11/08/2011
Summary:
The purpose of lab 6 was to (1) isolate and extracting total RNA from C. gigas gill tissue from all three dissolve oxygen acclimation groups and (2) produce cDNA.
Materials & Methods:
RNA Extraction
1) ~0.002-0.003g of gill tissue from each osyter (N=15) was homogenized with a sterile pestle in 500ul of TriReagent in a 1.5ml microfuge tube
2) Shortly vortex
3) Additional 500ul of TriReagent was then added to the test tube
4) Vortexed for 15s
5) Homogenized tissues were incubated at RT for 5min
6) In the fume hood 200ul of chloroform was added to each sample
7) Each sample was then vortex for 30s or until solution becomes "milky"
8) The samples were then incubated again at RT for 5min
9) The samples were spun in a refrigerated (4oC) microfuge for 15min at max speed
10) The tubes were gently removed
11) The aqueous phase (clear top layer) ONLY was carefully transferred into a separate 1.5ml microfuge tubes
12) 500ul of isopropanol was then added to each new tube containing the aqueous phase
13) The solution was mixed well by inverting the tubes several times (when solution no longer appeared viscous or lumpy, particularly near the edges)
14) The samples were then incubated at RT for 10min
15) Spun down at max speed for 8min with the TUBE HINGE away from the center
16) A small white pellet should have formed (if not visible, just continue)
17) The supernatant was removed
18) 1ml of 75% Ethanol (EtOH) was added to each pellet
19) Each pellet was vortex to dislodge pellet
20) Samples were spun in refrigerated microfuge (4oC) at 7500g for 5min
21) Supernatant was carefully removed, without removing the pellet
22) 15s of spinning to pool any remaining EtOH
23) Residual EtOH removed with small pipette tip (P10 or P20) and blotted with KIM wipes (if necessary)
24) Tubes left open to dry at RT (in the hood) for no more than 5min
25) Due to the larger size of the pellets, 200ul of 0.1% DEPC-H2O (instead of 100ul) was added to each tube to resuspend the pellets
26) The samples were then incubated in 55oC water bath for 5min to help solubilize the pellet
27) Each test tube was flicked a few time to mix. If flicking didn't work, the solution was pipetted.
28) The samples were then stored at -80oC
10/19/2011
Selected C.gigas primers from NCBI: GenBank
Primer pair 1: HIF-1a (accession: AB289857)
HIF-1a GACCCACCTGGCTCCGACCT
HIF-1a GCAGATCCGGGCGCTGAACA
Product: 130
Primer pair 2: hrIGF (accession: AJ535699)
hrIGF ATCCGGGCCATCTCCCTGGC
hrIGF GGCGATGAACCACACGCCGA
Product: 171
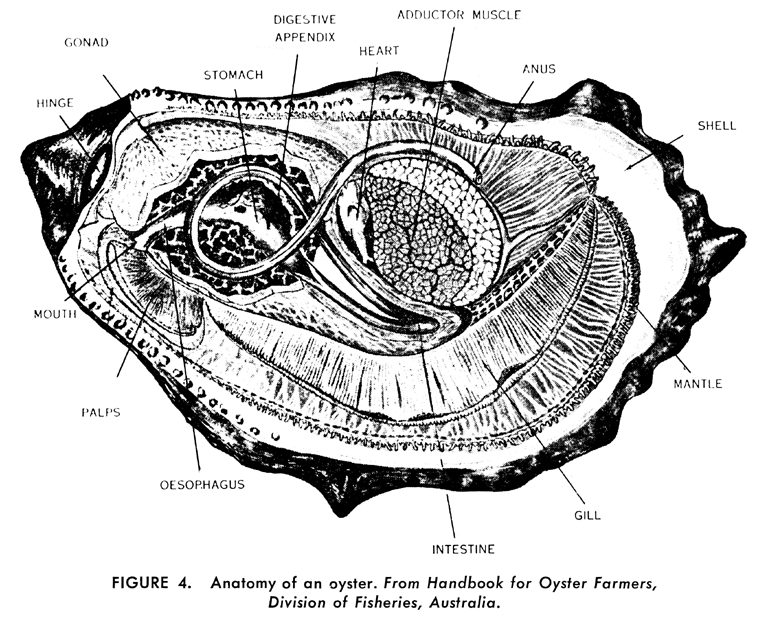
Anatomy of an Oyster
------------------------------
Question: Does acclimation to different dissolve oxygen (DO) concentrations result in different HIF-1α expression in Pacific oysters?
Importance: Hypoxia is becoming a regular feature in estuaries and semi enclosed seas, increasing in magnitude, extent, and frequency. If turnover rate of high oxygenated water is slowed or ceased will pacific oysters be able to acclimate to more permanent hypoxic conditions? In addition, how does this affect growth?
Hypotheses:
(1) When exposed to hypoxia, Pacific oysters acclimated to a lower DO environment (e.g., DO = 4mg/l) will express lower levels of HIF-1α mRNA than the conspecifics acclimated in more normoxic/hyperoxic conditions.
(2) When exposed to hypoxia, Pacific oysters acclimated to a lower DO environment (e.g., DO = 4mg/l) will express higher levels of hrIGF mRNA than the conspecifics acclimated in more normoxic/hyperoxic conditions.
Experimental Design: Oysters (N = 15; n = 5) will be acclimated to three different oxygen levels: high DO (9 mg/l), normoxia (6-7mg/l), and low DO (4mg/l) at 13± 1oC. The high DO saltwater will be prepared by pumping O2 gas into the water until DO = 9mg/l. The normoxic tank will consist of a simple flow through system, while the low DO acclimation will be prepared by bubbling nitrogen gas until DO = 4mg/l; the hypoxic treatment tank will be prepared in the same manner, but the DO will be reduced to < 2 mg/l. A total of five oysters will be placed in each acclimation tank for 24hrs. After acclimation all 15 oysters will be transferred into the hypoxic tank ( DO < 2mg/l) with two mesh dividers to separate each treatment group (Figure 1). After hypoxic treatment for 72hrs, gill tissue samples will be collected from each oyster for RNA analysis.
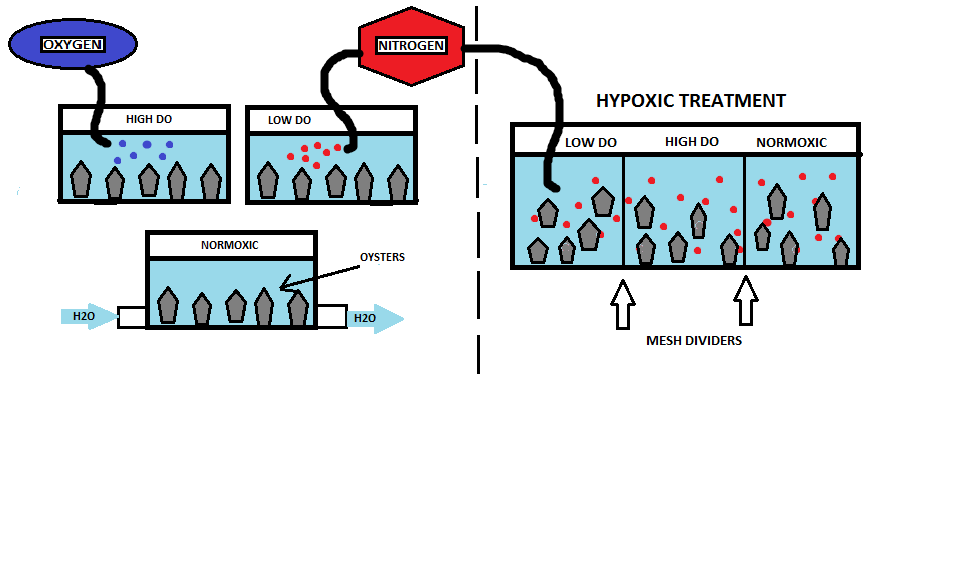 |
| Figure 1. Set up of the three acclimation tanks (left side) and hypoxic treatment (right side). |