November 27, 2012
Lab 11: qPCR and Experimental Results
Objective: QPCR was ran for astacin gene using the cDNA isolated from the previous weeks lab.
Methods and materials:
- A master ix was made for qPCR
| Reagent |
Master mix 1x |
Master mix 25x |
| Master mix |
12.5uL |
312.5ul |
| SYBR |
1uL |
25uL |
| Upstream Primer |
0.625uL |
15.625uL |
| Downstream Primer |
0.625uL |
15.625uL |
| Pure water |
8.25uL |
206.25uL |
- One thing to note is that the primer amount was cut in half from previous qPCR ran. This was due to early annealing to eachother.
- 23uL were pipetting into 24 wells (2 rows).
- The first row added 2uL of cDNA samples from pacific 1-10 and the last two wells were blank, using 2uL of pure water.
- The second row added 2uL of cDNA from cyclo 16-25 and the last two wells were blank
- The same process was ran for both astacin gene and pger gene.
- QPCR was then ran for each of the genes:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 30 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Results:
Figure 1:
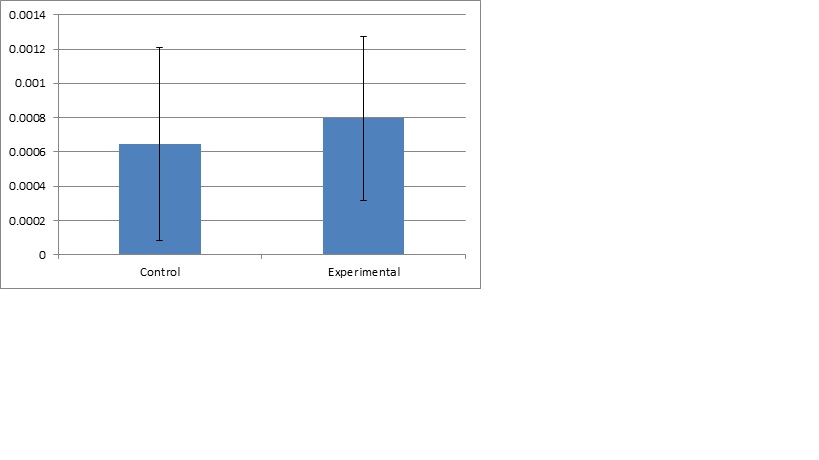
Figure 2:
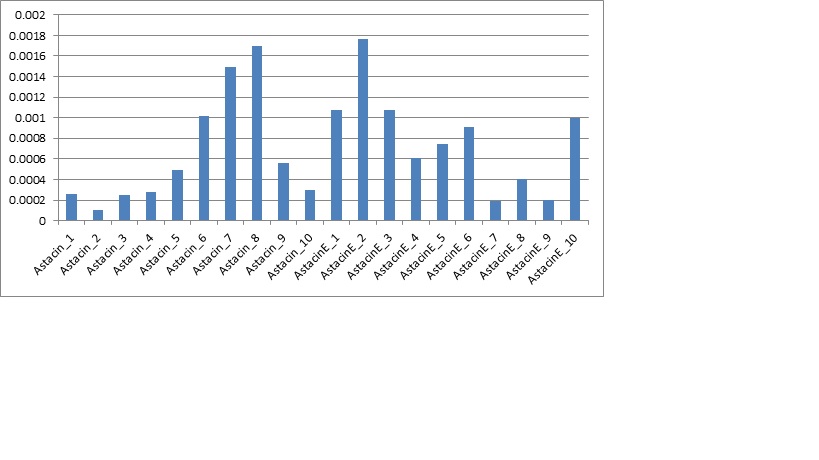
Figure 3:
Conclusion:
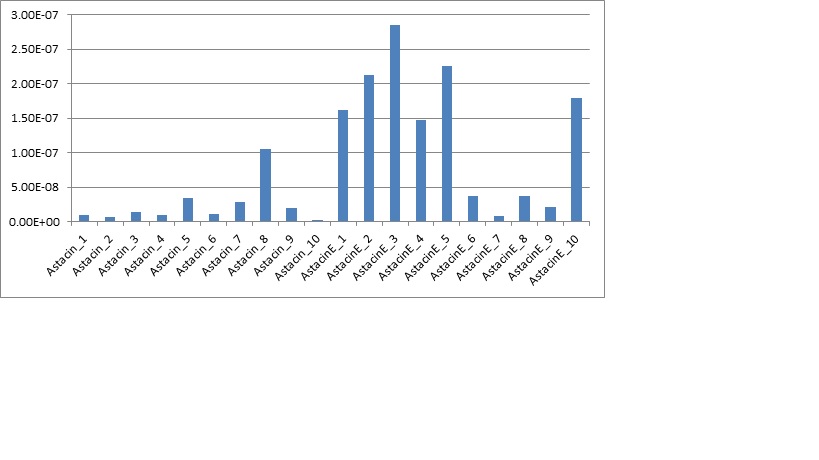
The qPCR ran for the gene astacin was the final step to the research project. This gave the hard data and numbers required for us to crunch and to actually see if it was significant. After the qPCR was ran for both astacin and PGER, only astacin produced any data worth taking. The PGER data had multiple melting points in the blank samples as well as multiple melting points in all of the cDNA samples. This could have been a result of contamination or it could have been due to genetic material. Overall, the data was messy and wouldn’t give anything reliable. Next was the gene expression for astacin, the immune gene for cytoskeleton development in the pacific oyster. The data looked clean from the qPCR, and had only one melt curve with blanks expressing nothing. After looking at it, the data was exported to a file, then ran through PCR miner. This gave us a gene efficiency and CT for the wells. Then using a normalizing gene (which doesn’t change expression from control to experimental) was compared to the gene expression of the astacin gene. This would give a unitless gene expression for both the control and experimental group. Then the averages were graphed with standard error bars, as along with a graph of all samples.
Looking at the data, the experimental group had a slightly higher average of gene expression, but due to the large standard error bars, the data ended up not having significance. The p-value was 0.51, which leads me to the conclusion of accepting the null hypothesis. What this actually means is that the control has a slightly lower average of gene expression compared to the experimental group, but when looking at the standard deviation bars it becomes clear there is a lot of variance. This variance becomes even more clear after graphing the individual plot (fig2), and seeing how spread out the data is. The control has low expression and high expression, as well with the experimental. These results are not what was expected, but leaves room for discussion.
Another point to note is that the astacin gene showed great expression and was clean data, on the other hand the pger gene was not so clean. Most wells had multiple melting points, some before and some after the projected. This could have been a result of either primer dimer or some sort of genetic material that got into the samples. Overall, the data was too messy to make anything of it and was then disregarded to go any farther.
Reflection:
Overall the purpose of this lab was to obtain final data for the experiment that was conducted throughout this quarter. The procedures we used (pcr) were used to measure overall gene expression of a specific gene by using primers made to amplify the region. Any research that is targeted towards specific genes or overall gene expression would use this methods of qPCR because of the fact that it can be quantified. This was the third time running qPCR so there was not many questions up for debate, but things that were unclear was the data. It makes sense to look at the two groups gene expression, but what else should be looked at to consider certain trends and other info in the data. This was the end of the project and leaves room for further research in this area or maybe even allowing me to test another gene before the end of the quarter.
November 20, 2012
Lab 10: Primer Rehydration, Testing Primers, Prepping cDNA
Objective: Primers were rehydrated and tested using qPCR from two immune compromised cDNA samples. Also, isolated RNA was placed into wells for preparation of cDNA to be made.
Methods and Materials:
Primer Rehydration:
- Based off the weight of the primers, water was added.
- For my primers:
| Primer name: |
Weight (nm) |
Water added (uL) |
| Cystatin B fwd |
33.4 |
334 |
| Cystatin B rvs |
30.9 |
309 |
| PGER-EP4 fwd |
37.0 |
370 |
| PGER-EP4 rvs |
28.9 |
289 |
| Astacin fwd |
32.4 |
324 |
| Astacin rvs |
33.0 |
330 |
- Both forward and reverse were vortexed to mix the primer into the water.
- Next a stock and working stock sample were made. The stock sample consisted of 10uL of primer solution and 90uL of water. The working stock was 10uL of the primer solution. These were made for both the forward and reverse, which resulted in 4 tubes for each primer.
- Working stock was labled “WS-fwd”,”WS-rvs” and the stock was labled “S-fwd”,”S-rvs.”
- These four samples for each primer were placed into the freezer for qPCR amplification later on.
Primer Testing:
- A master mix for qPCR was made for each of the three primers using this formula:
| Reagent |
For x1 |
For x7 |
| Master Mix |
12.5uL |
87.5uL |
| Syto 13 |
1uL |
7uL |
| Upstream primer |
1.25uL |
8.75uL |
| Downstream primer |
1.25uL |
8.75uL |
| Ultra-pure water |
7uL |
49uL |
- These master mix solutions provide enough for 6 wells to be filled in a qPCR.
- cDNA from two separate immune compromised hemocyte tissues were used to test the primers.
- Blanks had 23uL of master mix + 2uL of nuclease free water. cDNA samples had 23uL of master mix + 2uL of cDNA.
- Wells went as following:
| Primer |
Well |
2 |
3 |
4 |
5 |
6 |
7 |
| Cystatin B |
A |
(blank) |
(blank) |
(blank) |
(T2 cDNA) |
(PGN cDNA) |
(T2 cDNA) |
| PGER |
B |
(blank |
(blank) |
(blank) |
(T2 cDNA) |
(PGN cDNA) |
(T2 cDNA) |
| Astacin |
C |
(blank) |
(blank) |
(blank) |
(T2 cDNA) |
(PGN cDNA) |
(T2 cDNA) |
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 30 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Results:
Cystatin B showed no real indication of any expression. The blanks had expression which could be the result of primer dimer action, also the well of 5 and 7 showed different expression.
PGER showed expression but also had a couple melting points. Also, the blanks were showing some expression.
Astacin showed expression but also had a couple melting points. Also, the blanks were showing some expression.
Conclusion:
The results were not exactly what I was hoping for, but are manageable to work with still. The primer used for Cystatin B was pretty much a failed attempt, get no clear expression of the gene. PGER and Astacin primers worked to express the gene, but also had expression of the blank wells as along with another melting point, which could be the result of primer dimer action. This should be easily fixed by changing the amount of primer added to the master mix before running the qPCR. The expression showed in the for the second smaller melt temp in the cDNA wells was happening at earlier cycles but once the gene was found, amplification of this region began and the primer dimer action ended. By reducing the amount of primer added to the actual mix, the annealing should occur on the gene we want to express. These results are not exactly what I was expecting because the primers were already once tested and designed and published. The results give me a reason to tweak the primers in order to get max expression of the desired genes. By running this test qPCR I can be sure that I will get some expression of the genes. The next step is to run a qPCR for these genes with the exception of no cystatin B.
Reflection:
Overall this lab was a day to rehydrate primers and test them to ensure they were working properly. All of the RNA isolation was finished up today and placed into wells to be processed for cDNA. These techniques once again are common in a variety of fields that analyze gene expression. Making cDNA is a better alternative to using genomic DNA because there is splicing that can occur when transcription occurs. Testing the primers helped ensure that I was going to get some sort of results for the experimental and control oysters before actually testing it on them. By running an early qPCR, it gave me the ability to tweak the primers or amount of primer used in my master mix for the best result possible. There is only a few more steps to go until it is time to analyze the data. First, the cDNA will be made during the break as a class. Second, the cDNA will be used with our master mix from the desired primers to run a qPCR and determine overall gene expression. Lastly all of this data will then be interrupted and be used to write a research paper. Everything is falling into place and is almost ready to be making the last steps. The only thing I wish there was a little more information on was about primer re-design based off certain results that you get, which might not be desirable. Overall the TA did a great job explaining what to do, but it was hard to find much in literature and readings about how to re-design.
November 19, 2012
Lab 9: RNA Isolation (continued), RNA Quantification
Objective: Finish processing RNA from tissue samples and quantify them using a nanodrop spectrometer.
Methods and materials:
RNA Isolation:
• From the last lab session 11.13 there are 6 samples left to have RNA isolated. Pac cyclodextrin 17-22.
• These are pulled out of the freezer and placed on ice as along with the RNA that was isolated from last week to be quantified at the end of the isolation process.
• The sample was then incubated at room temp for 5 minutes.
• In the fume hood 200uL of Chloroform was added to the sample and then vortexed for 30 seconds at max speed. The sample turns a milky pink texture by the time the vortex is finished.
• Next the sample was incubated for another 5 minutes at room temp.
• Then 15 minutes in the refrigerated microfuge at maximum speed.
• The tube was removed carefully and the supernatant (top clear portion) was removed and placed into a separate tube labeled the same as above except for it saying “RNA o instead of RNA.”
• 500uL of isopropanol was added to the new tube. After, invert a few times to mix to ensure there is no lumpy texture.
• Next another incubation for 10 minutes at room temp.
• After incubation a 8 minutes spin in the refrigerated microfuge at max speed.
• When removed, a small white pellet of RNA is present. The supernatant was removed and placed in waste, leaving only the white pellet.
• 1mL of 75% EtOH was added and vortex briefly to dislodge the pellet.
• Next, spin the tube at 7500g for 5 minutes in the refrigerated microfuge.
• Removal of the supernatant was next, making sure to not remove the pellet.
• A brief 15 second spin was used to pool the rest of the EtOH.
• Using a P20 pipette, the rest of the EtOH was removed and then the pellet was left at room temp with the lid open for 5 minutes to dry.
• Using 100uL 0.1% DEPC H2O the pellet was suspended and then placed into a 55C water bath for 5 minutes to solubilize the RNA.
• After removing the tubes, a few flicks to mix the sample were given and then placed into -80C freezer to save for quantification.
• The RNA isolation protocol was repeated for each of the 6 samples at the same time.
RNA Quantification:
• Only two samples are quantified, one from last week’s lab and one from this weeks lab to ensure the quality of the RNA.
• 2uL of 0.1% DEPC H2O was pipetted onto the nanodrop pedestal.
• On the program “blank” was hit to zero the instrument.
• Next, 2uL of RNA sample was pipetted onto the pedestal.
• On the program “measure” was clicked to give us the data (written in results)
• After the two samples were quantified, they were placed on ice and stored back in a -80C freezer.
Results:
Sample [RNA] ng/uL A260/280 A260/230
Pac control 9 195.5 1.96 3.08
Pac cyclodextrin 18 1304.2 1.98 2.35
Conclusion:
The results achieved from this lab are pretty much what were expected. Realizing that I had added 1mL of 0.1%DEPC H2O instead of 100uL to the first batch showed a clear drop in the [RNA], but still had a high enough concentration to run for cDNA strands and qPCR. The ratios are a clear indication that this RNA is very close to pure with close to zero contaminants. Based off the nanodrop user manual,A260/280 of 2.0 is for pure RNA. http://www.nanodrop.com/Library/T009-NanoDrop%201000-&-NanoDrop%208000-Nucleic-Acid-Purity-Ratios.pdf This link shows that the ratios achieved from this lab fall in the guidelines of pure RNA. This lab sets me up for a few things in the upcoming week: testing primers with qPCR, create cDNA from the RNA isolated, and test for gene expression by running qPCR on the cDNA that we create.
Reflection:
Overall, this lab helped set us up for creating cDNA strands for everyone as a class. This will give everyone the ability to test for their specific genes. The techniques used in this lab were the same when we isolated RNA from the single tissue sample earlier in the quarter, except this time we batched multiple samples together to test more than one sample for gene expression. These techniques are a common first step in anyone trying to observe gene expression because cDNA can be made from the isolated RNA. To measure expression we want to use cDNA rather than DNA because this will be a better representation of the actual gene being expressed due DNA having introns and extrons in the genomic material. The only unclear part I had about this lab was when the A260/230 was above 2.2. This wasn’t explained in the nanodrop manual or anywhere else when I was searching. Other than that, everything was clear and was a repeat of the first time we isolated RNA.
November 13, 2012
Lab 8: Research Project (RNA Isolation)
Objective: RNA isolation of half of the samples (5 Pacific Control, 1 Pacific cyclodextrin) ) and preparation of the other half in tri-reagent.
Methods and Materials:
- Labeled snap cap 1.5mL tubes on the top with the sample number and tissue type (i.e. Pac 12-G) and on the side “JCD, 11.13, RNA and sample number and tissue type.” This was done for Pacific control: 6-10 and Pacific Cyclodextrin: 16-22.
- Next was to add 500uL of Tri-reagent to each of the labled tubes for preparation of tissue.
- Next, each of the tissue samples from the labeled tubes that was isolated 2 weeks back were brought out of the freezer and put on ice in order to get the right amount of each tissue.
- Only the first two samples were weighed: Pac control 6: 0.06g and Pac control 7: 0.068g. After weighing out two samples the rest were eyeballed to size and put into their respective tube with the tri-reagent. NOTE: After each use with the razor blade from a tissue sample, the instruments used to get them into the tube were sanitized using bleach then alcohol then DI water.
- Each of the samples were homogenized using a disposable pestle. After homogenizing, 6 samples were placed into the -80C freezer to be isolated at a later time (Pac cyclo 17-22).
- After homogenizing 500uL of Tri-reagent was added to the tube and then vortexed for 15s at max.
- The sample was then incubated at room temp for 5 minutes.
- In the fume hood 200uL of Chloroform was added to the sample and then vortexed for 30 seconds at max speed. The sample turns a milky pink texture by the time the vortex is finished.
- Next the sample was incubated for another 5 minutes at room temp.
- Then 15 minutes in the refrigerated microfuge at maximum speed.
- The tube was removed carefully and the supernatant (top clear portion) was removed and placed into a separate tube labeled the same as above except for it saying “RNA o instead of RNA.”
- 500uL of isopropanol was added to the new tube. After, invert a few times to mix to ensure there is no lumpy texture.
- Next another incubation for 10 minutes at room temp.
- After incubation a 8 minutes spin in the refrigerated microfuge at max speed.
- When removed, a small white pellet of RNA is present. The supernatant was removed and placed in waste, leaving only the white pellet.
- 1mL of 75% EtOH was added and vortex briefly to dislodge the pellet.
- Next, spin the tube at 7500g for 5 minutes in the refrigerated microfuge.
- Removal of the supernatant was next, making sure to not remove the pellet.
- A brief 15 second spin was used to pool the rest of the EtOH.
- Using a P20 pipette, the rest of the EtOH was removed and then the pellet was left at room temp with the lid open for 5 minutes to dry.
- Using 100uL 0.1% DEPC H2O the pellet was suspended and then placed into a 55C water bath for 5 minutes to solubilize the RNA.
- After removing the tubes, a few flicks to mix the sample were given and then placed into -80C freezer to save for quantification.
- The RNA isolation protocol was repeated for each of the 6 samples at the same time.
Only data were the two tissue weights.
| Species |
Weight |
| Pac control 6 |
0.06g |
| Pac control 7 |
0.068g |
Conclusion:
From this lab there was no real results, it is all in preparation for future labs. This was the first step to isolating all of the RNA samples for the experimental process. Quantification will be ran for all of the RNA samples when the other half are finished being isolated.
Reflection:
Overall, this lab is the first step in ultimately achieving the QPCR of the desired genes. Half of the RNA was isolated in this lab session and the other half will be finished in the next lab session. When the other half has been isolated, spectrometer quantification will be used to determine what amount of RNA was achieved in the isolation process. This lab will give other classmates the ability to use the isolated RNA for research in their experiments as well. For the future, the next batch of RNA isolation and quantification will occur and then cDNA will be made for each of the Pacific controls and Pacific Cyclodextrin samples. After the cDNA has been made, a qPCR can be ran for a few immune genes to see if there was a response in the cyclodextrin bathed oysters. Overall, this lab went very smoothly and it was the first lab were a large batch was processed at once.
November 6, 2012
Lab 7: Protein SDS/PAGE and Western blot, analyze cPCR and qPCR
Summary: The amount of hsp70 protein from the isolated proteins in week 6 lab was analyzed by using a SDS/PAGE and western blot technique. Also, gene expression was analyzed in two ways; by using a agarose gel for conventional PCR and quantitatively using qPCR.
Materials and Method:
Electrophoresis Procedure:
- Agarose gels were already made and prepped for this weeks lab.
- The first step was placing the gels into the electrophoresis boxes and pouring a 1X TAE buffer over them so that all of the wells are fully covered.
- To give a reading on base pair lengths 7uL of 100bp ladder was added to the far left in both the upper and lower wells. This gives us a scale to compare the results to qualitatively.
- Next 20uL of the PCR samples were places into the gel wells. My samples were on the bottom well of the Olympia Oyster gel and went in an order like so: 1: ladder 2:blank 3:blank 4:cDNA 5: genomic DNA.
- The box was then plugged in with a power supply with the cathode on the well side and the anode on the end of the ladder. This would pull the intact base pairs down the ladder based on how long they are.
- The gel was ran at 100V for 1 hour. Starting time: 3:00 p.m. Ending time: 4:00 p.m.
- After the gel was done running, it was carefully removed from the buffer solution and placed over a UV transilluminator that amplified the bands. The picture is in the data and results section.
- This is the process that actually separates proteins out based on molecular weight. The gel is ran through a reducing buffer that helps to get rid of the complications of the different protein structures and other charges they may have associated with them.
- Started by boiling water on a hot plate for preparation of placing the protein in them.
- In a fresh screw cap tube 15uL of protein stock and 15uL of 2X reducing sample buffer was added for each of the protein samples. A brief centrifuge was given to the sample for 10 seconds in order to get the mixture to the bottom of the screw cap tube.
- Next the sample was boiled for 5 minutes. This process denatures the proteins out of their secondary, tertiary, and quartenary states and leads to linear chains of amino acids.
- During this time the preparation of the gel box was put together and the wells were rinsed in preparation for the protein sample.
- After the samples were done boiling, they were immediately centrifuged for 1 minute in order to pool the liquid.
- Next both of the protein samples were loaded into separate wells on the gel.
- The lid was placed on and the power supply was turned on to 150V. The starting time was 2:30p.m. and ending time 3:05 p.m. for a total of 35 minutes.
- The power was turned off, box cracked open, gel was removed from the plastic casing carefully to expose the gel plates.
- Wells were trimmed and a notch was made at the first well position to indicate each position when transferring to a western blot.
- After the gel was removed. Filter papers, membrane and gel were soaked in Tris-Glycine transfer buffer for 10 to 15 minutes.
- After soaking had finished, the semi dry blotting apparatus sandwich was formed on the blotting machine.
- The order of the sandwich goes at followed: Anode base (+++), filter paper, filter paper, membrane, gel, filter paper, filter paper, cathode top (---). The apparatus was given a little bit a of buffer solution on the filter papers to keep moist.
- The blot was then ran for 30 minutes at 20V. Start time was 3:15 pm. And End time was 3:45 p.m.
- After finished running, the sandwich was removed and the gel and membrane were carefully placed into separate trays containing 20mL of pure water to wash.
- They were then placed in separate plastic boxes and mixed with 10mL of blocking solution and the gel was placed with Seeblue Plus 2 staining gel to stain where the blots were from the SDS page procedure. After this point the T.A. took care of the rest but is stated below:
- Decant the liquid.
- Rinse the membrane with 20mL of water for 5 minutes then decant. Repeat.
- Incubate the membrane in 10mL of Primary antibody solution (HSP70). Decant solution.
- Rinse the membrane with 20mL of Antibody wash for 5 minutes, then decant. Repeat 3 times
- Incubate the membrane in 20mL of Secondary Antibody solution for 30 minutes. Decant.
- Wash the membrane for 5 minutes with 20 mL of Antibody wash, then decant. Repeat 3 times.
- Rinse the membrane with 20 mL of pure water for 2 minutes, then decant. Repeat twice.
- Incibate the membrane in 5 mL of Chromogenic Substrate until a purple band appears. This will occur between 1-60 minutes after adding the chromogenic substrate.
- Dry the membrane on a clean piece of filter paper to the open air.
Oly Protein Gel:

Oly Western Blot:

Pac Protein Gel:

Pac Western Blot:

Electrophoresis Gel Images: http://imgur.com/a/2DHO8#1
QPCR: Data coming.
Conclusion:
There were three different sets of results that were gained from this weeks lab, cPCR, qPCR and protein blot.
The cPCR gel was made in this weeks lab by running a current from the top portion to the bottom portion, which separated the band lengths based by size. Based off the pictures on the given link, the data for my cPCR is in the oly gel (the horizontal image) and the only four on the bottom row. As seen the first well is the ladder which gives the scale for base pair lengths, the next to are the blanks where primer dimer action occurred, then cDNA and last genomic DNA. This PCR is a qualitative data set. So based off the gel there are a few things to take out of it. First, the two blanks bonded to each other to create a double stranded DNA with a relatively short length (around 50 bp based on the scale). This is known because the amplicon length was to be 140bp. Next, the cDNA copy looks to have some binding to the gene and also some primer dimer action. This conclusion is made because of the length of the band seems to go from around 140 to 50. It seems as if both bands length are being amplified. Lastly, the last well was the genomic DNA and it seems to have some small amounts of bind to larger bp strands, which is normal because of the introns and extrons included, but also there is a large amplification of the 50bp or so range which is due to primer dimer action. Based off this data it can be easily seen that the primers need to be redesigned in order to get full expression of the desired genes. This could be done by altering the melting point or even just the amount of primer put into the PCR solution.
The qPCR was the next data set gained from this week’s lab. From the graph is appears that there were three melting points. Two of which were a smaller hump and one in the middle was very large spike. The first melting point was most likely the primer dimer copy which would have a smaller melting point. Next was the large spike, which is the gene that was being expressed. Last was probably some of the genomic DNA which had a higher melting point due to being a larger strand of base pairs. The data hasn’t yet been posted so a larger conclusion will be written out at a later time.
The last part of the lab was the western blot procedure. This started by separating out the proteins based on molecular weight from a current then using a blotting machine to transfer the proteins from the gel to a membrane paper, which was then soaked with antibody wash for HSP70. This whole process would show exactly where the proteins would be expressed. Based off the images above the only band being express was at about 60 kD on the oly gel. This was actually a control, which makes sense. This data was expected because when the oyster is not under stress there is no need for the hsp70 to be active and there is larger amounts stored. When the oyster is stressed, it may be using up the hsp70 protein faster than it is being translated. Although, the data for the pacific gives no expression of the gene at all, which was not expected. Overall, this technique was useful to learn about how proteins can be separated and expressed.
Reflection:
The main purpose of this lab was data collection. Primarily the collaboration of what has been done so far this quarter came to an end during this lab. QPCR, CPCR and western blot results were all gained. This lab also served the purpose of how we take qualitative and quantitative data and make meaning of it. Also, the techniques used in this lab are three very useful tools that can be spread across multiple disciplines of science.
The procedures of this lab were used to measure a couple different things. The cPCR was used to measure gene expression based on band length size traveling down a gel because of current. This was compared to a ladder of known base pair lengths. The qPCR was done on the computer to give a nice log graph of the data. This was also used to measure gene expression. The western blot was used to measure the amount of HSP70 protein that was in each of the protein samples by using an anti-body specific to that protein. This was expressed on the gel and membrane by staining them.
All of these techniques can be used in studies that are trying to determine how much gene expression or protein expression is seen in specific genes and proteins. These are some of the most common procedures used in any molecular and cellular studies. Everything was pretty clear in the background from each section on the lab report. The only thing that could have had more information was how to redesign primers after getting results that were undesirable.
October 30, 2012
Lab 6: Conventional PCR, QPCR, and Protein Isolation
Objectives: Using genomic samples, cDNA samples and ordered primers, conventional and quantitative PCR tubes were prepped for thermo cycling. Also, protein was isolated from control and experimental oysters and quantified using a spectrometer.
Methods and Materials:
Conventional PCR:
- The first step to conventional PCR was to make a master mix for each of the PCR rubes. Enough was needed for 5 tubes: 1 cDNA, 1 genomic, 2 blanks, and one extra for pipetting error.
- The total amount of mix was made with:
| Reagent |
1x.reaction |
5x.reaction |
| 2x Apex Red |
12.5uL |
62.5uL |
| 10uM Primer F |
1uL |
5uL |
| 10uM Primer R |
1uL |
5uL |
| Nuclease Free water |
8.5uL |
42.5uL |
| Total: |
23uL |
115uL |
- After the mix was made, 23uL was pipetted into 4 tubes and then 2uL of cDNA was added to the first, 2uL of genomic DNA added to the second, and 2uL of nuclease free water was added to the two blanks.
- The tubes were vortexed and placed in pcr tray for thermocycling.
- Like conventional PCR, the qPCR required a master mix as well. For the qPCR enough mastermix for 8 was needed: 2 genomic DNA, 3 cDNA, 2 blanks, and one extra for pipetting error.
- The total amount of mix was made with:
| Reagent |
1x.reaction |
8x.reaction |
| Master Mix 2x (immomix) |
12.5uL |
100uL |
| Syto-13 dye (50uM) |
1uL |
8uL |
| Upstream Primer |
1.25uL |
7.5uL |
| Downstream Primer |
1.25uL |
7.5uL |
| Ultra Pure water |
7uL |
56uL |
| Total: |
23uL |
179uL |
- After the master mix was made 23uL was pipetted into 7 qPCR tubes. After the master mix was added 2uL of cDNA, genomic DNA, and nuclease free water were added. The initials “JCD” marks the start of the sequence. It goes: JCD|cDNA,cDNA,cDNA,DNA,DNA,blank,blank
- The tubes were capped and put into the PCR tray for thermocycling.
- Everyone was assigned 2 samples from the experiment and control group oysters to extract protein. The oyster I was assigned was the Olympia oyster control group numbers 51, and 52.
- 25mg of the tissue sample was weighed out into snap cap tubes labled “Oly-51 10.30” and “Oly-52 10.30”
- The actual weight of the sample was Oly51: 0.025g and Oly52: 0.025g
- 500uL of CellLytic MT solution was added to the 1.5mL snap cap tube and then homogenized using a disposable pestle.
- After breaking down the tissue and vortexing and then inverting a few times, the tube was spun in a refrigerated microfuge at 10 minutes at max speed. This step would help separate the protein out from the tissue.
- After removing from the tube, the supernatant was difficult to distinguish so the tubes were placed back into the microfuge and spun again for 10 minutes at max speed.
- When removed this time, the supernantant, which was the clearish liquid was extracted and placed into snap cap tubes labeled “Oly-51 10.30 Protein” and “Oly-52 10.30 Protein”
- While in the microfuge I labeled 3 screw top tubes “Oly-51 10.30 Protein”, “Oly-52 10.30 Protein”, and “Blank 10.30 Protein”
- 15uL of each of the protein sample was added to its selective tube. And 15uL of nuclease free water was added to the blank sample.
- In all the tubes 1000uL of Bradford Assay was added which is a colorimetric assay. When placed into protein solutions it was change different intensities of blue. This blue dye absorbs at 594nm and based off the absorbance curve that the Lab coordinator produced, the amount of protein could be determined.
- Next, another 500uL of B.A. was added to each of the tubes. The blank sample was then pipetted up and down in order to mix the solution.
- These were all incubated at room temp for 10 minutes before going to the spectrometer.
- A spectrometer was used to measure the absorbance of the samples we produced. It was first zero’ed using the blank sample that was created. This would give the base amount for zero protein and the dye from the Bradford Assay.
- After blanking the machine a cuvette was filled with the protein samples that we created and then recorded the absorbance two times and took the mean of these to get the absorbance.
Equation for spectrometer: y=996.52x-43.64
| Sample |
Absorbance |
|||
| Trial 1 |
Trial 2 |
Mean |
Concentration |
|
| Oly 51 |
0.524 |
0.518 |
0.521 |
475.547ug/mL |
| Oly 52 |
0.68 |
0.650 |
0.665 |
619.046ug/mL |
Conclusion:
In conclusion a lot was accomplished during this lab session. To start, the qPCR and conventional PCR tubes were set up and prepared to begin their thermocycling process. This will set us up for producing Agarose Gel Electrophoresis which will show us band lengths of the genes. The cDNA and DNA samples will be analyzed for the genes that we created primers for a couple weeks back.
The protein isolation section of the lab had a little bit more hands on data. The samples were taken from tissues extracted from the oysters used for the experiments and control groups. After the isolation process we were able to quantify it by using the spectrometer. The way this worked was a linear equation was produced for us using known proteins, which gave us an equation to use for the absorbance recorded. A Bradford assay was used in the supernatant that was extracted which changes different intensity of blue based off how much protein was in the sample. By setting the spectrometer to a blank sample with just water and the Bradford assay, we were able to get a reading for absorbance. Two readings were taken and averaged out to get a mean absorbance. Based off the graph the higher to a 1 absorbance or 952.88 ug/mL would result in a greater concentration of protein in the sample. The first Olympia oyster that was sampled had a mean absorbance of 0.521 which was about half the amount that would like to be achieved. The concentration was 475.547 ug/mL. This lower concentration could have resulted in error when pipetting the supernatant out of the microfuged tube. Some cell debris and tissue debris may have been accidentally sampled out which would have resulted in a lower amount of protein. The second Olympia oyster sample, 52, had a higher absorbance with a mean at 0.665. This is better, but still not up to the amount that would like to have been seen. This jump in absorbance resulted in a jump in concentration as well, at 619.ug/mL. This is a decent concentration, but still shows signs of error when transferring the supernatant over to the screw tube. This also may suggest that not all of the supernatant was extracted.
This data shows that there is a good amount of protein concentration, but could have been a larger amount. After isolating the protein sample from the tissues, a western blot could be produced to express the amount of the different proteins. Also, and SDS page could be made which would separate out the proteins based on size.
Reflection:
The purpose of this lab was to show us how to create a master mix for PCR amplification and the underlying steps behind that. This molecular technique can be used later on when we are trying to amplify our experimental oysters genes. Also, the purpose was to show how to isolate a protein sample from a tissue. This technique could be used across a wide spectrum of disciplines in biology, marine biology, and any other science that samples tissues for proteins. The first procedures performed in lab were used to amplify the genes that were chosen to express from the primers chosen. The protein isolation part measured the concentration of protein in the sample we created from the original tissue sample using a spectrometer. Any study that looks into what genes are being expressed is most likely going to use a qPCR to quickly amplify the DNA. Also, any study that looks at proteins are going to start with this first step of protein isolation and quantification. The only unclear aspect of this lab procedure was when making the master mix for the PCR. It was somewhat unclear about how many blanks and how much extra for pipetting error should be made. This part slowed most of the lab down the most, but after figuring it out, everything was straight forward. The lab provided the perfect amount of info needed for everything. It explained how both conventional and quantitative PCR both worked and the reagents within each of their master mixes. Also, the protein isolation gave great background on how the Bradford Assay worked to determine the absorbance of the protein sample. Overall the lab went smoothly and not many questions came up.
October 23, 2012
Lab 5: Tissue dissection, primer reconstruction, end-point PCR
Objective: Tissue samples were dissected from control and experimental oyster samples. Also, rehydrating primers was achieved.
Material and Methods:
Experimental Procedure:
- On Monday October 22, 2012 the experimental group for the cyclodextrin trial began.
- 15 oysters were added to a tank with 7 liters of sea water, which also contained a pump for water flow and an oxygen input.
- 1 more liter was added after dissolving 2.009 grams of beta-cyclodextrin in the sea water. Being a water soluble substance, stirring the water for 5 minutes completely dissolved the powder. After being completely dissolved, the water was dumped into the tank, resulting in 8 liters of sea water and 2.009 grams of beta-cyclodextrin.
- The official time the cyclodextrin went into the water was 1:30p.m. on Monday October 22, 2012
- The class split into two separate working groups, one for pacific oysters and one for Olympia oysters.
- A Chart was comprised with a numbering system:
| Sample |
Amount sampled |
Test tube identification |
| Pacific Control |
15 |
[1-15] |
| Pacific Cyclodextrin |
15 |
[16-30] |
| Pacific 35C |
10 |
[31-40] |
| Pacific 40C |
10 |
[41-50] |
| Olympia Control |
10 |
[51-60] |
| Olympia 35C |
10 |
[61-70] |
| Olympia gradual |
10 |
[71-80] |
| Olympia Pre-stress |
9 |
[81-89] |
- For each sample there were two gill samples and one mantel sample. For example the first control for pacific oyster would have three test tubes labeled “1G,1G,1M.” G=Gill, M=Mantel
- Four people for each group: 1) Measure length and width in centimeters and record 2)Shuck oysters 3 and 4) Dissect out 2 gill samples and 1 mantel sample
- Dissection took place by dissecting one sample group at a time and then when the tissues were all placed in tubes, they were moved into the freezer. For the pacific oyster group, we started with the cyclodextrin at 2:00p.m. then in order: pacific control, Pacific 35C, Pacific 40C
- After all of the tissue samples were collected, everything was washed and sanitized.
- Based off the weight of the primers, water was added.
- For my primers: Forward weight= 31.3nm so 313 uL of nuclease free water was added, Reverse weight=35.0nm so 350 uL of nuclease free water was added.
- Both were vortexed to mix the primer into the water.
- Next a stock and work stock sample were made. The stock sample consisted of 10uL of primer solution and 90uL of water. The working stock was 10uL of the primer solution. These were made for both the forward and reverse, which resulted in 4 tubes.
- Working stock was labled “WS-fwd”,”WS-rvs” and the stock was labled “S-fwd”,”S-rvs.”
- These four samples were placed into the freezer for PCR amplification next week.
Also, pictures that were posted can be found here: http://imgur.com/a/7m4i8/all
Conclusion:
The results of the length and width measurements don’t give us much information to go off of, but gives us some nice background data to continue on with the experiment. For the cyclodextrin bathed oysters, they had a little over 24 hours to bath in the sea water/cyclodextrin mixture. All of the tissue samples were dissected and stored for testing in the upcoming weeks. Also, the PCR primers were rehydrated and froze, which sets us up for the next step in the PCR analysis of the genes we selected. In conclusion, this lab ultimately set us up for the beginning of analyzing tissue samples from the experimental groups and taking the next step to starting PCR.
Reflection:
The purpose of this lab was simple, to collect and gather our experimental and control groups tissues for later processing. Also, the second purpose was rehydrating the PCR primers so they are ready to use for next week’s lab. The procedures in this lab were not necessarily used to measure much, with the exception of measuring the length and width of the oysters. Any study revolving research with oysters would use the techniques that this lab required. Measuring, shucking, and dissecting tissue samples. Also, rehydrating primers could be used in many fields of the scientific world. This lab was pretty straight forward and not many questions came to mind. When we were able to look at the gonad tissues under the microscope, it was very interesting to see how the two different oysters and how some people try to tell the different genders. More information revolving that would have been nice, but not necessary.
Lab 4 Reverse Transcription and primer design and prep for experiment
Objective: The objective of this lab was to create cDNA strand from the isolated DNA from lab week 3. Also, to learn how to create primers for these and set up or experimental procedure.
Methods and Procedures:
Reverse Transcription:
- First thaw out the RNA sample isolated from week 3 lab. Invert it several times to help speed the process.
- Using a 0.3mL PCR tube that was labled “JCD-cDNA” 5uL were added to the tube from the extracted RNA.
- Next 1 uL of oligio dT was added. This is a primer to help ensure everything stayed together.
- Next 4 uL of nuclease free H2O was added.
- After these had been added and mixed, the pcr tube was incubated for 5 minutes at 70 degrees celcius on a thermocycler then transferred straight to ice. The tube was then flicked to make sure the mixture was at the bottom of the tube.
- After heating, 5 uL of M-MLV 5x reaction buffer was added, which assists in pH change to not occur
- Next 5 uL of dNTPs were added. These are the bases that will actually bind to the RNA.
- Next 1 uL of M-MLV RT were added, which is the reverse transcriptase to actually make the copy.
- Lastly 4 uL of nuclease free H2O was added to the mixture.
- The pcr tube was then incubated for 60 minutes at 42 degrees celcius and then heat activated at for 3 minutes at 70 degrees celcius. After removing the tube was centrifuged and put on ice.
- NCBI is the national database of genes as along with UW own database that has been comprised for the Olympia oyster.
- After finding a gene of interest the sequence is then copied into a primer blast where it goes through algorithms to ensure the best quality primer for the sequence
- It takes into account self-complimentary strands and if they were to form dimers.
- Also melting point was taken into account.
- Worked with one oyster at a time to shuck them.
- First inserted the shucker into the hinge and twisting it and then slicing the adductor muscle.
- After carefully opening the oyster, the mantel needed to be scraped off the valve
- Two separate tanks were set up for the controls which have a pump and O2.
- These will contain the two oyster species.
- For our experiment, two spate tanks for the cyclodextrin bathed oysters were assembled. These contained a pump for water flow and O2.
- There will be 5 oysters in each of the tanks in order to prevent any cross contamination.
- The cyclodextrin will then be administered 24 hr. prior to next weeks lab.
No raw data this week.
Conclusion:
The cDNA strand was made by mixing a bunch a bases and reverse transcriptase together and using the thermocycler to incubate the mixtures at its proper heat to copy.The cDNA was prepped for next weeks PCR test. Also, we learned how primers are made and the significance behind them. This involves going into databases and searching for the desired sequence and then creating the primer from that. The experimental design was set up, which included separate tanks, pumps, O2 and marking the tanks for next weeks experiments.
Reflection:
Overall this weeks lab was not too difficult, but several important step relating to the rest of the quarters research were achieved. Starting off we learned how to dissect the oysters using a shucker and was given an anatomy lecture of the oyster. After this, a revised experimental design was achieved by narrowing down exactly what each person needed for the question they wanted answered. From this we were able to set up our experiments for next week taking into account flow of water and O2. After this portion we went about the cDNA aspect of the lab. This portion was quick and simple. Mixing bases and RT together and putting through a thermocycler. Overall, the lab was pretty straight forward and didn’t have any huge question that I reflected upon.
October 9, 2012
Lab 3: RNA Isolation, Part 2
Summary: In this lab RNA was isolated after sitting in TriReagent for a week and then quantified using a nanodrop machine.
Materials and Method:
- The tube from the previous week’s lab labeled “RNA 17-M” was taken out of the ice and incubated at room temp for 5 minutes. The tissue sample was homogenized and had been soaking in TriReagent. This works in three ways: Guanidine isothiocyanate denatures proteins, phenol acts to keep the RNA soluble and the proteins and other inorganic material insoluble, and pH to keep DNA out of the solution.
- After sitting for 5 minutes 200uL of chloroform was added to the sample, which is an organic solvent that works by separating the phenol and other insoluble components from the RNA.
- Tube was vortexed for 30 seconds to make a milky texture to the sample.
- The microfuge tube was spun for 15 minutes at max speed in the refrigerator.
- The spinning helped to separate the material into three layers: organic phase (bottom portion), the interphase (layer of cell debris), and the aqueous portion (top). The aqueous portion is the part that contains the RNA.
- A micropipette was used to transfer the aqueous portion into a new tube. This was done carefully to ensure no cell debris got into the new tube.
- 500uL of isopropanol was then added to the aqueous solution. RNA is insoluble in isopropanol, and as a result, will form a pellet of RNA. It also acts to remove alcohol soluble salts.
- The tube was inverted five times and then incubated for 10 minutes at room temp. This step allows for the RNA to separate out from the isopropanol.
- The tube was then spun for 8 minutes at max speed to create a small pellet on RNA on the bottom of the tube.
- The supernatant was removed using a micropipette and then 1mL of 75% EtOH was added to the tube.
- The tube was the vortexed for 30 seconds to help dislodge the RNA pellet from the bottom of the tube.
- Next, the tube was spun at 7500g for 5 minutes in order for the pellet to settle at the bottom again.
- The supernatant was then removed, and the tube was spun again briefly for about 30 seconds. This enabled the small amount of ethanol to separate. A small micropipette was used to remove the small amount left.
- Then the tube was left open for 5 minutes, which allowed the RNA pellet to dry.
- After the pellet had dried, 100 uL of 0.1% DEPC water was added to the tube and micropipetted up and down in order to solubilize the RNA.
- Lastly the sample was put into a hot water bath of 55 degrees Celsius for 5 minutes.
- The tube was removed and put back into ice where the quantification process would begin.
- To set up the nanodrop machine 2uL of 0.1% DEPC water was added to the nanodrop pedestal in order to calibrate the instrument.
- Next the RNA measure button was pressed.
- 2 uL of the sample RNA was added to the nanodrop to measure our results.
Results:
| RNA Concentration |
575.3 ng/uL |
| A260/280 |
1.95 |
| A230/260 |
1.12 |
The data received from the nanodrop were results that were slightly unexpected. The range for the A260/280 should result anywhere from 1.8-2.0, which means that the RNA is separated from proteins. The results for this absorbance ratio was in range. For A260/230 the range is anywhere from 1.5-2.0. The results don’t fall between this range and could be a result of not fully extracting all of the ethanol from the RNA pellet. Overall it had a significant amount of RNA, just the ratio between 230/260 was not desirable. Based off these results I need to focus on being more careful about removing all excess fluid. This could be done by using a smaller micropipette or just taking more time to ensure quality.
Reflection:
The purpose of this lab was to learn how to extract RNA from a tissue sample and to quantify it using a nanodrop machine. The procedures in this lab were used to measure how much RNA could be extracted from the tissue sample. Any study that wants to measure how much of a certain gene is being transcribed could use this technique. Measuring certain sequences of RNA being expressed can tell us about how the DNA is being read and what genes are being turned on and off. Overall this technique can be used in a variety of studies surrounding multiple disciplines. The procedure was pretty clear and straight forward. More information about surrounding the RNA concentration levels and what is a general range for certain tissues would be nice.
Assignment:
Hypothesis/question: How are growth factor hormones effected by temperature stress and the introduction of cyclodextrin to Olympia oysters.
Gene of interest: Transforming Growth Factor-β or TGF-β
October 2, 2012
RNA Extraction Part 1 and DNA Isolation (DNazol)
Summary:
The first section of RNA extraction was started by isolating and homogenizing a portion of the Olympia oyster mantel tissue. DNA was also isolated from Olympia oyster mantel tissue using DNazol. The total DNA of this concentration was determined by using a nanodrop to evaluate the wavelengths of the DNA.
Material and Methods:
- Selected tissue sample 17- M; Olympia oyster mantel tissue. Kept on ice throughout the sampling process.
- Obtain 4 microcentrifuge tubes and label with initials (JCD) on the top along with the date (10.2) and on the side with one that says RNA and the other three that say DNA. Each have the species and tissue type on the side as well (17-M; Olympia oyster mantel tissue)
- Next the sample was trimmed down by razor blade to get the desired amounts for microcentrifuge tubes for both the RNA extraction and DNA isolation. Desired ranges for DNA: 25-50mg and RNA: 50-100mg.
- Measurements achieved:
| Weight (mg) |
|
| DNA 25-50 mg |
29.0 mg |
| RNA 50-100 mg |
50.0 mg |
- After weighing out the desired amounts, both were put back on ice to avoid RNases from destroying our sample.
- Add 500 uL of TriReagent to the tube that has RNA labeled on the side. This TriReagent works in three ways: 1) guanidine isothiocyanate, to denature the proteins 2)phenol, to keep proteins insoluble 3) pH, to keep DNA out of the solution
- Using a disposable pestle, the sample was carefully homogenized. Then it was vortexed on high for 30 seconds to help break up the sample. After the vortex the pestle was used again to ensure it was fully homogenized.
- Another 500 uL of TriReagent is added to the tube labled RNA and then was put back on the vortex for another 15 seconds on high.
- The tube with the label RNA 17-M was given to the TA to store on ice for the next stage of RNA extraction.
- Take the tube labeled DNA on the side to the fume hood and add 0.5 mL of DNazol. DNazol uses a guanidine-detergent to hydrolyze the RNA and precipitate the DNA from the cell. In order for the DNazol to work, a pestle is used to homogenize the tissue. After it has been broken up another 0.5 mL of the DNazol is added to the tube.
- The tube sits at room temperature for approximately 5 minutes.
- After the 5 minutes is up the tube is transferred to the centrifuge and spun at 10,000g for 10 minutes at room temp.
- After pulling the tube out, the supernantant is extracted by a micro pipette into one of the other tubes labled DNA on the side. Only the top layer of supernanant is extracted, not the cell debris on the base of the tube.
- In the new tube, 0.5 mL of 100% Ethanol is pipetted is added to the supernanant. By using the ethanol it will precipitate the DNA.
- After mixing the tube 8 times by inverting it, the precipitant was not able to be seen. Small fragments could be seen but not well enough to extract.
- Next the tube was placed back in the centrifuge, this time at 5,000 * g at 5 minutes.
- After removing the tube, a precipitant of DNA was clumped together at the base of the tube.
- With a micro pipette, the DNA was transferred to the next tube labeled DNA on the side.
- 1 mL of 75% ethanol was then added to wash the sample. Close the cap and inverted 6 to 8 times.
- Then removal of the ethanol and added another 1 mL of the 75% ethanol to the tube. Once again inverted the tube 6 to 8 times and the ethanol was removed. To get the last remained ethanol out trapped by the DNA sample, a 200 uL micropipette was used. This ensured that most of the ethanol was off the DNA sample.
- The last step was to add DEPC water to the tube to solubilize the DNA.
- 150 uL of 0.1% DEPC water was added to the DNA and then to solubilize the sample, it had to be micropipetted up and down multiple times.
- Using the solubilized DNA sample, it could be measured using the nanodrop in the lab.
- Used 2 uL of 0.1% DEPC water on the nanodrop pedestal first and lowered the arm.
- “dsDNA” was selected on the computer and “blank” was then clicked.
- After the blank 2 uL of the DNA sample was put onto the nanodrop pedestal and arm was lowered.
- Clicked “measure” for the DNA concentration results (ng/uL), A260/280 ration and A260/230 ration. The nanodrop uses the Beer- Lambert law for calculations.
- Where A is absorbance (no units, since A = log10 P0 / P )
e is the molar absorbtivity with units of L mol-1 cm-1
b is the path length of the sample - that is, the path length of the cuvette in which the sample is contained. We will express this measurement in centimetres.
c is the concentration of the compound in solution, expressed in mol L-1 - Using Chem wipes, the nanodrop is wiped off and the sample was placed on a rack with the name JASON DIAZ taped onto it. This rack is in the freezer at -20 degrees Celsius
- The results from the computer were as follows:
| DNA concentration |
231.6 ng/uL |
| A260/280 |
1.88 |
| A260/230 |
1.01 |
Results:
Weights of samples:
| Weight (mg) |
|
| DNA 25-50 mg |
29.0 mg |
| RNA 50-100 mg |
50.0 mg |
Nanometer recordings:
| DNA concentration |
231.6 ng/uL |
| A260/280 |
1.88 |
| A260/230 |
1.01 |
Conclusion:
The lab guide states that the purified DNA should have a A260/280 ration of 1.7-1.9 which indicates a good quality DNA. The data recorded falls in this range which means that there is a low amount of proteins compared to the DNA, about 1:9 ratio. This was an expected result to achieve because of using the DNazol to hydrolyze the RNA and selectively precipitate the DNA. Based off the results this sample of DNA was good quality. It can be stored and used to compare other 17-M (Olympia oyster mantel tissue) tissues that have had an environmental stress applied.
Reflection:
The purpose of this lab was to begin to get a feel for using molecular techniques in the laboratory setting and to start preparing us to think about our projects. More specifically, tissue was homogenized and then DNA was isolated by using DNazol. This lab technique will help us isolate DNA later on during our projects.
The procedures used to measure in this lab was the quality of the DNA and the ratios of DNA compared to to salts, ethanol, phenol and proteins. The goal was to get in a range of 1.7-1.9 ratio for the A260/280 spectrum of light. This specifically compares proteins to DNA ratio of the sample.
Anything related studying an organisms genome could use this technique of DNA isolation. During the procedure, when extracting the supernatant, it was difficult to get all of it without the cell debris. A little of the supernatant was left in in the microphuge tube. The information that was provided for the lab explained everything in great detail. More detail relating to the Trireagent for the pH would have been nice.