Tissue Extraction I
I. RNA extractions conducted by Tri-Reagent.II. Protein extraction and concentration measurement.
III. Think about primers to design.
Tissue sample: Rainbow trout (Oncorhynchus mykiss) white muscle frozen at -80°C.
I. RNA Extraction
RNA extraction was performed with TriReagent (extraction by means of: 1- guanidine isothiocyanate, 2- phenol, and 3- low pH).Protocol
1. Turn on heating block to 55C. Also turn on spectrophotometer.
2. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.
3. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.
4. Carefully homogenize the tissue using a disposable pestle.
5. Add an additional 500uL of TriReagent to the tube and close the tube.
6. Vortex vigorously for 15s.
7. Store at -80°C until lab next week.
My samples
Sample #R1: 79 mg of white muscle tissue
Sample #R2: 78 mg of white muscle tissue
Comments
Homogenization: tissue was mushed up as much as possible with pestle for about 1 minute, but not completely homogeneous in liquid.
II. Protein Extraction & Quantification of Concentration
Protein extraction was conducted with CelLytic MT (mixture of salts and detergents supplemented with a cocktail of protease inhibitors to: disrupt lipid membranes, lyse cells, and buffer the cellular proteins at the appropriate pH). Quantification of protein concentration determined using Bradford Assay (calorimetric assay using Coomassie Blue reagent). When Coomassie dye binds to protein, color changes from brown to blue.Protocol
1. Add 0.5mL of CelLytic MT solution to a 1.5mL snap cap tube.
2. Add 25mg of your tissue to the tube.
3. Homogenize the tissue with a disposable pestle.
4. Close the tube and invert the tube several times.
5. Spin the tube in a refrigerated microfuge for 10mins. @ max speed.
6. While spinning, label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
7. Carefully transfer supernatant to labeled tube and store tube on ice.
8. To a fresh tube, add 1.5mL of Bradford reagent.
9. To this same tube, add 30uL of your protein extract.
10. Invert the tube several times and then incubate at RT for 10mins.
11. Mix the tube several times and transfer 1mL to a plastic, disposable cuvette.
12. Measure the absorbance at 595nm and record the value.
13. Remove the cuvette from the spectrophotometer. Using a P1000 set to 1mL, carefully pipette the solution in the cuvette up and down a couple of times to mix.
14. Measure the absorbance at 595nm and record the value.
15. Repeat steps 13 and 14.
16. Average the three absorbance values you recorded.
My sample
Sample #P1: 35 mg of white muscle tissue
Spectrophotometer readings:
- 1.532
- 1.524
- 1.522
Average ABS = 1.526
Comments
- Used Amy’s blank sample (30uL of CellLytic MT without any protein, added to 1.5 mL Bradford reagent) to zero spectrophotometer.
- Jon measured the Bradford reagent without CelLytic MT solution, and it was a negative value measured in the spectrophotometer.
- Note that Coomassie kit is non-linear with higher levels of protein concentration, and thus a standard curve should be run with each assay (Coomassie Protein Extraction Kit Instructions). For standard curve: With the spectrophotometer set to 595 nm, zero the instrument on a cuvette filled only with water. Subsequently, measure the absorbance of all the samples.Subtract the average 595 nm measurement for the Blank replicates from the 595 nm measurements of all other individual standard and unknown sample replicates. Prepare a standard curve by plotting the average Blank-corrected 595 nm measurement for each BSA standard vs. concentration in μg/ml. Use the standard curve to determine the protein concentration of each unknown sample.
E.g. Typical color response curves for bovine serum albumin (BSA) and bovine gamma globulin (BGG) using the Standard Test Tube protocol of the Coomassie Assay.
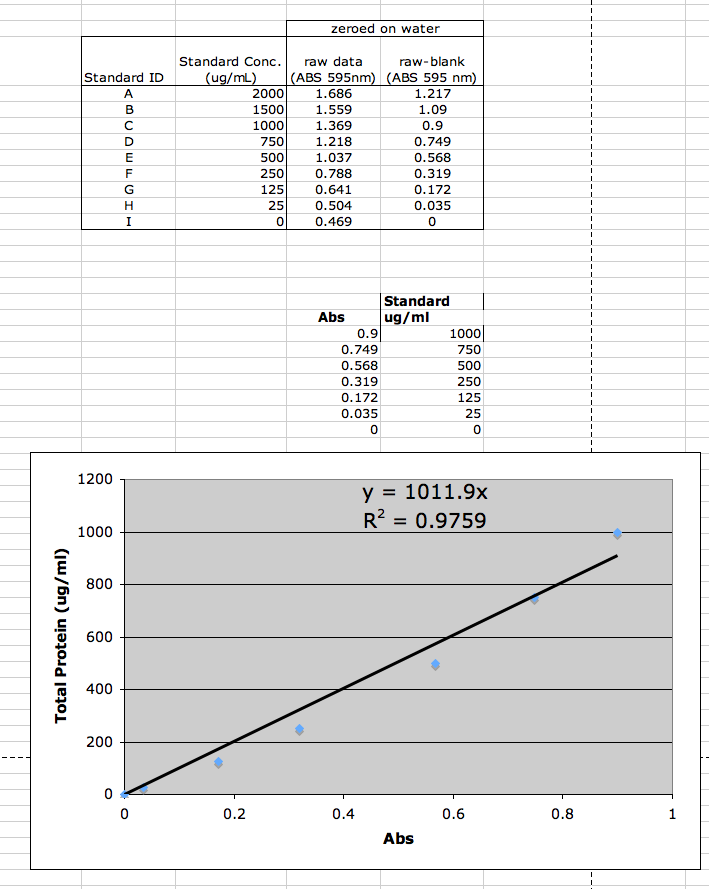
Standard curve for our samples
Blank (water): spectrophotometer reading of 0.469
Linear regression
Protein concentration vs. ABS
Determined for first 7 of 9 datapoints (i.e. standard concentrations from 0 to 1000 ug/mL), and forced through point (0,0).
Equation: y = 1011.9x
where y = total protein (ug/mL) and x = ABS at 595 nm


Log-linear regression
Ln(protein concentration) vs. Ln(ABS)
Just trying a different equation to use all 9 datapoints (i.e. standard concentrations from 0 to 2000 ug/mL) and not forced through point (0,0) because there might be error in readings by the spectrophotometer.... just assuming.
Equation: y = 7.07 + 1.20x
where y = Ln(protein concentration in ug/mL), and x = Ln(ABS at 595 nm)
Therefore, our samples were too highly concentrated to accurately estimate concentration of protein. We will need to dilute our samples and measure again.
In Lab 2, I measured three diluted samples:
| Sample dilution |
protein:H20 (uL) |
Replicate |
Readings |
| One half |
15 uL : 15 uL |
1 |
1.423 |
| |
|
2 |
1.420 |
| |
|
3 |
1.418 |
| |
|
Average |
1.420 |
| One third |
10uL : 20 uL |
1 |
1.160 |
| |
|
2 |
1.157 |
| |
|
3 |
1.156 |
| |
|
Average |
1.158 |
| One fifth |
5uL : 25 uL |
1 |
0.702 |
| |
|
2 |
0.679 |
| |
|
3 |
0.667 |
| |
|
Average |
0.683 |
| Blank |
0 uL : 30 uL |
|
Used to zero spectrophotometer |
Linear regression y = 1011.9x
One half dilution: y = 2(1011.9*1.420) = 2*1436.898 = 2873.8 ug/mL
One third dilution: y = 3(1011.9*1.158) = 3*1171.78 = 3515.3 ug/mL
One fifth dilution: y = 5(1011.9*0.683) = 5*691.128 = 3455.6 ug/mL
Average: 3281.6 ug of protein / mL
Comments: At one half dilution and one third dilution, the concentration may still be a little high for estimation. The linear regression was run on samples </= 1000 ug/mL. Thus, the estimate at one fifth dilution may be better than the average of these three dilutions.
Log-linear regressiony = 7.07 + 1.20x
where y = Ln(protein concentration in ug/mL), and x = Ln(ABS at 595 nm)
One half dilution: 3582.9 ug/mL
One third dilution: 4207.6 ug/mL
One fifth dilution: 3721.7 ug/mL
Average: 3837.4 ug of protein / mL
(Don't use this estimate. I was just exploring.)
III. Primer Design
Used Primer
i. Species and tissue type: Guppy (Poecilia reticulata) liver
Target gene/EST: superoxide dismutase
EST accession number: ES381482
Forward primer (left primer): TTTGGCAACTTCGTCTCACA
Reverse primer (right primer): CTCATGCTGCGTATTCCAGA
Expected size: 231
Annealing temperature: 60'C
ii. Species and tissue type: Guppy (Poecilia reticulata) muscle
Target gene/EST: hsp70
EST accession number: AB298594
Forward primer (left primer): TGAGATCGTGCTTGTTGGAG
Reverse primer (right primer): TTCTGGGGATCAGTTTGGTC
Expected size: 248
Annealing temperature: 60'C
iii. Species and tissue type: Guppy (Poecilia reticulata) liver
Target gene/EST: glycogen phosphorylase
EST accession number: ES375965
Forward primer (left primer): TGTTAACGACTTGGCCTTCC
Reverse primer (right primer): GTTCTTCCTGATGCGGTTGT
Expected size: 208
Annealing temperature: 60'C
iv. Species and tissue type: Guppy (Poecilia reticulata) gills and brain
Target gene/EST: Na+/K+ ATPase
EST accession number: ES375881
Forward primer (left primer): TGGTTTGACAACCAGATCCA
Reverse primer (right primer): TTCATAAGCCAGGGAGATGG
Expected size: 207
Annealing temperature: 60'C
v. Species and tissue type: Guppy (Poecilia reticulata) brain and heart
Target gene/EST: glutathione peroxidase
EST accession number: ES373060
Forward primer (left primer): TAAACAAGGATGGGGAGGTG
Reverse primer (right primer): ATATGGGCAGGTCCTTTTCC
Expected size: 244
Annealing temperature: 60'C
vi. Species and tissue type: Guppy (Poecilia reticulata) liver
Target gene/EST: hsp90
EST accession number: ES381313
Forward primer (left primer): TTCTACGAGGGCTTCTCCAA Reverse primer (right primer): AGCAGAGTTTGCCACCTGAT Expected size: 210
Annealing temperature: 60'C
Lab 2 - Jan 14, 2009
Tissue Extraction II
I. Re-spec dilution of protein sampleII.a Redo RNA extractions conducted by Tri-Reagent with guppy (Poecilia reticulata) liver muscle tissues
II.b Continue with RNA extraction protocol
III. Run SDS-PAGE protein gel
IV. Order Primers
I. Re-spec dilution of protein sample
Results & Comments written up in Lab I section above.II. RNA ISOLATION PROTOCOL (continued)
Tissue samples:
Guppy sample #2: muscle tissue (22 mg)
- From guppy pilot study: challenged at ~30'C. Sampled 01/09/09 and stored in RNAlater in -80'C freezer
- From guppy pilot study: challenged at ~30'C. Sampled 01/13/09 and stored in RNAlater in -80'C freezer
1. Turn on heating block to 55C. Also turn on spectrophotometer.
2. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.
3. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.
4. Carefully homogenize the tissue using a disposable pestle.
5. Add an additional 500uL of TriReagent to the tube and close the tube.
6. Vortex vigorously for 15s.
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube. NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
17. Incubate at RT for 10 mins.
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
- Very small pellet for sample #2 (muscle tissue), and substantial sized pellet for sample # 9 (liver tissue).
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
- Used kimwipes to remove EtOH from sides and bottom of tube.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer.
PROTEIN GEL PROTOCOL
Tissue: Rainbow trout white muscle frozen in -80'C from last week's lab.
1. Begin boiling water on hot plate.
2. Thaw you protein extract from last week. Mix well by inverting tube several times.
- Note: Supernatant collected from last week looks slightly milky with some small solids floating around.
- I tried to collect liquid without any solid pieces.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer .
- Added 60 uL of protein sample and 60 uL of 2X Reducing Sample Buffer in one tube (#1). And again in another tube (#2).
4. Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly as demonstrated.
- Added 10uL of ladder solution in wells #1 and #12 on Back side. A little bit of the 10uL came out of the well as I pulled the pipette tip from the well.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well (30 uL into each well) using a gel loading tip.
- Loaded solution from tube #1 into wells # 3 and 4. Loaded solution from tube #2 into wells #5 and 6.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
- Don't forget to add more 1X Tris-HEPES SDS buffer on the outsides of the gels before turning on power supply. D'oh.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL (does not have to be measured - just need enough to cover the gel) of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Be careful not to dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible. This may need to incubate O/N. If so, cover container with plastic wrap and leave on shaker/rocker.
- Changed out the acetic acid after 15 minutes, and then left overnight. Mac took a picture of the gels the next day. Thanks Mac!
Lanes: 12.Blank --- 11.Ladder--- 10.Blank --- 9.8.Bob --- 7.Blank --- 6.5.4.3. Jenn (O. mykiss white muscle) --- 2.Blank --- 1.Ladder
Lane 3. Tube #1, replicate 1; Lane 4. Tube #1, replicate 2
Lane 5. Tube #2, replicate 1; Lane 6. Tube #2, replicate 2
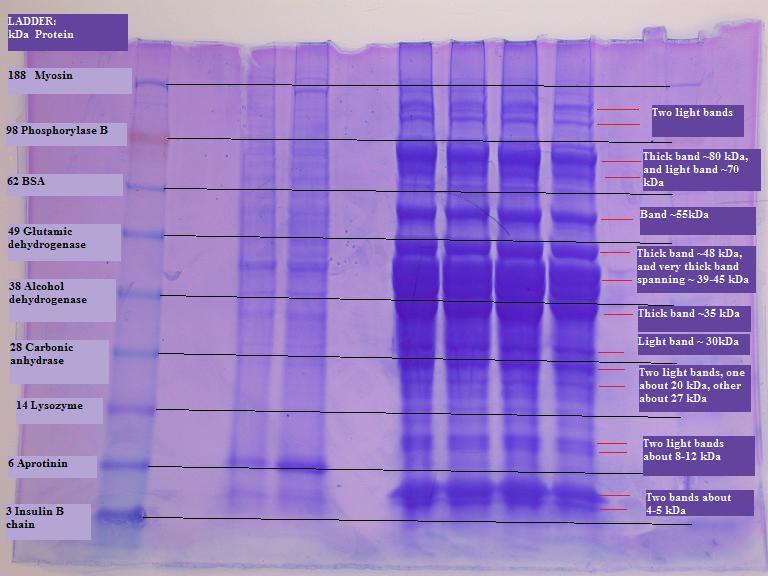
I drew lines between the ladders on the left and right to compare to my bands. My bands do not seem to fall right where proteins in the ladder delineate. I would have expected to see a lot of myosin because my tissue sample is white muscle. With a quick online search, I found that myosin is 210 kDa and actin is 42 kDa (http://www.astro.hr/s3/2005/Fish.pdf). Not sure why I don't see a really thick band at the top of my gel for myosin, but the really thick bands at about 30-45 kDa is probably actin. I would have also expected to see some phosphorylase B. Phosphorylase B in trout as a dimer is 193 kDa and with a subunit of 87 kDa (Mehrani and Storey 1993). There is a band at about 87 kDa. But who knows if these really are the proteins. I'd be more convinced with sequencing data.
From the protein quantification results, I've got a lot of protein relative to some other samples in the class. I can see here in the gel that my samples have a lot of protein relative to Bob's samples.
IV. Primers
Primers were ordered Thursday morning.
Lab 3 - Jan 21, 2009
Reverse Transcription and PCR
I. Quantify RNAII. Reverse transcribe RNA to cDNA
III. Perform PCR
I. Quantify RNA
Used nanodrop spectrophotometer in Steven's lab.Sample #2: guppy muscle
RNA concentration: 71.78 ng/uL
A260 = 1.794
A280 = 0.934
260:280 = 1.92
260:230 = 1.21
Sample #9: guppy liver
RNA concentration: 403.25 ng/uL
A260 = 10.081
A280 = 5.332
260:280 = 1.89
260:230 = 1.86
A260:A280 is between 1.8 and 2.0 => No protein; Pure RNA
A260:A230 is between 1.5 and 2.0 => No phenol, ethanol, high salts; Pure RNA
REVERSE TRANSCRIPTION PROTOCOL
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
If sample is not more than 25 ug, then no need to dilute up to 5 uL.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix
PER RXN
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
Should make 2 replicates for each tissue sample, and a negative control for a total 3 samples. I didn't make a replicate. Oops. I only prepared one sample for the guppy #2 muscle tissue, one sample for the guppy #9 liver tissue, and a blank.
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave on ice or store at –20C
PCR
Polymerase Chain Reaction involved amplifying a DNA (genomic or complementary) target using a polymerase, primers (short oligonucleotide), and dNTPs (A, C, T, Gs). In general the reaction is placed in a machine (thermocycler) where a series of temperature changes are performed [Denature (~94), Anneal (primer specific ~50-60), and Extention (~72)].
For this lab you will be using Promega's GoTaq Product. Please Read!
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
For a 50μl reaction volume:
| Component |
Volume |
Volume used |
Final Conc. |
| GoTaq®Green Master Mix, 2X |
25μl |
25μl |
1x |
| upstream primer, 10μM |
0.5–5.0μl |
1μl 10μM |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
1μl 10μM |
0.1–1.0μM |
| DNA template |
1–5μl |
2μl |
<250ng |
| Nanopure water |
21μl |
Made 100μL 10μM solution for hsp70 primer R and F.
Made replicates of guppy #2 sample and guppy #9 sample, and also replicates of negative controls (water).
Load reactions into thermocycler.
Thermocycler set to:
1 minute at 95'C
1 minute at 55'C
2 minute at 72'C
For 40 cycles
Then, 10 minutes at 72'C
Then, stays at 4'C.
Make Agarose Gels
Mix 2 g agarose with 150 mL Buffer TRIS-TAD
Microwave for 3 minutes
Let it cool down
Add 12 mL ethidium bromide (TOXIC!)
Pour into gel box with well rack in place
Load samples into gel
15uL 100 bp ladder/well
25uL sample/well
Voltage: 100V
Next Day:
Run out PCR products on agarose gels and photograph.
Bottom of gel (Top is John & Amy's)
| Lane |
Sample/blank |
| 1-3 |
Blank |
| 4 |
Ladder |
| 5 |
Negative control 1 |
| 6 |
Negative control 2 |
| 7 |
Guppy #2 muscle, replicate 1 |
| 8 |
Guppy #2 muscle, replicate 2 |
| 9 |
Blank |
| 10 |
Ladder |
| 11 |
Negative control 3 |
| 12 |
Negative control 4 |
| 13 |
Guppy #9 liver, replicate 1 |
| 14 |
Guppy #9 liver, replicate 2 |
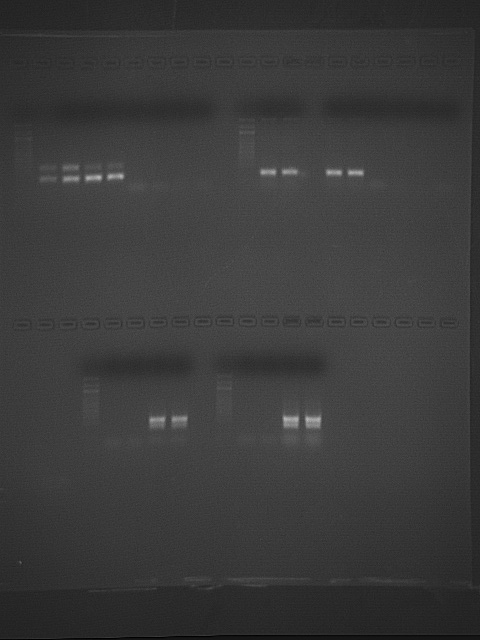
When I looked at the gel in lab, it did not look like there were any primer dimers, but in this photo on-screen, it looks like I've got something very small in my negative controls and samples. Runninga higher annealing temperature may help with this because higher temperature increases specificity of binding. Nothing else showing up in the negative controls. Yay!. But, there are two bands showing up in my samples. Maybe because these are isoforms of hsp70? hsp71 and hsp72? Again, increasing the annealing temperature from 55'C to 60'C may help. I cannot see the ladder very well, but it shows that the mRNA target sequence is small. Expected size is 248 bp. It looks like the right size.
Bands in lane 14 (guppy #9 liver, replicate 2) collected separately into two tubes for sequencing.
Lab 4 - Jan 28, 2009
Western Transfer - Immunoblots
I. Run out protein samples as done previously
II. Transfer proteins from gel to nitrocellulose membrane
III. Probe membrane with antibody (HSP)
II. Transfer proteins from gel to nitrocellulose membrane
1. Cool the transfer buffer to 4°C.
2. Soak the filter paper, membrane and gel in Transfer Buffer for 15 minutes.
3. Assemble the blotting sandwich in a semi-dry blotting apparatus as follows:
• Anode (+++)
• Filter paper
• Nitrocellulose Membrane
• Gel
• Filter paper
• Cathode (– – –)
4. Transfer the blot for 30 minutes at 20V.
5. Remove the gel from the sandwich and rinse with transfer buffer.
6. Use a cotton swab to remove any adhering gel from the membrane.
III. Probe membrane with antibody (HSP)
Western Blotting Protocol Western Breeze Manufacturer's Protocol
General Guidelines
• Avoid touching the working surface of the membrane, even with gloves.
• Work quickly when changing solutions as membranes dry quickly. If the membrane dries, re-wet the membrane with methanol and rinse with water before proceeding.
• Add solutions to the trays slowly, at the membrane edge, to avoid bubbles forming under the membrane. Decant from the same corner of the dish to ensure complete removal of previous solutions.
1. Prepare 20 mL of Blocking Solution
Ultra filtered Water 14 ml
Blocker/Diluent (Part A) 4 ml
Blocker/Diluent (Part B) 2 ml
Total Volume 20 ml
2. Place the membrane in 10 ml of the appropriate Blocking Solution in a covered, plastic dish provided in the kit. Incubate for 30 minutes on a rotary shaker set at 1 revolution/sec.
3. Decant the Blocking Solution.
4. Rinse the membrane with 20 ml of water for 5 minutes, then decant. Repeat once.
5. Prepare 10 mL of Primary Antibody Solution (1:3000 dilution)
Blocking Solution 10 ml
HSP 70 antibody 3.3 µl
Total Volume 10 ml
6. Incubate the membrane with 10 ml of Primary Antibody Solution for OVERNIGHT
Next Day:
7. Prepare Antibody Wash
Ultra filtered Water 150 ml
Antibody Wash solution (16X) 10 ml
Total volume 160 ml
8. Decant Primary Antibody and save in a Falcon tube at 4 degrees C.
9. Wash the membrane for 5 minutes with 20 ml of prepared Antibody Wash, then decant. Repeat for a total of 4 times.
10. Incubate the membrane in 10 ml of Secondary Antibody Solution for 30 minutes, then decant.
11. Wash the membrane for 5 minutes with 20 mL of Antibody Wash, then decant. Repeat for a total of 4 times.
12. Rinse the membrane with 20 ml of water for 2 minutes, then decant. Repeat for a total of three times.
13. Incubate the membrane in 5 ml of Chromogenic Substrate until purple bands develop on the membrane. Development is complete in 1 to 60 minutes.
14. Rinse the membrane with 20 ml of water for 2 minutes, then decant. Repeat for a total of three times.
15. Dry the membrane on a clean piece of filter paper to open air, by a stream of slightly warm air or under an infrared lamp.
Protein gel:
Ladder, John's 2 samples, Amy's 2 samples, my 2 samples, Bob's 2 samples
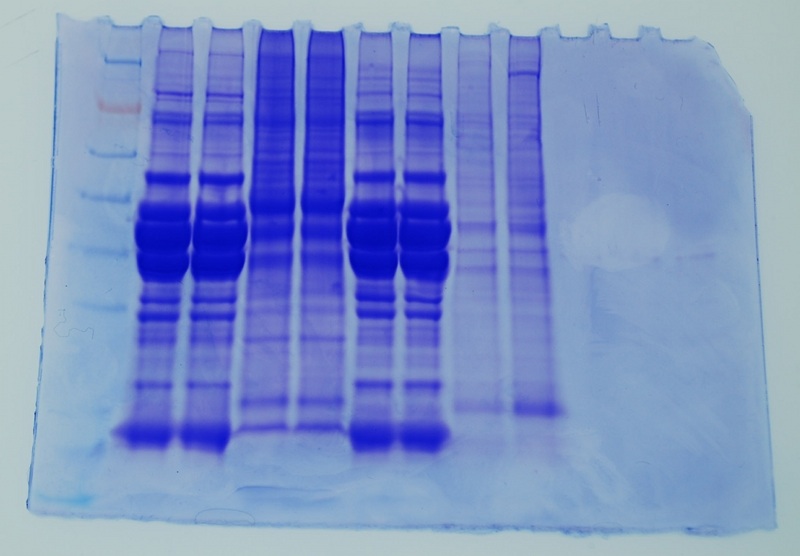
Western: (1.5 hours after Chromogenic Substrate was added to membrane)
Ladder, John's 2 samples, Amy's 2 samples, my 2 samples, Bob's 2 samples
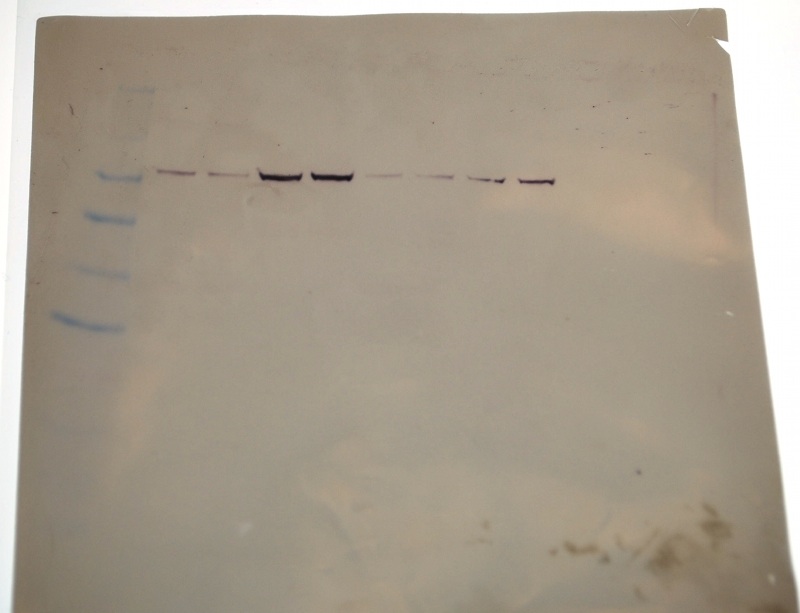
I seem to have the faintest bands compared ton John, Amy and Bob's bands.
Protein gel overlaying Western membrane:
Ladder, John's 2 samples, Amy's 2 samples, my 2 samples, Bob's 2 samples
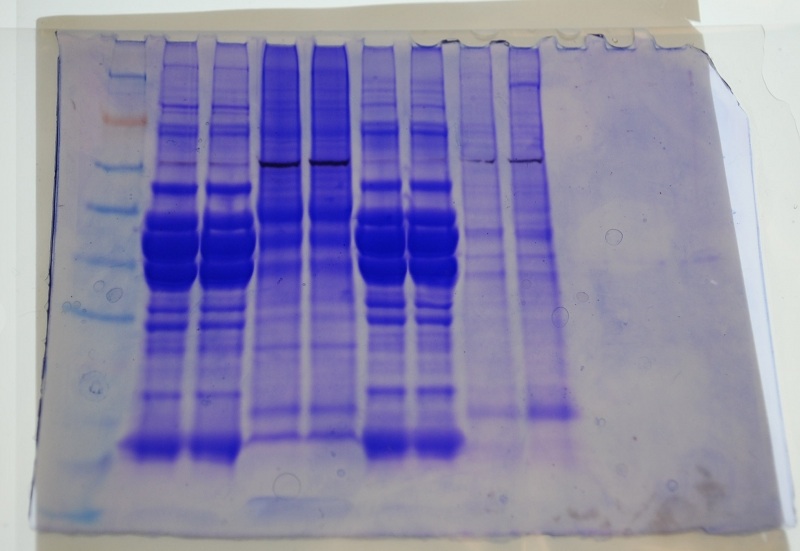
There doesn't seem to be any or much protein where the Western blots are for my sample. This seems to be the same case for John and Bob's samples.The Coomasie blue-stained gels are not as sensitive as the Western.
Lab 5 - Feb 4, 2009
METHODS
PCR
Polymerase Chain Reaction involved amplifying a DNA (genomic or complementary) target using a polymerase, primers (short oligonucleotide), and dNTPs (A, C, T, Gs). In general the reaction is placed in a machine (thermocycler) where a series of temperature changes are performed [Denature (~94), Anneal (primer specific ~50-60), and Extention (~72)].
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
1. Prepare master mix: Prepare enough reactions to run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template). Add 1 additional reaction to the total to ensure sufficient volume recovery.
For a 50μl reaction volume:
| Component |
Volume |
Volume used |
Final Conc. |
| Master Mix, 2X (Immomix) |
25µL |
25 µL |
1x |
| Syto-13 dye (50uM) |
2-5µL |
2 µL |
2 - 5µM |
| upstream primer, 10μM |
0.5–5.0μl |
2 µL |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
2 µL |
0.1–1.0μM |
| Ultra Pure Water |
to 48uL |
17 µL |
NA |
2. Add mastermix to wells of a white PCR plate
3. Thaw cDNA samples.
4. Add 2uL cDNA template to each reaction.
5. Add 2uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. If necessary, spin the strips to collect volume in the bottom of the wells.
8. and ensure the lids are clean (wipe with a Kim Wipe) before going into the Opticon.
9. Load the plate, verify the PCR conditions and start the run.
RESULTS
Quantitation image
| Sample |
C(t) |
Average C(t) per 11mg tissue |
| G2 muscle, R1 |
27.44 |
13.8075 |
| G2 muscle, R2 |
27.79 |
|
| G9 liver, R1 |
18.65 |
19.1733 |
| G9 liver, R2 + 2 uL H20 by accident |
18.65 |
|
| Negative control 1 |
N/A |
|
| Negative control 2 |
38.47 |
|
| G9 liver, R3 |
20.22 |
|
Not sure why I have such a high C(t) for my 2nd negative control, but at least I have nothing in my 1st negative control.Interestingly, I got identical results for replicates 1 and 2 even though I accidently added 2 uL of water to replicate 2.Also, my guppy #9 replicate #3 sample seems to be quite a bit higher than replicates 1 and 2. I think there was a little bit of extra cDNA solution stuck to the outer side of my pipet tip, or I somehow did not pipet as much for replicates 1 and 2.
Although I did not use any primers of constitutively expressed genes to normalize with, I do know the weight of my tissue samples. The muscle tissue was 22 mg and the liver tissue was 11 mg. When adding cDNA template to each reaction, it would have been better if I had only added 1uL of my Guppy #2 liver sample + 1 uL H20 to compare with 2uL of Guppy #2 muscle sample. But I can still figure it out mathematically. For 11 mg of tissue, I expect I would have gotten C(t)=13.88 for muscle and C(t)=19.17 for liver. With this difference in C(t) of 5.29, this corresponds to 39-fold more hsp70 mRNA in liver than in muscle.
19.17 - 13.88 = 5.29
2^5.29 = 39
February 11, 2009
Started RNA extraction on guppy liver samples: #2, 3, 16, 17, 18, 20, 21, 31, 38
Tissue weights are approximately 0.5 mg to 3 mg.
These samples are in -80'C.
For these samples, I have been drying the tissue samples on kimwipes to remove the RNAlater. But, to minimize transfer and contamination of the samples, I could microcentrifuge the tubes, remove the RNAlater, add the TriReagent, and continue the RNA extraction protocol from there. In some of the 1.5 mL tubes with the liver samples in RNAlater had salt precipitate. But after defrosting, shaking the tube helped dissolve the salts. Perhaps it's because I had a lot of RNAlater in the tube relative to the amount of tissue sample, and it was in the -80'C freezer. Samples in RNAlater are now in the -20'C freezer.
Because the tissue samples are so small, I used only one quarter of the volumes listed in the RNA extraction protocol.
250 uL TriReagent
50 uL chloroform
125 uL isopropanol
250 uL 75% EtOH
25 uL 0.1% DEPC H2O
February 12, 2009
Continued RNA extraction with above samples, and with samples # 8, 10, 11, 12, 13, 14, 15.
RNA samples are now stored in -80'C freezer.
Did not quantitate yet.
February 18, 2009
Quantitated RNA for the 16 samples extracted Feb 11 & 12.
DNAsed samples: Ambion-Turbo DNA free (DNAse procedure to eliminate genomic carry-over)
12 uL RNA
2 uL 10X buffer
1 uL DNAse
5 uL H20
20 uL TOTAL
Quantitated RNA again with Nandrop reader
| Sample |
RNA before DNAsed (ng/ul) |
RNA after DNAsed (ng/uL) |
| L2 |
646.9 |
277.0 |
| L3 |
837.5 |
351.9 |
| L8 |
584.9 |
406.3 |
| L10 |
214.0 |
71.7 |
| L11 |
156.5 |
47.8 |
| L12 |
419.0 |
421.3 |
| L13 |
187.0 |
76.9 |
| L14 |
472.2 |
200.7 |
| L15 |
1709.2 |
516.8 |
| L16 |
61.2 |
21.3 |
| L17 |
458.0 |
299.8 |
| L18 |
76.4 |
27.7 |
| L20 |
669.5 |
299.7 |
| L21 |
105.9 |
45.1 |
| L31 |
659.2 |
316.3 |
| L38 |
152.1 |
63.6 |
February 19, 2009
Reverse transcribed the 16 samples above.
Master Mix solution:
140 uL 5X Buffer (AMV RT Buffer)
280 uL dNTPs
35 uL AMV RTranscriptase
35 uL Oligo dT Primer
35 uL RNAse free water
525 uL TOTAL
Made 2 replicates for each of the 16 samples. Don't do this anymore! Steven will senesce more rapidly than usual! Replicates were okay during lab to test our technique, but don't do it for the project.
Stored in -20 degrees C freezer.
Homogenized 7 samples (# 7, 19, 23-27) in TriReagent (250 uL) and stored in -80 degrees C freezer.
February 23, 2009
Ran real-time qPCR on 16 samples above with hsp70 primers.
Master mix solution: (35 reactions)
875 uL Master Mix, 2X Immomix
70 uL Syto-13 dye
70 uL upstream primer (10uM)
70 uL dowstream primer (10uM)
595 uL Ultra pure water
1680 uL TOTAL
| Well |
Sample |
Replicate |
Efficiency |
C(t) |
| G1 |
2 |
1 |
96.22% |
25.08 |
| H1 |
2 |
2 |
101.38% |
24.65 |
| E1 |
3 |
1 |
114.18% |
23.65 |
| F1 |
3 |
2 |
98.55% |
23.29 |
| C1 |
8 |
1 |
72.20% |
24.24 |
| D1 |
8 |
2 |
72.37% |
24.35 |
| G2 |
10 |
1 |
89.56% |
29.15 |
| H2 |
10 |
2 |
74.36% |
29.18 |
| E2 |
11 |
1 |
N/A |
1.86 |
| F2 |
11 |
2 |
N/A |
1.91 |
| C2 |
12 |
1 |
177.29% |
19.88 |
| D2 |
12 |
2 |
110.85% |
20.29 |
| A2 |
13 |
1 |
91.91% |
26.8 |
| B2 |
13 |
2 |
60.75% |
26.07 |
| G3 |
14 |
1 |
90.93% |
24.38 |
| H3 |
14 |
2 |
111.39% |
24.64 |
| E3 |
15 |
1 |
98.85% |
20.14 |
| F3 |
15 |
2 |
113.98% |
20.05 |
| C3 |
16 |
1 |
81.80% |
28.09 |
| D3 |
16 |
2 |
107.87% |
28.35 |
| A3 |
17 |
1 |
136.66% |
2.41 |
| B3 |
17 |
2 |
147.70% |
2.17 |
| G4 |
18 |
1 |
87.53% |
31.92 |
| H4 |
18 |
2 |
111.79% |
31.74 |
| E4 |
20 |
1 |
93.90% |
33.41 |
| F4 |
20 |
2 |
100.54% |
33.14 |
| C4 |
21 |
1 |
72.82% |
32.69 |
| D4 |
21 |
2 |
89.96% |
32.33 |
| A4 |
31 |
1 |
85.38% |
34.36 |
| B4 |
31 |
2 |
88.98% |
35.04 |
| A1 |
Neg |
1 |
N/A |
N/A |
| B1 |
Neg |
2 |
N/A |
N/A |
February 23, 2009
Removed liver tissue from guppies (#47 - 61) preserved in RNAlater. Did not blot the liver on kimwipes. There may have been a little bit of RNAlater still on the liver.
RNA extraction for samples #47 to #61. For each sample, used one quarter of amount of reagents listed in protocol like I did on Feb 11, 2009. Stored in -80'C freezer.
After adding chloroform, vortexing, and allowing sample to sit for 5 min., the TriReagent portion looked "bubbly". I have never noticed this before with my other samples if this does ocur. When solution was vortexed, it looked milky, so I hope it's fine.
February 28, 2009
Set of primers for normalizing gene:
Species and tissue type: Guppy (Poecilia reticulata) liver
Target gene/EST: beta-actin
EST accession number: EU143772
Forward primer (left primer): GAGCGTGGCTACTCCTTCAC
Reverse primer (right primer): GCAGGACTCCATACCAAGGA
Expected size: 234
Annealing temperature: 60'C
Ran qPCR on samples #2 and #3 with replicates and negatives (i.e.6 reactions) with beta-actin and glycogen phosphorylase primers (total 12 reactions).
Master Mix solution for each set of primers:
(7 reactions)
175 uL Immomix
14 uL Syto-13 dye
14 uL upstream primer
14 uL downstream primer
119 uL ultra pure water
336 uL TOTAL
| Well |
Sample |
Replicate |
Target gene |
Efficiency |
C(t) |
| F9 |
2 |
1 |
Actin |
85.92% |
31.06 |
| F10 |
2 |
2 |
Actin |
78.58% |
30.96 |
| F11 |
3 |
1 |
Actin |
105.89% |
19.37 |
| F12 |
3 |
2 |
Actin |
90.36% |
19.27 |
| G9 |
Neg_Actin |
Actin |
N/A |
N/A |
|
| G10 |
Neg_Actin |
Actin |
N/A |
N/A |
|
| G11 |
Neg_GP |
GP |
N/A |
N/A |
|
| G12 |
Neg_GP |
GP |
N/A |
N/A |
|
| H9 |
2 |
1 |
GP |
92.84% |
32.86 |
| H10 |
2 |
2 |
GP |
87.05% |
32.04 |
| H11 |
3 |
1 |
GP |
48.30% |
19.11 |
| H12 |
3 |
2 |
GP |
36.14% |
17.59 |
RNA quantification of samples #47 to #61 before DNAsing.
DNAsed samples #47 to #61.
Ambion-Turbo DNA free (DNAse procedure to eliminate genomic carry-over)
For each sample DNAsed:
12 uL RNA
2 uL 10X buffer
1 uL DNAse
5 uL water
20 uL TOTAL
| Sample ID |
RNA before DNAsed (ng/uL) |
RNA after DNAsed (ng/uL) |
| L47 |
328.48 |
47.96 |
| L48 |
322.95 |
170.62 |
| L49 |
175.63 |
59.68 |
| L50 |
269.36 |
124.23 |
| L51 |
217.79 |
81.74 |
| L52 |
237.29 |
90.61 |
| L53 |
130.6 |
61.55 |
| L54 |
168.89 |
79.08 |
| L55 |
509.63 |
241.62 |
| L56 |
97.95 |
38.35 |
| L57 |
129.16 |
59.93 |
| L58 |
153.63 |
73.29 |
| L59 |
220.52 |
97.19 |
| L60 |
263.07 |
118.85 |
| L61 |
26.01 |
17.91 |
March 3, 2009
RNA quantification of samples #47-69 after being DNAsed.
Reverse transcriptiong of DNAsed RNA samples #47-#67.
Master mix (16 reactions):
64 uL 5X Buffer (AMV RT Buffer)
128 uL dNTPs
16 uL AMV RTranscriptase
16 uL Oligo dT Primer
16 uL RNAse free water
15 uL TOTAL
RT qPCR of samples #2, 3, 8-10, 12-16, 18, 20, 21, 31, 38, 47-61 with set of actin primers +2 negatives, and samples #47-61 with set of hsp70 primers +2 negatives.
Master Mix solutions:
For set of actin primers: (in each of the two 1.5 uL tube)
750 uL Immomix
60 uL Syto-13 dye
60 uL F primer
60 uL R primer
510 uL ultra pure water
For set of hsp70 primers: (in each of the two 1.5 uL tube)
425 uL Immomix
34 uL Syto-13 dye
34 uL F primer
34 uL R primer
289 uL ultra pure water
| Well / Set |
Content |
Replicate |
Efficiency |
C(t) |
| A1 |
8 |
1 |
76.00% |
21.08 |
| A2 |
8 |
2 |
94.58% |
20.84 |
| A3 |
9 |
1 |
95.44% |
21 |
| A4 |
9 |
2 |
102.89% |
21.27 |
| A5 |
10 |
1 |
99.51% |
24.55 |
| A6 |
10 |
2 |
85.20% |
24.27 |
| A7 |
12 |
1 |
98.28% |
18.21 |
| A8 |
12 |
2 |
56.73% |
13.48 |
| A9 |
13 |
1 |
85.54% |
24.28 |
| A10 |
13 |
2 |
93.05% |
24.25 |
| A11 |
14 |
1 |
91.58% |
20.2 |
| A12 |
14 |
2 |
47.65% |
17.86 |
| B1 |
15 |
1 |
57.90% |
14.65 |
| B2 |
15 |
2 |
145.39% |
15.67 |
| B3 |
16 |
1 |
75.03% |
23.84 |
| B4 |
16 |
2 |
71.57% |
23.03 |
| B5 |
18 |
1 |
42.10% |
30.62 |
| B6 |
18 |
2 |
65.99% |
30.07 |
| B7 |
20 |
1 |
61.76% |
31.43 |
| B8 |
20 |
2 |
67.71% |
31.17 |
| B9 |
21 |
1 |
80.12% |
29.55 |
| B10 |
21 |
2 |
70.32% |
31.41 |
| B11 |
Neg_Actin |
1 |
18.74% |
32.44 |
| B12 |
Neg_Actin |
2 |
13.29% |
37.08 |
| C1 |
31 |
1 |
54.77% |
31.37 |
| C2 |
31 |
2 |
57.80% |
31.48 |
| C3 |
38 |
1 |
69.31% |
28.58 |
| C4 |
38 |
2 |
57.46% |
29.05 |
| C5 |
47 |
1 |
131.65% |
20.47 |
| C6 |
47 |
2 |
104.37% |
21.68 |
| C7 |
48 |
1 |
92.05% |
23.09 |
| C8 |
48 |
2 |
99.79% |
23.57 |
| C9 |
49 |
1 |
108.35% |
24.48 |
| C10 |
49 |
2 |
116.04% |
23.73 |
| C11 |
50 |
1 |
114.54% |
22.22 |
| C12 |
50 |
2 |
117.74% |
22.67 |
| D1 |
51 |
1 |
96.92% |
22.78 |
| D2 |
51 |
2 |
90.78% |
22.36 |
| D3 |
52 |
1 |
117.64% |
22.1 |
| D4 |
52 |
2 |
103.38% |
22.79 |
| D5 |
53 |
1 |
113.08% |
23.22 |
| D6 |
53 |
2 |
89.66% |
23.14 |
| D7 |
54 |
1 |
104.08% |
23.36 |
| D8 |
54 |
2 |
112.49% |
24.14 |
| D9 |
55 |
1 |
150.96% |
22.37 |
| D10 |
55 |
2 |
144.59% |
22.48 |
| D11 |
56 |
1 |
101.82% |
23.23 |
| D12 |
56 |
2 |
95.05% |
23.63 |
| E1 |
57 |
1 |
105.13% |
23.89 |
| E2 |
57 |
2 |
93.13% |
23.61 |
| E3 |
58 |
1 |
93.58% |
24.47 |
| E4 |
58 |
2 |
98.15% |
24.16 |
| E5 |
59 |
1 |
10.01% |
34.45 |
| E6 |
59 |
2 |
18.61% |
33.71 |
| E7 |
60 |
1 |
97.60% |
24 |
| E8 |
60 |
2 |
127.65% |
24.39 |
| E9 |
61 |
1 |
12.59% |
33.23 |
| E10 |
61 |
2 |
13.83% |
33.46 |
| E11 |
Neg_hsp70 |
1 |
17.86% |
33.56 |
| E12 |
Neg_hsp70 |
2 |
13.97% |
34.09 |
| F1 |
47 |
1 |
72.30% |
28.95 |
| F2 |
47 |
2 |
64.76% |
29.05 |
| F3 |
48 |
1 |
75.85% |
28.75 |
| F4 |
48 |
2 |
72.01% |
28.49 |
| F5 |
49 |
1 |
55.03% |
31.03 |
| F6 |
49 |
2 |
66.86% |
31.81 |
| F7 |
50 |
1 |
66.63% |
30.72 |
| F8 |
50 |
2 |
76.62% |
29.47 |
| F9 |
51 |
1 |
52.52% |
30.22 |
| F10 |
51 |
2 |
53.75% |
30.04 |
| F11 |
52 |
1 |
76.55% |
28.39 |
| F12 |
52 |
2 |
62.32% |
28.81 |
| G1 |
53 |
1 |
60.01% |
30.3 |
| G2 |
53 |
2 |
70.63% |
30.82 |
| G3 |
54 |
1 |
55.89% |
32.1 |
| G4 |
54 |
2 |
58.68% |
32.25 |
| G5 |
55 |
1 |
73.16% |
29.77 |
| G6 |
55 |
2 |
79.84% |
29.4 |
| G7 |
56 |
1 |
53.23% |
30.86 |
| G8 |
56 |
2 |
62.43% |
31.26 |
| G9 |
57 |
1 |
53.12% |
32.27 |
| G10 |
57 |
2 |
40.59% |
31.72 |
| G11 |
58 |
1 |
68.72% |
28.58 |
| G12 |
58 |
2 |
62.66% |
28.59 |
| H1 |
59 |
1 |
N/A |
N/A |
| H2 |
59 |
2 |
N/A |
N/A |
| H3 |
60 |
1 |
48.84% |
33.2 |
| H4 |
60 |
2 |
49.70% |
33.01 |
| H5 |
61 |
1 |
N/A |
N/A |
| H6 |
61 |
2 |
N/A |
N/A |
March 3, 2009
Ran qPCR with set of actin primers on sample #2 and 2 negatives, and with set of glycogen phosphorylase primers on samples #2, 8-10, 12-16, 18, 20-21, 31, 38, 47-58, 60 and 4 negatives. Ran qPCR at 60 degrees C annealing temperature.
| Well / Set |
Content |
Replicate |
Target gene |
Efficiency |
C(t) |
| A1 |
2 |
1 |
GP |
66.52% |
26.75 |
| A2 |
2 |
2 |
GP |
73.63% |
26.65 |
| A3 |
8 |
1 |
GP |
75.82% |
27.74 |
| A4 |
8 |
2 |
GP |
75.96% |
27.34 |
| A5 |
9 |
1 |
GP |
89.63% |
24.71 |
| A7 |
Neg_GP |
1 |
GP |
N/A |
N/A |
| A8 |
Neg_GP |
2 |
GP |
N/A |
N/A |
| A9 |
Neg_GP |
3 |
GP |
N/A |
N/A |
| A10 |
Neg_GP |
4 |
GP |
N/A |
N/A |
| A11 |
Neg_Actin |
1 |
Actin |
N/A |
N/A |
| A12 |
Neg_Actin |
2 |
Actin |
N/A |
N/A |
| B1 |
9 |
2 |
GP |
73.17% |
24.6 |
| B2 |
10 |
1 |
GP |
84.79% |
27.74 |
| B3 |
10 |
2 |
GP |
73.31% |
28.1 |
| B4 |
12 |
1 |
GP |
74.63% |
26.22 |
| B5 |
12 |
2 |
GP |
75.71% |
26.24 |
| C1 |
13 |
1 |
GP |
87.33% |
30.78 |
| C2 |
13 |
2 |
GP |
72.91% |
30.5 |
| C3 |
14 |
1 |
GP |
87.84% |
25.51 |
| C4 |
14 |
2 |
GP |
87.19% |
26.58 |
| C5 |
15 |
1 |
GP |
72.53% |
24.07 |
| D1 |
15 |
2 |
GP |
96.83% |
23.8 |
| D2 |
16 |
1 |
GP |
70.80% |
32.65 |
| D3 |
16 |
2 |
GP |
71.76% |
33.05 |
| D4 |
18 |
1 |
GP |
78.71% |
35.12 |
| D5 |
18 |
2 |
GP |
72.37% |
36.33 |
| E1 |
20 |
1 |
GP |
80.01% |
32.22 |
| E2 |
20 |
2 |
GP |
85.17% |
31.62 |
| E3 |
21 |
1 |
GP |
66.57% |
33.97 |
| E4 |
21 |
2 |
GP |
113.87% |
32.96 |
| E5 |
31 |
1 |
GP |
77.20% |
34.29 |
| E7 |
53 |
1 |
GP |
91.37% |
32.71 |
| E8 |
53 |
2 |
GP |
84.53% |
32.76 |
| E9 |
54 |
1 |
GP |
69.05% |
33.83 |
| E10 |
54 |
2 |
GP |
83.74% |
33.34 |
| F1 |
31 |
2 |
GP |
68.41% |
35.08 |
| F2 |
38 |
1 |
GP |
66.06% |
34.74 |
| F3 |
38 |
2 |
GP |
78.58% |
33.93 |
| F4 |
47 |
1 |
GP |
86.08% |
30.97 |
| F5 |
47 |
2 |
GP |
77.04% |
31.4 |
| F7 |
55 |
1 |
GP |
69.92% |
31.16 |
| F8 |
55 |
2 |
GP |
68.20% |
30.25 |
| F9 |
56 |
1 |
GP |
69.82% |
34.44 |
| F10 |
56 |
2 |
GP |
102.41% |
33.07 |
| G1 |
48 |
1 |
GP |
77.13% |
30.52 |
| G2 |
48 |
2 |
GP |
76.40% |
30.49 |
| G3 |
49 |
1 |
GP |
78.58% |
30.79 |
| G4 |
49 |
2 |
GP |
85.01% |
30.08 |
| G5 |
50 |
1 |
GP |
74.48% |
30.59 |
| G7 |
57 |
1 |
GP |
78.53% |
35.29 |
| G8 |
57 |
2 |
GP |
79.27% |
32.44 |
| G9 |
58 |
1 |
GP |
73.16% |
33.1 |
| G10 |
58 |
2 |
GP |
45.14% |
30.87 |
| H1 |
50 |
2 |
GP |
71.42% |
31.16 |
| H2 |
51 |
1 |
GP |
70.18% |
31.86 |
| H3 |
51 |
2 |
GP |
71.03% |
31.13 |
| H4 |
52 |
1 |
GP |
89.63% |
30.67 |
| H5 |
52 |
2 |
GP |
69.12% |
30.31 |
| H7 |
61 |
1 |
GP |
75.49% |
32.64 |
| H8 |
61 |
2 |
GP |
78.87% |
30.62 |
| H9 |
2 |
1 |
Actin |
94.49% |
18.43 |
| H10 |
2 |
2 |
Actin |
99.25% |
18.28 |