapidly.
find_repeat_families.java
*Of course anyone looking at my code will forgive the messiness of it as it is still pretty beta. Expect more on this program.
August 6,2012
If I look at one of the entries of Family 6 that is not the key I get some hits that suggest that the family may be another retrotransposon. The large question is whether it is acceptable to use TE's defined from other taxa to call the ones found via De novo discovery TE's as well ( We have had matches to mosquitos, corn, fruit flies, parasitic wasps). Nevertheless, here is the link,
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=19127&lib=root
All of the entries for Family 9 were queried in the repBase database and none of the entries yielded any hits.
I will update the GFF file accordingly.
August 4, 2012,
If I ever decide to return to the TE topics this file will be useful. It is like the coordinates.csv file earlier except here I have not only removed matches to the same region but also duplicate pairs of matches so the file is much smaller. Thus, it would be easier/faster to make groupings.
halfsize_coordinates_file.csv
August 1, 2012,
A short diversion to see if any of the unidentified regions could potentially be the Pearl transposable element. To check this I ran the palindrome algorithm to find palindromes,
which would mean it could potentially be the Pearl element.
Family9:
palindrome9.palindrome
Some palindromes of the appropriate size are here.
Family6:
palindrome6.palindrome
Ditto.
Family18:
palindrome18.palindrome.txt
July 31, 2012,
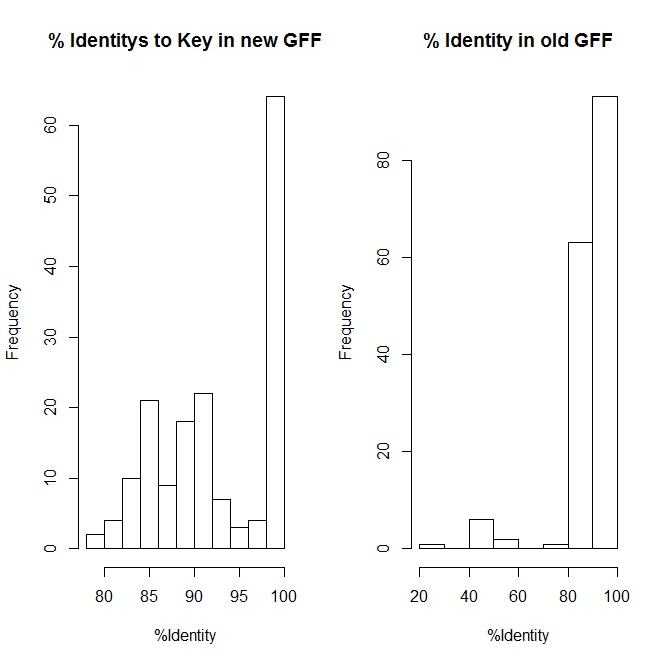 |
| The improvement of repeat regions identity back to keys with new revisions. |
Found the link to query my repeat regions against the Genetic Information Research Institutes, repBase database here,
http://www.girinst.org/censor/index.php
Family1:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=4198&lib=root
Family2:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=4734&lib=root
Family3:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=4941&lib=root
Family4:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=6061&lib=root
Family5:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=6314&lib=root
Family6:
No hits.
Family7:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=6949&lib=root
Family9:
No hits:
Family10:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=7913&lib=root
Family11:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=8168&lib=root
Family12:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=8543&lib=root
Family13:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=8798&lib=root
Family14:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=9006&lib=root
Family15:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=9189&lib=root
Family16:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=9556&lib=root
Family17:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=9850&lib=root
Family18:
No hits.
Family19:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=10204&lib=root
Family20:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=10724&lib=root
Family21:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=10918&lib=root
Family22:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=11171&lib=root
Family23:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=11362&lib=root
Family24:
http://www.girinst.org/cgi-bin/censor/show_results.cgi?id=11719&lib=root
Now some of these features have names which is exciting. It seems most of the families were simple sequences/satellite regions. However, other features came up that will require more research. I will compile these into the gff file.
Group IV,GroupV and GroupVII appear to be retrotransposon groups.
Updated GFF
finalNest.names.txt
July 30, 2012,
With the new family list I need to verify the groupings I made earlier (i.e. Group I, Group II, Group III, etc). Many of the families have not changed so I should be able to do this pretty quickly.
Using the new Keys here is Group I's multiple alignment
badAlignmentchoice.clustal
Clearly some issues with the new keys Family 16 hardly matches to this old framework, so it should be omitted. Also after looking at the multiple sequence alignment I see that some families should be included. Also, since we are 1 family less than the old framework perhaps some of the old ones got new names?. Either way, I am much more happy with these alignments in conjunction with the new GFF file.
Multiple Alignment of all new keys (note the minor error described at the bottom of todays entry)
largeClustal.clustal
Group I: (1,2,3,4,11,12*,14*,21,22,24) [* denotes a new addition to the family]
newGroupI.clustal
Group II: (5, 7, 19, 20, 23)
newGroupII.clustal
Group III: (9, 18)
newGroupIII.clustal
Group IV: (15, 16)
newGroupIV.clustal
Group V: (10, 17)
newGroupV.clustal
Group VI: (6)
newGroupVI.fasta
Group VII:(13)
newGroupVII.fasta
Just found a slightly embarassing error in the above data. There is no Family 8. Nevertheless, the groupings are accurate and this is an easy fix I can do later.
Here is a link to a infographic/contingency table of the distribution of repeat regions in the C. gigas genomic sequences.
https://docs.google.com/spreadsheet/ccc?key=0Au4jubvnN-e5dFM2NmhtSjl2bnJiUTVpRFVXSUFBRFE
July 25, 2012
Working on new GFF file by hand.
It appears I erroneously classified 2 and 4 as the same family in actuality Family 4 is a derivative of Family 2 +/+.
Also, Family 2 is now defined by a new key (the old key ended up being derivative and I can better define this region using a new one)
FAMILY2.FASTA
Family 3 is supposed to be a derivative of Family 19
Looks like I should change Family 3's key as well,
Having trouble uploading family 3 key but it will go here
Family 19 should be possibly split into two small families 19a, 19b
again the fasta files are not uploading. I will be sure to get them up eventually.
Family 4 didnt require any further changes
Family 5 has been included into Family 1 lets just verify all of the scores relative to family1's key
Family 1 and Family 5 are NOT the same
Family 5 needs a new key
Family6 is getting a new key but it should be inconsequential because the key being used has 100% identity to the old one
Family7 Done
Family8 done
Family9 done
Family10 done
Family 11 done
Family 12 done
Family 13 done
Family 14 done
Family 15 done
Family 16 done
Family17,18,20 done
Family 21 needs a new key
Family 22,23,24 done
New family key fasta file. This should make everything concerning family names unambiguous.
familykeys.fasta
The keys now map to many of the repeat regions must better but there are still 6 regions less than 79% identity to the key. Perhaps some of these are just issues with orientation.
All of these regions were fixed in this way. Hence, here is the new updated GFF file that I am very happy with as it does a very good job explaining the relationships of these regions. Howver, it would be nice to show how these families relate to each other.
finalNest.txt
July 24, 2012
Back from field work let me try and characterize those repeat region using the Institute for Systems Biology's repeat masking based on protein similarity.
Lets start with Family 19 as it forms the backbone of Group I which is the largest.
<span style="color: #000000;">pValue Score Method SeqID Begin End Repeat Type Begin End - 1574 TRF ?_GU207425__cgiga 1 1065 + Tandem_Repeat - - </span>
Group II (Family 7)
<span style="color: #000000;">pValue Score Method SeqID Begin End Repeat Type Begin End - 1220 TRF Family7 10 1297 + Tandem_Repeat - - - 1066 TRF Family7 10 1297 + Tandem_Repeat - -</span>Group III (Family 9)
No hits yielded
That was actually a mistake because Family 23 defines Group III
Group III (Family 23)
No hits yielded
Group IV (Family 14+15)
No hits yielded
Group V (Family 8)
No hits yielded
Group VI (Family 13)
No hits yielded
That was with simple regions masked. The masked sequences are all N's so I think I need to take the simple repeat option off.
MASKING OFF
Group I
No hits yielded
Group II
No hits yielded
Group III
No hits yielded
Group IV
No hits yielded
Group V
No hits yielded
Group VI
No hits yielded
I am concerned that the first two families were masked because of simple regions. This suggests that because they are low complexity they may have an elevated alignment score which may be the cause of its identification as a repeat family. The next thing I will be doing is updating the GFF file to fix the following
1. Try to identify what to do with low scoring repeats (They may belong in a different family or possibly deleted altogether)
2. Make sure that the alignment is correct for every family.
July 10, 2012,
Here is a distribution of repeat families throughout the genome.
P.S. This is just cut out from my computer so there are some goofy clustering forays in the other sheets of the excel file. I will work on making all of the data quickly and convienently located tomorrow and then do some work further characterizing these repeats.
repeatDistribution.xlsx
July 8, 2012,
family7Key.fasta
family13Key.fasta
family8Key.fasta
July 6, 2012
Same work as yesterday...
Family 8 is NOT a derivative of Family 25
Family 10+12 is NOT a derivative of Family 25
Family 2+4 is a potential derivative of Family 25 (100% coverage)(Score = 183 bits (202), Expect = 4e-51 Identities = 183/234 (78%), Gaps = 9/234 (4%) Strand=Plus/Minus)
Family 14+15 is NOT a derivative of Family 17
Family 1+5 may be a derivative of Family 17 (100% coverage)(Score = 459 bits (508), Expect = 2e-133 Identities = 404/496 (81%), Gaps = 26/496 (5%) Strand=Plus/Plus)
Family 9 is NOT a derivative of Family 17(13% coverage)
Family 23 is NOT a derivative of Family 17(13% coverage)
Family 9 is NOT a derivative of Family 18 (8% query coverage)
Family 23 is NOT a derivative of Family18 (8% query coverage)
Family 1+5 is a possible derivative of Family 18 (100% coverage)(Score = 398 bits (440), Expect = 5e-115 Identities = 399/506 (79%), Gaps = 43/506 (8%) Strand=Plus/Plus)
Family 14+15 is not a derivative of Family 18
Family 13 is NOT a derivative of Family 10+12
Family 1+5/8/25/14+15 are NOT derivatives of Family 9
##Family 9 is a potential derivative of Family 23 (100% coverage)(Score = 783 bits (868), Expect = 0.0 Identities = 505/552 (91%), Gaps = 9/552 (2%) Strand=Plus/Plus) (interesting)
Family 1+5/14+15/25 are NOT derivatives of Family 23
Family1+5 is a potential derivative of Family 11 (100% coverage)(Score = 244 bits (270), Expect = 7e-69 Identities = 347/473 (73%), Gaps = 35/473 (7%) Strand=Plus/Minus)
Family 9/14+15/23 are NOT derivatives of Family 11
Family 14+15 is NOT a derivative of Family 1+5
Famiyl 25 is a possible derivative of Family 1+5 (100% coverage)(Score = 260 bits (288), Expect = 4e-74 Identities = 245/312 (79%), Gaps = 19/312 (6%) Strand=Plus/Plus)
Family 8 is NOT a possible derivative of Family 1+5
Family 8/Family 25 are NOT possible derivatives of Family 14+15
Family 13/10+12 are NOT derivatives of Family 2+4
Family 2+4 is NOT a derivative of Family 8
Family 10+12 is NOT a derivative of Family 8
I just verified all possible matches than are greater than half the length of the subject.
Did a multiple sequence alignment of some interesting group of related families to create the following clustalW file. Very interesting. Check it out.
GroupIMutlipleAlignment.clustal
Testing some of the minor repeat families,
Family2+4 is a partial match to Family 19 (87% coverage)(Score = 241 bits (266), Expect = 6e-68 Identities = 196/234 (84%), Gaps = 9/234 (4%) Strand=Plus/Minus) (Probably should be added to Group I)
Family 3+24 is a partial match (53% coverage)(Score = 147 bits (162), Expect = 3e-40 Identities = 83/84 (99%), Gaps = 0/84 (0%) Strand=Plus/Plus) I suspect that this should be added to Group I
Family 7 is larger than Family 19
Family 8 is NOT a derivative of Family 19 (1% query coverage)
Family 9 is NOT a derivative of Family 19 (11% query coverage)
Family 10+12 had no similarity to Family 19
Family 13 is had no similarity to Family 19
Family 14+15 had no similarity to Family 19
Family 23 is NOT a derivative of Family 19 (11% query coverage)
Family 25 should be a derivative of Family 19(99% coverage)(Score = 356 bits (394), Expect = 2e-102 Identities = 277/326 (85%), Gaps = 3/326 (1%) Strand=Plus/Plus)
Updated ClustalW, less consensus hits but it still is very cool to see.
GroupI.clustal
Lets see if there are any other relationships between the remaining families,Starting with the largest region we already checked some of the regions
#Copied from earlier
Family 19 is NOT a derivative of Family 7
Family 22+16 is NOT a derivative of Family 7
Family 11 is NOT a derivative of Family 7
Family 18 is NOT a derivative of Family 7
Family 20 is NOT a derivative of Family 7
Family 21 is NOT a derivative of Family 7
Family 17 is NOT a derivative of Family 7
or in other words all of Group I,
Family 8 is NOT a derivative of Family 7
Family 9 is NOT a derivative of Family 7
Family10-12 is NOT a derivative of Family 7
Family 13 is NOT a derivative of Family 7
Family14+15 is NOT a derivative of Family7
Family 23 is NOT a derivative of Family7
Just to check Family 1+5 is NOT a derivative either
Thus,these results suggest that Family 7 is a novel repeat family.
The next largest family is Family 23,
Family 8 is NOT a derivative of Family 23
Family 9 may be a derivative of Family 23(99% Query Coverage)(Score = 783 bits (868), Expect = 0.0 Identities = 505/552 (91%), Gaps = 9/552 (2%) Strand=Plus/Plus)
Family 10+12 is NOT a derivative of Family 23
Family 13 is NOT a derivative of Family 23
Family 14+15 is NOT a derivative of Family 23.
Hence, Family 23 and Family 9 appear to make another Group. Which is incidentaly the next largest group so the next group after that is Sequence 14+15
Family 8 is NOT a derivative of Family 14+15
Family 10+12 may be a derivative of Family 14+15 (98% Coverage)(Score = 190 bits (210), Expect = 3e-53 Identities = 154/186 (83%), Gaps = 13/186 (7%) Strand=Plus/Plus)
Family 13 is NOT a derivative of Family 14+15
So Family 14+15 and Family 10+12 appear to make up another group, finally we must check and see if Family 8 and Family 13 are related which they are not so we have finally grouped all the repeat families together into relevant groups
Group I: Family 1,2,3,4,5,6,11,16,17,18,19,20,21,22,24,25
Group II: Family 7
Group III: Family 9,23
Group IV: Family 10,12,14,15
Group V: Family 8
Group VI: Family 13
Group I is characterized in the above ClustalW file
Group II,V and VI are just the given sequence/repeat family.
Group III ClustalW file,
GroupIII.clustal
Group IV ClustalW file,
GroupIV.clustal
July 5, 2012,
Continuing to characterize derivatives in the repeat families. Wrote a program in biojava to test if certain families were derivative of others. It relies on Smith-Waterman local alignments which will return the most conserved region betwen two sequences (longest common substring with modification), not extremely confident in how robust it is but it should point me in the direction of families that are particularly similar allowing me to condense my family list further.
BLAST verification of Top Hits (using blastn)
1. Family 16 and 22 are the same (Checked with BLAST 100% Identity, E-value=0)
Family 21 is likely a derivative of 20(nearly 100% coverage (1 nucleotide short), 89% identity, 4% gaps)
Family 1+5 is NOT a derivative of Family 14+15
Family 16+22 may be derivatives of Family 19 (100% coverage) (Score = 1317 bits (1460), Expect = 0.0 Identities = 920/1040 (88%), Gaps = 21/1040 (2%), Strand=Plus/Minus)
Family 6 may be a derivative of 16+22 (by extension and BLAST yields Score = 1109 bits (600), Expect = 0.0 Identities = 849/965 (88%), Gaps = 34/965 (4%), Strand: Plus/Minus)
Family 11 is NOT a derivative of Family 18
Family 23 is NOT a derivative of Family 11
Family 19 is NOT a derivative of Family 7
Family 22+16 is NOT a derivative of Family 7
Family 6 is NOT a derivative of Family 7
Family 11 is NOT a derivative of Family 7
Family 18 is NOT a derivative of Family 7
Family 20 is NOT a derivative of Family 7
Family 21 is NOT a derivative of Family 7
Family 17 is NOT a derivative of Family 7
Family 20 is a possible derivative of Family 22+16 (100% coverage)(Score = 1249 bits (1384), Expect = 0.0 Identities = 838/929 (90%), Gaps = 16/929 (2%) Strand=Plus/Plus)
Family 21 is another obvious possible derivative of 22+16 (100% coverage)(Score = 1204 bits (1334), Expect = 0.0 Identities = 807/894 (90%), Gaps = 16/894 (2%) Strand=Plus/Plus)
Family 17 is a possible derivative of Family 22+16 (100% coverage)(Score = 937 bits (1038), Expect = 0.0 Identities = 630/700 (90%), Gaps = 3/700 (0%) Strand=Plus/Minus)
Family 18 is a possible derivative of Family 22+16 (100% coverage)(Score = 731 bits (810), Expect = 0.0 Identities = 610/733 (83%), Gaps = 40/733 (5%) Strand=Plus/Minus)
Family 11 is a possible derivative of Family 22+16 (99% coverage) (Score = 553 bits (612), Expect = 2e-161 Identities = 542/696 (78%), Gaps = 30/696 (4%) Strand=Plus/Plus)
Family 23 is NOT a possible derivative of Family 22+16 (11% coverage)(Score = 31.9 bits (34), Expect = 2e-04 Identities = 19/20 (95%), Gaps = 0/20 (0%) Strand=Plus/Minus) *[short sequence possible low complexity
Family 9 is NOT a possible derivative and has the same problem with Family 23 (Score = 31.9 bits (34), Expect = 1e-04 Identities = 19/20 (95%), Gaps = 0/20 (0%) Strand=Plus/Minus)
Since Family 16+22 is a derivative of Family 19 we expect 20,21,17,18,6 and 11 to be derivatives of Family 19 I wont check this but i think it is a reasonable assumption.
Family 23 is not a possible derivative of Family 19(%query coverage=11%)
Family 20 is likely a derivative of 6(100% query coverage)(Score = 1153 bits (1278), Expect = 0.0 Identities = 807/913 (88%), Gaps = 33/913 (4%)Strand=Plus/Minus)
By extension Family 21 should be a derivative of Family 6 (100% query coverage)(Score = 1151 bits (1276), Expect = 0.0 Identities = 806/912 (88%), Gaps = 33/912 (4%) Strand=Plus/Minus)
Family 17 may be a derivative of Family 6 (Query Coverage=100%)(Score = 940 bits (1042), Expect = 0.0 Identities = 656/747 (88%), Gaps = 18/747 (2%) Strand=Plus/Plus)
Family 18 may also be a derivative of Family 6 (Query coverage=99%)(Score = 663 bits (734), Expect = 0.0 Identities = 601/748 (80%), Gaps = 53/748 (7%) Strand=Plus/Plus)
[perhaps 17 and 18 are closely related]
Family 18 may be a derivative of 17 (82% identity, 7% gaps, some large gaps missing from the middle)
Family 11 may be a derivative of Family 6 (100% query coverage)(Score = 486 bits (538), Expect = 2e-141 Identities = 537/710 (76%), Gaps = 44/710 (6%) Strand=Plus/Minus)
Family 23 is NOT a derivative of Family 6 (8% query coverage)
Family 9 is NOT a derivative of Family 6 (8% query coverage)
Family 17 is a possible derivative of Family 20 (100% query coverage) (Score = 948 bits (1050), Expect = 0.0 Identities = 654/734 (89%), Gaps = 17/734 (2%) Strand=Plus/Minus)
Family 18 is a possible derivative of Family 20 (100% query coverage) (Score = 740 bits (820), Expect = 0.0 Identities = 612/727 (84%), Gaps = 36/727 (5%) Strand=Plus/Minus)
Family 11 is a possible derivative of Family 20 (99% query coverage) (Score = 544 bits (602), Expect = 1e-158 Identities = 540/687 (79%), Gaps = 23/687 (3%) Strand=Plus/Plus)
Family 23 is NOT a derivative of Family 20 (8% query coverage)
Family 9 is a potential derivative of Family 23 (87% coverage)(Score = 783 bits (868), Expect = 0.0 Identities = 505/552 (91%), Gaps = 9/552 (2%) Strand=Plus/Plus)
Family 14+15 is NOT a derivative of Family 20
Family 17 is potentially a derivative of Family 21(query 100%)(Score = 948 bits (1050), Expect = 0.0 Identities = 654/734 (89%), Gaps = 17/734 (2%) Strand=Plus/Minus)
Family 18 is potentially a derivative of Family 21(100% query coverage)(Score = 740 bits (820), Expect = 0.0 Identities = 612/727 (84%), Gaps = 36/727 (5%) Strand=Plus/Minus)
Family 11 is a potential derivative of Family 21(99% coverage)(Score = 544 bits (602), Expect = 9e-159 Identities = 540/687 (79%), Gaps = 23/687 (3%) Strand=Plus/Plus)
Family23(9% coverage)
Family9(9% coverage)
Family 1+5 is a potential derivative of Family 21 (100% coverage)(Score = 533 bits (590), Expect = 1e-155 Identities = 417/492 (85%), Gaps = 22/492 (4%) Strand=Plus/Minus)
Family 14+15 is NOT a derivative of Family 21
Family24+3 is NOT a derivative of Family 13(49 % coverage)(Score = 111 bits (122), Expect = 7e-30 Identities = 77/85 (91%), Gaps = 2/85 (2%) Strand=Plus/Plus)
June 27, 2012,
Preparing for presentation and doing data analysis. Namely I am interested in which families are related to one another. I would like to do some contingency tables to see if I can find some families that are spatially related.
I already know that Family 1 occupies,
| Contig_156220_v032 |
| Contig_157058_v032 |
| Contig_157898_v032 |
| Contig_159174_v032 |
| Contig_160834_v032 |
| Contig_166837_v032 |
| Contig_167004_v032 |
| Contig_167024_v032 |
| Contig_167557_v032 |
| Contig_167665_v032 |
| Contig_167795_v032 |
| Contig_168856_v032 |
| Contig_169358_v032 |
Also,
Family 24 and Family 3 are part of the same family just different directionality
Family 21 is a derivative of Family 20
Family 10 and Family 12 are part of the same family just different directionality
Family 7 may be a tandem repeat (etandem yielded a possible repeat unit of size 75 repeated 16 times with a %identity of 77.9%)
Family 14 and Family 15 are part of the same family just different directionality
Family 2 and Family 4 are part of the same family just different directionality
Family 1 and Family 5 are part of the same family just different directionality
Hence, we can reduce the 25 families into 20 families.
The most numerous families were,
Family 10+12 (22 repeats)
Family 14+15 (19 repeats)
Family 1+5 (19 repeats)
June 22, 2012,
Just finished debugging/creating a new nested interval removal program. And this SHOULD be that last posting of repeat_regions because I ran my interval through a very thorough/robust algorithm to remove nested intervals. Enjoy.
nestsRemoved3.txt
BED file,
cleanNest.bed
(not the same as the above file, as of now there are 2 additional derivative regions, 2 apparently slipped by the first run through my algorithm)
Now that the locations and reliability of the regions has been determined within the genome. It would be interesting to characterize each family. That is do a multiple sequence alignment of each family, the families distribution within the genome, classifying families as derivatives of another( I strongly suspect some of the families are derivative) and other things to understand the families themself. (Look at my notes for June 7-8 to understand what I am interested in pursuing).
Additionally, I believe it would certainly be worth looking at some of the minor repeat regions (repeats<10 times).
Doing some data exploration,
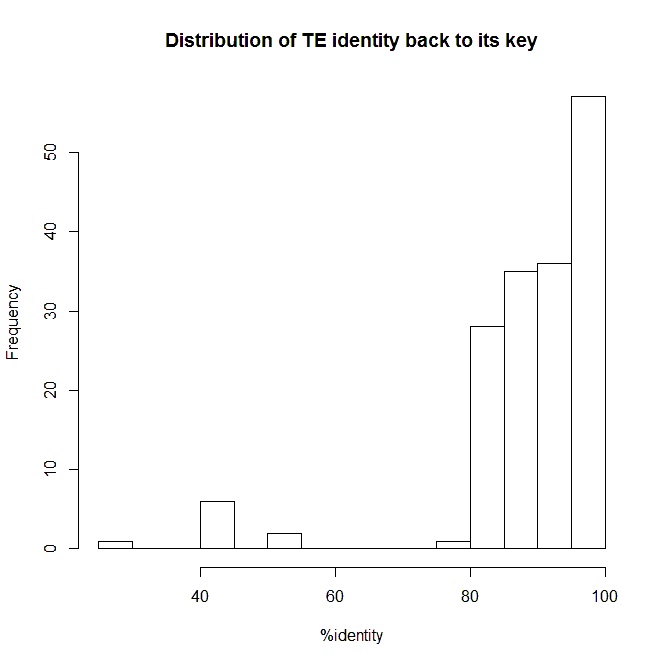 |
| Figure 1. A Histogram of the percent identity of each identified repeat region back to the keys defined by nonMSrepeat_regions file. Notice the unexplained low valued outliers. Perhaps they are another unidentified family, problems with directionality though I have tried that as a fix.n= 166. |
June 21, 2012
I noticed some errors in the GFF and have edited them. I also have noted that though most of the regions are an excellent fit. There is a very small subset that are relatively low (40<%identity<60). There orientation is in the opposite direction but changing that does not change the identity score in Galaxy. I will publish it just the same and at the very least anyone looking at my GFF file will be able to tell these regions are not a good fit. Everything else, however, is a good fit back to the keys. So the REAL final GFF file is here,
,scoredFamily1.txt
BED file (soon)
June 20, 2012,
Debugged the nested interval remover program. Cleanest GFF to date.
cleanFamilyGFF1.txt
BED File,
cleanBED.bed
Still have to add scores to the GFF and investigate the possibility of correlation between repeat_regions and tandem_repeats.
See the following
(Repeat_Region:GU207407 160393-161111 (Family 19) and Tandem_Repeat: GU207407 161058-161114)
(Repeat_Region:GU207422 160401-161119 (Family 19) and Tandem_Repeat: GU207407 161066-161122)
(Repeat_Region:GU207441 11739-12456 (Family 19), Flanking No Tandem)
(Repeat_Region: GU207449 50990-51707 (Family 19) No Flanking Tandem)
Interesting but im not sure that they are related.
To find the scores, I would like to do one of 2 different things. Either
1.) Use the original keys to calculate scores
2.) Use a multiple sequence alignment to determine identity or alignment scores.
The former should be easier and also more representitive of the particular algorithm I used. The latter is nice because it treats the regions as families and I suspect should maximize the identity.
I will pursue the former method for now but afterward I will check on the feasability of the other method.
The method I used on June 8th for exploring the realiability of the algorithm will work fine and can be implemented in Galaxy and a short Java program.
Didn't complete the score interval work today but I wrote a java program that may serve as a framework for Friday. Moreover, im running into trouble getting a %identity for the regions. I will leave that for Friday.
*UPDATE*
Completed %identity of the regions for a complete GFF3 file
scoredFamily.txt
(This version is obsolete use the above version)
June 15, 2012,
Cleaning up the data set. The first easy thing I can do is sort all of the regions by name and by start sequence and only take tthe largest size region where there are multiple regions on the same start position. This should help eliminate derivative repeat regions). Next, we can remove regions that are complete derivatives, by which I mean repeat regions whose position is located within another identified repeat region. Using this pseudo-algorithm a much cleaner GFF file of the repeat regions has been created. Although now some of the regions may occur less than ten times they are derivatives of larger regions and are still worth identifying as transposable elements may fragment. Anyways, the cleaned file is posted here and was created via 2 Java programs and some hand cleaning,
cleanFamilyGFF1.txt
Family 8,22 and 25 only occurs once suggesting it may be completely derivative of the other regions. Family24 occured only twice.
June 8, 2012,
Lets verify the scores of each family (make sure that my algorithm worked at least reasonably well). Unlike before where I wanted to get scored of paired data, right now I will be interested in how well the "key" of the family map,aligns to each of the "values" in the family. Thus, I will be able to easily implement this in Galaxy since I will not need to loop anything.
Using the needle command and pair format, the quality of each alignment can be easily seen,
Family1: is certainly a repeat family as everything matched back to the key with 100% identity
Family2: lowest identity is 85.8% which seems reasonable most of the other members of the family are greater than 90% identity
Family3: lowest identity 88.3%, most in the 90's
Family4: lower identity family lowest identity 79.8% most in the 80's
Family5: lowest identity 82.6% but most in upper 80's and 90's
Family6: not a large family in terms of members lowest identity 79.2 and members are spread pretty uniformly from this to 100% identity
Family7: lowest identity 82.3%, most in the 80's.
Family8: all have an identity >90% compared to the key
Family9: all have an identity >90% compared to the key
Family10: all between 84.2% and 86% identity to the key
Family11: lowest identity 88%
Family12: lowest identity 89% most in the 90's
Family13: lowest identity 86.5%, identities vary.
Family14: lowest identity 93.3%, values well matched to keys
Family15: lowest identity 83.7%, most values in the 80's
Family16: lowest identity 86.1%, most values in the 80's
Family17: lowest identity 88.1%
Family18: lowest identity 88.1%
Family19: lowest identity 83.9%
Family20: lowest identity 89.0%
Family21: lowest identity 91.4%
Family22: lots of repeats in this family, actual sequence size is small, lowest identity 95.3%
Family23: another small sequence, lowest identity 94.0%
Family24: lowest identity 85.2%, most in the 80's
Family25: lowest identity 91.7%, very long sequence
Hence, though I have not gone through every family yet, my algorithm appears to work reasonably well as the values have relatively high identities to the keys. It may be worth my time to check and see if any of these families are derivatives of another (i.e. suppose sequence A is GATCTTTCGA and sequence B is GATC and sequenceB is a derivative of sequence A).
Moreover, because we are dealing with such a significantly smaller data set I may be able to reopen the idea of using RECON to group these families, though I believe that having derivative sequencesavailble is still important, as though there is aggregation of the repeat regions in certain areas, smaller chunks seem of these regions are scattered throughout the genome (splitting of transposable elements?)
June 7, 2012
I thought I would try to BLAST some of the repeat regions. Many of them have already been identified as repeat_regions, however, some of the repeat regions have not been identified as so
(i.e.GU207404cgigasBAC_149215_149697, GU207426cgigasBAC_100873_101906,GU207429cgigasBAC_43458_44755). Created a beta_version of the GFF file (converted it a a BED in galaxy to visualize). The repeat regions families appear to cluster around certain regions. I will post for others to see, I don't know what the significance of this is (repeat_regions forming genes, transposable element is actually a larger element made up of the families). I will post and research.
familyGFF.txt
For whatever reason I can only visualize BED files in galaxy,
repeat_regions_beta.bed
It should be noted that the scores have not been put in here I am going to look into multiple sequence alignments of each family to achieve a score. I am also uncertain of some of the directionality but that will come up in later analysis. The GFF and BED files are at least interesting to look at for the time being, as like I said earlier many of the families cluster around certain regions.
GU207408cgigasBAC_149215_149697_+---->None of the high scoring BLAST hits identified this as a repeat region
GU207418cgigasBAC_75043_75275_+---->A pairwise BLAST between this region and another region found by BLAST suggests that this is actually part of another repeat region (accession number gb|GU207418.1|)
GU207455cgigasBAC_8229_8313_+---->No one identified this relatively small repeated region as such, though other repeat regions were identified in the BLAST
June 6, 2012
These memory bugs are a serious obstacle. I am going to try a different approach. Firstly, in Java's defense I should have trimmed the MSP file down as I have done now. Here only repeat regions that had >10 hits are shown. This alone reduced the number of regions from >77,000 to about 15,000. From here, I would like to group these sequences into families, this requires me to construct a "workflow". The program I have written has identified 208 "families" whose sequence is repeated more than 10 times (resulted in a BLAST hit elsewhere in sequence). The following code was used, if anyone is anyone is skeptical of the algorithm used,
getFamily.java
List of "repeat families" (I expect some deviation from these sequences but they represent the "family" of repeats) given here, Looks like most (not all) of them are microsatellite regions.
repeat_families_alpha.fasta
Luckily this file is small enough I can go through by hand and pick out what are microsatellites as opposed to "interesting" repeat regions. So here is a list of sequences that have been referenced more than ten times in a self local alignment with mummer and are not microsatellites
nonMS_repeat_families.txt
(its text but if you save it as .fasta it will turn into a fasta file). Again, of course note that this EXACT sequence may not be found more than 10 times but mummer has identified that a sequence very close to is has occured more than 10 times (think of BLAST hits, all will have %identity>79.14 more than 96% of the sequences had an identity greater than 90% the alignment scores however are unknown). This is also nice because this is only 25 sequences/families. From this I can readily construct a GFF file of repeat regions, this is a little involved but nothing I cant do. Already have a partial program to create the GFF file but it wont be finished until later in the week. For now it is nice to have a reasonable number of sequences to work with.
CURRENT CODE: (currently non-functional)
##Open reference file
##a.)Upload subjectList
##b.)Upload query list (a and b are pairwise data)
- Cycle through each paired FASTA and get alignment score of each(Large file)
from Bio import pairwise2
##BATCH ITERATOR METHOD (not my code)
def batch_iterator(iterator, batch_size) :
entry = True #Make sure we loop once
while entry :
batch = []
while len(batch) < batch_size :
try :
entry = iterator.next()
except StopIteration :
entry = None
if entry is None :
#End of file
break
batch.append(entry)
if batch :
yield batch
##MAIN PROGRAM
##Here is our paired list of FASTA files
subject = open("C:\\Users\\Harry\\Documents\\cgigas\\subjectFASTA.fasta", "rU")
query =open("C:\\Users\\Harry\\Documents\\cgigas\\queryFASTA.fasta", "rU")
##Query Iterator and Batch Subject Iterator
query_iterator = SeqIO.parse(query,"fasta")
record_iter = SeqIO.parse(subject,"fasta")
##Writes both large file into many small files
print "Splitting Subject File..."
for j, batch1 in enumerate(batch_iterator(record_iter, 10)) :
filename1="groupA_%i.fasta" % (j+1)
handle1=open(filename1, "w")
count1 = SeqIO.write(batch1, handle1, "fasta")
handle1.close()
print "Done splitting Subject file"
print "Splitting Query File..."
for k, batch2 in enumerate(batch_iterator(query_iterator,10)):
filename2="groupB_%i.fasta" % (k+1)
handle2=open(filename2, "w")
count2 = SeqIO.write(batch2, handle2, "fasta")
handle2.close()
print "Done splitting both FASTA files" print " "
##This file will hold the alignment scores in a tab deliminated text
f = open("C:\\Users\\Harry\\Documents\\cgigas\\alignScore.txt", 'r+')
i=0
for x in range(k):
##Open the first small file
subjectFile = open("C:\\Users\\Harry\\Documents\\cgigas\\BioPython Programs\\groupA_" + str(x+1)+".fasta", "rU")
queryFile =open("C:\\Users\\Harry\\Documents\\cgigas\\BioPython Programs\\groupB_" + str(x+1)+".fasta", "rU")
smallF = open("score_" + str(x+1) + ".txt",'w')
##Make small file iterator smallQuery=SeqIO.parse(queryFile,"fasta")
##Cycles through both sets of FASTA files
for base in SeqIO.parse(subjectFile, "fasta"):
i=i+1
currentQuery=smallQuery.next()
##Verify every pair is correct
print " "
print "Pair: " + str(i)
print "Subject: "+ base.id
print "Query: " + currentQuery.id
a = pairwise2.align.globalxx(base.seq, currentQuery.seq, score_only=True)
score=str(a)
print "Score: "+ score
smallF.write(score+"\n")
print "New file"
May 29, 2012
Just finished writing a program in BioPython that will get all of the scores needed to run RECON. Need to implement something that will take the scores write them into a tab deliminated file then I can copy and paste it into my MSPs file and run RECON.
##Open reference file
##a.)Upload subjectList
##b.)Upload query list (a and b are pairwise data)
- Cycle through each element and get alignment score of each
from Bio import pairwise2
i=0
##Here is our paired list of FASTA files
subject = open("C:\\Users\\Harry\\Documents\\cgigas\\shortSeqSet.fasta", "rU")
query =open("C:\\Users\\Harry\\Documents\\cgigas\\shortSeqSet.fasta", "rU")
##Iterate over the query
query_iterator = SeqIO.parse(query,"fasta")
#This file will hold the alignment scores in a tab deliminated text
f = open("C:\\Users\\Harry\\Documents\\cgigas\\alignScore.txt", 'r+')
##Cycles through both sets of FASTA files
for base in SeqIO.parse(subject, "fasta"):
i=i+1
currentQuery=query_iterator.next()
##Verify every pair is correct
print "Pair: " + str(i)
print "Subject: "+ base.id
print "Query: " + currentQuery.id
a = pairwise2.align.globalxx(base.seq, currentQuery.seq, score_only=True)
score=str(a)
print "Score: "+ score
print " "
f.write(score+"\n")
Next I get to run with the larger files and put the output into RECON. Program currently running with large (>77,000 sequence pairs) data file.
Quickly got a memory error but I should be able to get around it. BioPython recommends that I split my gigantic FASTA file into smaller "batches".
Dealing with Large Python Files
May 26,2012
(From Galaxy)
Example
Typing the following values in the form:
Chromosome: chrX Start position: 151087187 End position: 151370486 Name: NM_000808 Strand: minus</span>will create a single interval:
chrX 151087187 151370486 NM_00080Working on workflow in galaxy to get pairwise alignment scores for MSPs file.
Taking file gigasMSP to construct 2 intervals of regions from this I will use my workflow to calculate the alignment score using the Smith-Waterman algorithm to get the most exact score. From this I will take the scores plug them back into the original file to have a finished MSPs file to run in RECON.
1.gigasMSP
2.queryInterval and subjectInterval used to calculate alignment scores with Smith-Waterman algorithm
3.use scores to make MSP file with gigasMSP
Looks like galaxy is being a little buggy. I get as far as extracting the sequences from the reference genome and then when it performs the Smith-Waterman algorithm it only considers the first subject interval. I will try again with the Needleman-Wunsch global alignment algorithm. I just need the score to use for RECON.
May 24, 2012
Now that the repeat regions have been identified the challenging part has begun which is classifying the repeat regions into specific families. I will need to go back to RECON to do this. I already made 1 of the input files necessary (seqnames) earlier. I need to construct a MSPs file which is similar but not exactly the same as the output from the MUMmer3.23 software.
Should be of form,
Score %iden start1 end1 query_name start2 end2 subject_name
I have constructed/know how to construct all of the fields except the first one. But the following link was illuminating in the type of data needed to use the RECON program. My mistake in using before was trying to have it define repeats with a sequence. Actually, the program looks for adjacent repeat regions (that I already found) to cluster repeat families together (no reference sequence). Now I think I understand the mechanics of the algorithm better and hopefully can get it running before long.
BLAST info
May 22, 2012
Have a list of repeat regions that has been shortened by removing paired repeats that have the same start and end position. Admittedly, this means that if a repeat region happened to start and end at the same position on two different segments it would not be considered. However, this should not be too common. After this step, I need to cluster pairs of aligned DNA segments into repeat families and filter out non-TE repeats (ie: Tandem repeats which was already seen with the shortest repeat section and nested repeats which will intrinsically be repeated in the larger repeat sequence). Can be implememented in RECON or in GROUPER.
coordinatesEX.txt
Attached is the beta GFF file of repeat regions, however it has not been verified
cgigas_repeats_beta.txt
May 17, 2012
Submitting the FASTA sequences to the reputer website for analysis. Also I have contacted some people across the street in the Genomics Department. For now I am sending the data to the online submissions at REPuter 5MB at a time (as is the maximum). Found a "w" in the sequence at line 367, "s" at line 874, "m" at 890, a "y" at 894 I replaced all with an N. Going to write a quick program to make sure the file is nothing but
A,C,G,T,U,N or else I may be doing this song and dance all day.
Ran into new error REPuter doesn't take multiple sequence FASTA files? To get around it I am going to submit the FASTA as a single sequence, seems to be working although it is unsatisfying to do that to the data. Yet if it finds a repeat I think it would be unlikely that it would be at the border between to sequences although it certainly is possible. Here is the code I used to fix the FASTA file. I made no attempts to make it concise the code was made to work and thats all. I also had to add N's at the end of each incomplete line as a quick fix to make sure that the file is a single block of sequence.
REPuter Online Submissions
import java.util.*;
import java.io.*;
publicclass charVer{
private String path;
publicstaticvoid main(String[] args)throws IOException {
Open the file
File mastaFasta = new File("C:\\Users\\Harry\\Documents\\cgigas\\firstForty5.txt");
Scanner console = new Scanner(mastaFasta);
Output file
PrintStream output= new PrintStream("riley1.txt");
Now we need to scan through console
int filenum=0;
int i=0;
int m=45;
int k=0;
while (console.hasNextLine() && filenum<=m) {
String line=console.nextLine();
k++;
if(line.charAt(0)!='>'){If it is not a title line
for(int j=0;j<line.length();j++){//Test each character of the line
char test=line.charAt(j);
if(test!='A' && test!='C' && test!='T' && test!='G' && test!='N'){
line=line.replace(test,'N');
}
}
output.println(line);
}else{
System.out.println("We are on sequence " + i + " of " + m);
i++;
}
}
}
}
Quite unhappily I keep getting this error message
reputer - ERROR (-700)
Using reputer on BiBiServ creates an error (Code : -700) with following error message :Calling WebService reputer::request generates an error org.apache.xmlrpc.XmlRpcException: an general error occurred running reputer (bibiserv_1337282613_6039)
Contacting the people at REPuter to see what this could be. Meanwhile, I will try yet another transposable element software RepeatMatcher/MUMmer. While this program isn't explicitly for repeat searching it has some charateristics
that allow for easy identification of repeats. I need to look at this a little more but it seems very promising.
From website
"Although MUMmer was not specifically designed to identify repeats, it does has a few methods of identifying exact and exact tandem repeats. In addition to these methods, the nucmer alignment script can be used to align a sequence (or set of sequences) to itself. By ignoring all of the hits that have the same coordinates in both inputs, one can generate a list of inexact repeats. When using this method of repeat detection, be sure to set the --maxmatch and --nosimplify options to ensure the correct results.
To find large inexact repeats in a set of sequences seq.fasta, type the following and ignore all hits with the same start coordinate in each copy of the sequence:
nucmer --maxmatch --nosimplify --prefix=seq_seq seq.fasta seq.fasta
show-coords -r seq_seq.delta > seq_seq.coords"
*UPDATE*
coordinates.txt
I may finally have found a file full of repeats as described above all that is left (for the first step of de novo TE discovery) is to remove hits that have the same start coordinates. This is extremely exiciting to be finally start collecting repetitive elements in the sequences. Certainly, there is more to come. The text file is above to verify, obviously there is lots of junk info in this file but it shouldn't be to hard to find a sequence with 2 different start positions. However, admittedly I do not know what all of the fields are immediately. Still, very interesting to have available.
May 15, 2012,
After trying to run the full FASTA file, I realized that doing some of the operations on it will take prohibitively long ( I started one operation at 11 pm last night and it ran all night into this morning and it still hadnt finished). I am going to try the program on a piece of the sequence (The first 5 sequences). Then I will consider running the program on the cluster in the labs.
Got around to emailling the lead programmer explaining my troubles converting the FASTA file to the correct format. I have tried different variants of this command with little success
"grep >AB259818mod_ER_mRNA ~/Documents/Bioinformatics/testSeq | sort > output"
Im going to look into what other software options are available as well ie: I am trying the abbreviated sequence on the RepeatMasker Web Server to see how well that works. The problem however with repeatmasker is it is not a de novo algorithm so it must rely on a library of know transposable elements and would thus fail the "Platypus test".
Using the first 7 sequences and the repeat masker algorithm the output is given by
Analysis of first 7 sequences
May 11, 2012,
For future reference the command line for opening the seqnames file and MSPs file is
(while in home directory defined by recon.pl in the demos case the "bin" directory)
~/Documents/RECON1.05/bin$ ~/Documents/RECON1.05/scripts/recon.pl ~/Documents/RECON1.05/Demos/input/seqnames ~/Documents/RECON1.05/Demos/input/elegans.msps
Now I need to convert my FASTA file into a seqnames file and an MSPs file.
Having difficulty with the conversion I will email the lead programmer tommorrow.
May 8, 2012
Downloaded RECON from
http://selab.janelia.org/recon.html
and completed the demo to familarize myself with the program. This program is used for de nove transposable element discovery. Next step is to use our FASTA file. May run in to trouble later depending on what an msps file is. Also, this is not so straight forward as tandem repeat searches as this is only a preliminary step and more will be needed afterward to clean up the data according to what I have read about TE discovery.
May 3, 2012
Wrapping up the Microsatellite GFF file. As of now I have an excel file with all of the repeats identified by the tandem algorithm and a couple repeats that were identified with equicktandem because etandem failed to
work on those particular sequence for whatever reason. As well as about 11 repeats that were identified visually and not picked up by the etandem algorithm. GFF file will be up tonight for viewing.
*UPDATE: GFF3 complete only thing that can be improved on is adding an Sequence Ontology Accession number to the type field.
. MicroSatellite_GFF_Final1.txt
May 1, 2012
Working GFF!! I dont know why I had so much trouble on Thursday but I have verified it with the website I used Thursday.
Cgiga_tandems.txt
Now I will spend today looking for ways to filter this file and leave only relevant reads. EMBOSS recommends that I use tandem to verify these equicktandem reads.
However, this was extremely time consuming and I would like to do something else if possible. I am also toying with the idea of writing my own program to analyze these repeats.
I think this may be a good application of Shannon Information Theory to quantify how much uncertainty is in this potential repeat region, but I need to read more into this. I believe BioPython
will allow me to isolate the tandem regions. Then I can analyze the sequence and have use some uncertainty metric to decide if the region is a bona fide repeat or not.
According to the EMBOSS API, equicktandem (the algorithm I used to find the tandemrepeats), looks for segments in which each base matches the base before it. That is it does not look
for a consensus sequence for the whole block (etandem does that). The given score is +1 for a matched base and -1 for a mismatch. Looking at the sequences Thursday it appeared that larger sequences
may have a misleadingly large score but may not be a bona fide tandem repeat. (That is we would intrinsically expect that a repeat that is 700 bases long to have an inflated score even if it really did not
repeat often.
Alternatively, I have figured out how to use Galaxy's workflow to automate what I was doing by hand a couple weeks ago. This is in conjunction with me downloading an FTP software to further speed up this process.
Now I am having issues with etandem reading my sequence files, but I wasn't having this trouble earlier today. Once thats figured out though I can very quickly go through with what took me days of work, which is exciting
April 26, 2012
Trying to validate my GFF file. Have Integrative Genomic Viewer which is not displaying my GFF file. Could be an issue with the FASTA file needing to be indexed.Just the same, the formatting looks correct. So here is the draft of tandem repeats. I am able to upload the GFF3 file to ARGO but it is a mess which leads me to believe it is FASTA formatting issues.
Today I will try make a trimmed version of this that will remove bad reads. Looking for pattern between score and repeat length that will yield "good" repeats
- Lets concentrate on tandem repeats where the repeat units are <9 (Microsatellites)
- Repeat units should be larger than 1(That is omit sequences like AAAAAAAAAAA; These 2 conditions alone knocks our 4000 hits down to less than 1000 but we still have some bad reads)
- More to come...
I noted today that some of the "repeats" on MacKenzie's data may have been bona fide repeats but were to large to be noticed visually. I need to verify that these GFF files were made correctly but Steven and Mac are out today.
http://modencode.oicr.on.ca/cgi-bin/validate_gff3_online
Is a MUCH better GFF verifier (despite the fact they look exactly the same, I have had better luck verifying my GFF files here).
Still, I am getting error messages and without a way to visually check that this GFF works, I dont know if the following text is correct or not but here it is anyway and the verifier also yields the following message.
Line Number Error/Warning ----------- ------------- 3565
[ERROR] illegal character in field (seqid: Contig_54493_v032
Which doesn't make much sense to me. Everything should be properly formatted, hopefully Steven or Mac can shed some light on this.
April 24, 2012
First, very happy with UNIPRO as it will go find all the tandem repeats and annotate them for me. I am unsure now where to take this, some of the "repeats" flagged by the algorithm I am unsure of but leaving things on the microsatellite setting leads to nice searches.
Lets convert my massive data set to a GFF3 file.
From there I can later try and clean up the data and leave only the relevant data.
Validating the GFF3 file at
http://modencode.oicr.on.ca/cgi-bin/validate_gff3_online
Which is taking a long time. Im just going to see if they will work on gBrowse. While that is installing I noted that UNIPRO also had a nice feature that will upload annotations in CSV format and will let the client be able to pick the important data fields. Very user friendly. I would like to have GFF3 files before Thursday and then clean up the data. Then move on to finding transposable elements or evidence of miRNA.
April 20, 2012
It is apparent to me that to continue doing my research I will need to be downloading open source software. Windows is notoriously anti- open source so I am taking today to install ubuntu linux virtual machine. This will let me get software to find repeats and hopefully even get an instance of jbrowse going.
Trying UNIPRO bioinformatics software out, to see if they have any TE tools I can use.
April 19, 2012
First thing, lets find out what the seemingly unnecessary data on the equicktandem searches looks like.
A size 1, length 28 equicktandem repeat looks like this.
EU342886_hexokinase (bp 6995-7022)
CCCCCCCCCCCCCATAAAAAAAAAAAAA
Another example is,
AM905317_gnrhr (bp 11842-12072) (Size 1, Count 231)
AAAAATANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGAAGGTGGATTTTAATGAAAATTTACCTTCGGGATTTTTCCATTTTGGAATAGTGTTTTTGAAAAGGTTCAAGGCAATTAAAGTGGGGATTCCATTTGTAAATGGCGTTTTTTATTTTTTTCCC
Which does not look like a tandem repeat.
However,
GU207436cgigasBAC (bp 12629-12650)(Size 1, repeats 22) looks like this
AAAAAAAAAAAAAAAAAAAAAA
which is a reasonable hit.
Some single repeat examples
GU207422cgigasBAC (bp 135445-135487) (Size 22, Count 1)
AAAGAACATCATGGGGGATATTATTGCCCATGCAGCGGAAAA
While I notice after this the sequence may appear slightly similar to the original sequence above
ACGAAAGAGCATT
I hardly think this justifies a tandem repeat.
Contig_29545_v032 (bp 299-352) (Size 29, Count 1)
TCCCTTATTACCCCAGGGTCAGTGTCCGATCCCTTCTTACCCCAGGGTCAGTGT
plus the following chain of sequence
GAGAGTTCCCTTCTTACCCCAGGGTCAGTGCCCGAGTTCCCTTCTTACC
Again, pretty close. Hence, I may have erroneously interpreted count 1 to be a single chain when it is actually 1 repeat. Just the same I will add a trimmed set of tandem repeats excluding single repeats and single nucleotide sequences.
Raw data had 4139 entries, trimmed data has 2167.
cgigas_tandemrepeats_trim.xlsx
As of now I don't know what else to do with this catalog so I will begin pursuing a different topic. I would like to try to find some evidence of miRNAs in the sequence file, however my laptop is dead and winzip is not on the lab computer so I will put this off until later. Alternatively, I will try to look for transposable elements.
For future reference,
G. Benson, "Tandem repeats finder: a program to analyze DNA sequences" Nucleic Acid Research(1999) Vol. 27, No. 2, pp. 573-580.Will find tandem repeats of multiple sequence FASTA files, much to my chagrin and embarrassment.
To search for Transposable elements (TE's) I have several software options.
- RepeatFinder- takes output from Reputer/Repeat Match and then will merge overlapping and neighboring repeats to detact non-exact repeats. There are faster methods using hashing and k-mer approaches. However, the data set we are using is relatively small and I will take a thorough algorithm in light of a more efficient one. Can be used for FINDING repeats but further analysis will be required after this.
- After repeat regions have been identified, the next step is to filter TE repeats from non-TE repeats. This can be done with RECON or GROUPER tool from BLASTER suite.
- PILER program may do all of this without using 2 programs, RepeatFinderToolKit, RepeatGluer and RepeatScout are other options for De Novo TE searches.
Unfortunately, without my laptop working and without winzip on this computer. There is little else for me to do here at the labs today. I will try to get started on my search for TE's today and continue more tomorrow here at the labs.
April 17, 2012
Continued cataloging microsatellites have completed 222 of the 272 sequences. Will have complete catalog completed before thursday. I need to look into the accuracy of the equicktandem as i think i may have read something about it being a unrobust algorithm.
*UPDATE*
First draft of catalog I could potentially remove uni-nucleotide repeats and single multi-nucleotide "repeats" but for now here is the raw catalog of microsatellites using Galaxy's default settings ( Max repeat size=600, Threshold score=20)
cgigas_tandemrepeats.xlsx
Enjoy.
April 12, 2012
Wrote a short java program to split up larger fasta file into smaller text files that I can feed into Galaxy's equicktandem to find microsatellites. Run in galaxy and SUCCESS!
equicktandem on >GU207417__cgigasBAC yielded 36 hits
equicktandem-GU207417_cgigasBAC.table
equick tandem on > putative_vtg wont upload but it did not have much of a hit
SeqName Start End Score Strand Size Count
putative_vtg 4522 4663 39 + 75 1
equick tandem on the other wont upload so here they are
SeqName Start End Score Strand Size Count
FJ347754_LAP 309 373 63 + 2 32
FJ347754_LAP 3651 3692 40 + 2 21
FJ347754_LAP 5443 5495 21 + 2 26
FJ347754_LAP 4769 4950 21 + 157 1
SeqName Start End Score Strand Size Count
EU342886_hexokinase 6995 7022 21 + 1 28
EU342886_hexokinase 6725 6794 20 + 44 1
EU342886_hexokinase 5396 5875 23 + 431 1
EU342886_hexokinase 7102 7579 20 + 456 1
EU342886_hexokinase 4660 5160 22 + 467 1
SeqName Start End Score Strand Size Count
AY780357_timp 3678 3814 23 + 94 1
>AY660003_caa NO HITS
SeqName Start End Score Strand Size Count
AM905317_gnrhr 2312 2411 99 + 1 100
AM905317_gnrhr 4258 4371 101 + 1 114
AM905317_gnrhr 7807 7916 101 + 1 110
AM905317_gnrhr 8723 8822 99 + 1 100
AM905317_gnrhr 9358 9522 112 + 1 165
AM905317_gnrhr 11842 12072 104 + 1 231
AM905317_gnrhr 2583 2617 21 + 10 3
AM905317_gnrhr 6915 7025 25 + 36 3
AM905317_gnrhr 6485 6885 279 + 52 7
AM905317_gnrhr 6375 6477 22 + 57 1
AJ564739_GS NO HITS
SeqName Start End Score Strand Size Count
AJ543432_Tld 7854 7937 43 + 5 16
SeqName Start End Score Strand Size Count
AJ512213_pgm 2565 2620 34 + 2 28
AJ512213_pgm 3583 3619 31 + 2 18
AJ512213_pgm 6667 6721 26 + 9 6
AJ512213_pgm 3449 3488 20 + 20 2
AJ512213_pgm 1460 1506 21 + 22 2
SeqName Start End Score Strand Size Count
AJ309316_ALR1 1866 1930 57 + 2 32
AJ309316_ALR1 1590 1699 22 + 78 1
>AJ305315_hsc70 NO HITS
>AB259818mod_ER_mRNA NO HITS (which makes sense in light of the problems I had Tuesday)
While I am pleased to be getting some results unlike Tuesday. I am unhappy that there are only 12 sequences here. After looking it up there is a limit of the number/size of characters you copy and paste so this represents a small portion of the sequences. So the first sequence will need to be redone but the rest should be okay.
*UPDATE* Fixed I now have 272 sequence txt files on my computer expect more searches to be posted
April 10, 2012
Searching for microsatellites earlier on Galaxy did not yield any results though I suspect that it should have. Currently, troubleshooting the problem by trying to get a FASTA file with an obvious/known tandem repeat.So I made one up.
>gi|129295|sp|P01013|OVAX_CHICK GENE X PROTEIN GAGAGAGAGAGAGAGAGACGCGCGCGGCCGGCCGGCCGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGAGTCGA
Sequence: OVAX_CHICK from: 1 to: 102
- HitCount: 2
# Threshold: 20
- Maxrepeat: 9
#=======================================
Start End Score Size Count
1 26 20 2 13
38 102 60 5 13
So the algorithm worked for this short made up sequence but not for our collection of 271 sequences. Perhaps a formatting error ended up in the text file somehow? Isolating the sequences does not yield any hits. I have posted a discussion post on the Galaxy message board to see if they can shed some light on the problems I am having. I am also considering using other sftware such as Sputnik or Msatcommander. Also, im going to head over to the lab computers now and see if the software on those computers may locates some microsatellites.
Had better luck finding microsatellites with CLC genomics find function and just typing a tandem repeat obviously thats not how I will continue searching for them but that confirms the rather obvious suspicion that there is microsatellites and for whatever reason I am not identifying them with Galaxy's equicktandem.
*UPDATE*
Just got a mildly reassuring email from the people at Galaxy.
"Galaxy exposes those old emboss tools but they don't get a lot of use AFAIK. Checking the documentation at http://emboss.bioinformatics.nl/cgi-bin/emboss/help/equicktandem seems to show that the tool accepts only a single sequence - not a fasta file with multiple sequences - so maybe the code from emboss is stopping after reading your first sequence?"
Hence, I may need to change file formats to make it work, but it seems promising.
April 5, 2012
Lets try and install Jbrowse,
Downloaded Perl 5.14.2 Win64bit.
Next looks like I need BioPerl
http://www.bioperl.org/wiki/Installing_Bioperl_on_Windows
ppm> repo add uwinnipeg/trouchelle yielded the following message
ppm repo failed:constraint failed
repo.packlist_uri may not be NULL
Regardless, it looks like BioPerl installed.
Other needed packages added:
- JSON 2
- Heap:Simple
- Heap:Simple:XS
Can't find and apparently may need
- Heap::Simple::Perl
- PerlIO::gzip
- Devel::Size
Having difficulty installing requesting the following article from the UW libraries to help me out.
Current Protocols in Bioinformatics: Setting up the JBrowse genome browser
Skinner, M
Paper:
http://cl.ly/0D2A1K2W1U3C0Y1W042L
Also, It may be easier to use Amazon's machine image
http://biowiki.org/view/JBrowse/AmazonMachineImage
Additonally, running on a Linux/Unix Virtual Machine may simplify installation. Not much luck installing today. I am going to come back to this tonight/this weekend.