12/9/2013
Analysis of arbitrary expression shows no significant differences between treatments and other variations of grouping| T-test p-value |
|
| Treatment: pre vs. post |
0.301 |
| Control: pre vs. post |
0.934 |
| Treatment post vs. Control post |
0.333 |
| Treatment pre vs. Control pre |
0.239 |
| Control pre vs. Treatment post |
0.448 |
12/4/2013
Success!| Sample |
Efficiency |
C(t) |
Expression Value |
Average Expression |
|
| Treatment (pre) |
U4 |
0.637 |
27.03 |
1961.390 |
1477.697 |
| U3 |
0.712 |
28.01 |
994.004 |
||
| Treatment (post) |
U4(2) |
0.657 |
29.11 |
463.519 |
393.350 |
| U3(2) |
0.642 |
29.63 |
323.180 |
||
| Control (pre) |
U2 |
0.625 |
29.42 |
373.849 |
292.041 |
| U1 |
0.640 |
30.25 |
210.233 |
||
| Control (post) |
U2(2) |
0.712 |
30.18 |
220.691 |
277.637 |
| U1(2) |
0.757 |
29.58 |
334.583 |
||
| Negative Control |
blank |
N/A |
N/A |
N/A |
N/A |
| Treatment post Treatment post Treatment pre Treatment pre Control post Control post Control pre Control pre Negative control |
Most likely there's another source of stress that's making the controls and baseline samples amplify.
12/3/2013
Secondary qPCR with new primersmaster mix 10x using the first protocol
Per PCR tube:
Sso Fast Eva Green-- 12.5 ul
F primer-- 0.5 ul
R primer-- 0.5 ul
Nuclease free water-- 10.5
cDNA-- 1 ul (or 1ul nuclease free water for negative control -- Labeled "J")
Ran all 8 samples this time, hopefully a trend will appear from this data using more but we'll see!
12/1/2013
Primers either failed or there was no gene expression; my samples were A1-3.Found new primers to order this week to run another qPCR and see if the results differ. These are species specific for gp96 so they may be more viable. If no amplification occurs with the new primers might be able to conclude that there was no expression
Primer pair 5
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
|---|---|---|---|---|---|---|---|---|---|
| Forward primer |
ATTCTTGGTTGCTGAGCGTG |
Plus |
20 |
448 |
467 |
59.12 |
50.00 |
5.00 |
0.00 |
| Reverse primer |
GCCGAGTCAGATTCCCAGAT |
Minus |
20 |
525 |
506 |
59.24 |
55.00 |
5.00 |
2.00 |
| Product length |
78 |
||||||||
11/26/2013
Reverse transcription RNA--> cDNAqPCR
| RNA quantification |
|||||
| Label |
Sample |
Concentration |
A 260/280 |
A 260/230 |
|
| Control |
U1 |
U1 |
19.5 |
1.67 |
0.32 |
| U2 |
U2 |
31 |
1.56 |
0.36 |
|
| U7 |
U1 (2) |
26.7 |
1.52 |
0.27 |
|
| U8 |
U2 (2) |
38.8 |
1.43 |
0.26 |
|
| Treatment |
U3 |
U3 |
32.8 |
1.64 |
0.43 |
| U4 |
U4 |
51.6 |
1.62 |
0.58 |
|
| U6 |
U3 (2) |
20.7 |
1.66 |
0.82 |
|
| U5 |
U4 (2) |
35.1 |
1.55 |
0.31 |
|
| cDNA quantification |
|||||
| Label |
Sample |
Concentration |
A 260/280 |
A 260/230 |
|
| Control |
U1 |
U1 |
1455.8 |
1.59 |
1.93 |
| U2 |
U2 |
1548.3 |
1.59 |
1.95 |
|
| U7 |
U1 (2) |
1581.4 |
1.60 |
1.96 |
|
| U8 |
U2 (2) |
1630.8 |
1.60 |
1.91 |
|
| Treatment |
U3 |
U3 |
1620.5 |
1.60 |
1.97 |
| U4 |
U4 |
1648.9 |
1.60 |
1.97 |
|
| U6 |
U3 (2) |
1607.5 |
1.59 |
1.93 |
|
| U5 |
U4 (2) |
1607.4 |
1.60 |
2.01 |
|
11/25/2013
RNA extractionAfter thawing each of the samples I added 200 ul chloroform, vortexed for 30 sec-- created a milky emulsion.
Incubated for 5 minutes, centrifuged in thefridge for 15 min at 16.3 rpm
transfered aqueous phase-- appeard tan/brown liquid; interphase was only obviously present in U4, not in the rest of the samples
To the aqueous solution I added 500 ul ispropoanol (to each tube)
Inverted 3X to mix-- no "lumps" visible
Incubated for 10 min
spin in microfuge for 8 minutes at 16.3 rpm with hinge up-- no pellet was conspicuous. *this is of concern for pipetting off supernatant and resuspending the "pellet"
After removing the supernatant I added 1 mL 75% EtOH and votexed for about 5 sec each
spun in refrigerated centrifuge for 5 min at 7500 g
removed supernatant (though I don't know where the RNA was since there is no pellet.. I just avoided the very bottom)
Spun tubes for about 15 seconds in centrifuge in fridge
Pipetted off EtOh, allowed to sit while open for about 5 min (small trace amount of EtOH remained to avoid pipetting the pellet)
resuspended in 0.1 % DEPC H2O, allowed to sit at room temperature for less than 5 minutes then stored on ice.
11/22/2013
Beginning RNA isolationAfter removing excess sea water from each tissue tube I added 1000 ul TriReagent (500 ul at a time with homogenizing between additions with a sterile pestle)
Reconstituted the primers with 276 ul nuclease free water in the Forward primer and 305 ul for the reverse. Next I diluted them to a 10:1 by adding 90 ul water to 10 ul primers (did so for each)
Stored in the -80 freezer over the weekend until I have more time to extract.
11/20/2013
Sampling!Unfortunately I wasn't able to do the removing of tube feet as i had for the baseline data but luckily Danny was able to. While he didn't get as much tissue as we initially extracted for the first sampling, there should still be enough for something to appear when amplified (hopefully). His labeling was a little different than the initial sample; "U" means they were in the treatment "C" means they were control and 1 and 2 correspond to each urchin to be determined by pictures and matched to the other labeling system. "S2" indicated it was for RNA extraction "S1" is for protein. all samples were stored in the -80 freezer in a white cardboard box labelled "Team Heat Shock"
11/18/2013
Day 6 of treatmentI fed the urchins and checked on the anemones, more spines were in the urchin treatment tank as well as dark colored "balls" that may have been from the suface of the urchins.
11/16/2013
Day 4 of treatmentI began the heat shock at around 11:15 AM and concluded at 1:15. I noticed when I came back to check on the specimens around 12:45 that the thermometer furthest from the wall was slightly higher than the other, it read about 24 degrees rather than the target 23 degrees C. Other than that, everything else went as planned. Heat stressed for 2 hours with lamps in and acclimated back to the aquarium temperature/water conditions by splashing water over the animals about once every thirty seconds or so for about 3 minutes. I noticed there were quite a few loose spines in the treatment urchin tank but not as many in the control. I fed the urchins a piece of kelp from the stock drawers (will need to find out what that actually was) both the controls and treatments the same amount (about a square inch per urchin) I fed them the day prior as well.
11/13/2013
Day 1 of treatment and sampling
After talking things over with Claire our group decided to severely cut back on our sample sizes and organisms. Now we will only be testing Aggregating anemones and Green urchins, 2 each for treatment and 2 of each for control. Collected tissue from anemones and urchins prior to heat exposure and placed each sample in microcentrifuge tubes--labelled appropriately as shown below.
| Control |
Treatment |
||
| Urchins |
Anemones |
Urchins |
Anemones |
| U1 G |
A1 G |
U3 G |
A3 G |
| U1 P |
A1 P |
U3 P |
A3 P |
| U2 G |
A2 G |
U4 G |
A3 G |
| U2 P |
A2 P |
U4 P |
A3 P |
A= Anemone
G= for genetic expression
P= for protein expression
The urchins were kept in the large flow through container and placed under the heat lamp with a small amount of water on the bottom (about 1 cm in depth across the container-- probably too be recorded as volume later), the anemones were placed in individual containers on rocks and exposed to the heat without any water.
The act of pulling off tube feet and snipping tentacles is likely to be a source of stress, I'm interested to see how our controls turn out ( also just to see if our primers work..)
To find out:
temperatures in the aquariumtemperatures once exposed to heat
how much mass is needed for RNA and Protein extraction-- urchins are proving to be difficult.
To DO: type out exact protocol (need volumes of TriReagent for DNA extraction, reverse transcriptase, gels, buffers, antibodies, etc etc etc)
11/12/2013
Timeline:| Date |
11/12 |
11/13 Day 1 of thermal stress |
11/14 Day 2 |
11/15 Day 3 |
11/16 Day 4 |
11/17 Day 5 |
11/18 Day 6 |
11/19 Day 7 |
11/20 Final sample, heat shock, secondary sample |
11/21 |
| Goal |
sample Urchin tube feet and anemones (test primers?); Get heat lamp, thermometers, set up platforms/ containers |
Extract tissue sample before heat stress, 2 hours of heat |
Heat 2 hours |
Heat 2 hours |
Heat 2 hours |
Heat 2 hours |
Heat 2 hours |
Heat 2 hours |
Sample, heat both control and treatment, sample again? |
Start processing samples |
| Notes |
Limited sample sizes, now only using 2 individuals for treatment and 2 for control for 2 different species. |
=
=
11/5/2013
Specimen CollectionPrimers came in! I removed two tube feet from a green urchin in the FSH aquarium to test the primers. Unfortunately I didn't get a chance to test them because we spontaneously decided to collect my green urchins from Shilshole. I left the tube feet in salt water in the freezer in the yellow tube rack in the freezer door. It's labelled JB. I need to lyse the tissue to proceed with DNA extraction then PCR.
We collected 17 green urchins of various size from the southernmost dock in the north bay of the Shilshole marina. Urchins were removed from the sides of the dock using nets and/or just our hands. They're held in tank within the aquarium flow through table-- they'll be separated later to be placed in something that can be raised for heat exposure while not damaging the tube feet.
11/1/2013
Specimen collection -- Southern AlkiAnthopleura everywhere! Those were pretty easy to spot. Just under the surf there were a few purple Pisaster as well. Those were collected immediately-- into two separate buckets. The anemones still had their rocks as well (will help minimize stress).
Collected about 16 ochre stars-- though they were collected by more than one group member and when released they were in a ball so no complete count as of now. The nudibranchs were harder to find. The southernmost part of the beach where we were collecting had a few larger rocks clustered together as well as a few nudibranchs. We collected 5 shaggy mouse nudibranchs-- though this poses a problem for having even sample sizes.
Unfortunately we didn't see any urchins where we were. I'd like to go further south towards Emma-Schmitz park or talk to Greg Jensen. Dr. Jensen mentioned being able to collect a few for me if need be.
Though the primer I put on the Google Doc was for purple sea urchins I'd like to use green sea urchins since they're more likely found in the areas we sampled.
To Do:
Get sea urchins!
Order primers-- see if I can find one for S. droebachiensis
10/29/2013
Team Nematocyst is now Team Heat Shock ProteinExperimental Set Up:
We will have four different organisms to work with; Sea Urchins, Aggregating Anemones, Nudibranchs, and Sea Stars. I personally will be using Sea urchins.
The treatment will consist of lifting the organisms from the bottom of the tank to a shallow covering of water while exposed to a heat lamp for two hours a day over the course of one week. A little more research should be done to ensure accuracy of tidal timelines and shallow water temperatures.
The control and treatment groups will be in the same tank in the aquarium in the basement of the Fisheries Building on the far left tank when entering the room.
As of right now it sounds like we will be taking samples at three stages of the experiment- once at the beginning, one at the mid-point, and one at the end-- for both PCR and protein analysis (Western Blot)
I would also like to have an additional experiment where the control group is exposed to the same high heat conditions as the treatment group had and then take tissue samples from all the specimens to see how their protein expression differs. I'm hoping this will give an example of "priming" a system
To Do:
Collect specimens (hoping for n=8 per treatment)
Order primers for each organism
Set up platform to raise critters
Get heat lamp (or more than one to make sure all treatments get heat)
Lab 4: Protein SDS/PAGE and Western blot, analyze conventional PCR via agarose gel and qPCR data
Summary:
The purpose of this lab is to complete protein extraction and start run on SDS-PAGE Gel/Western Blot as well as to run conventional PCR samples on an agarose gel.
PCR ELECTROPHORESIS PROCEDURE
- Place gel in gel box and fill with 1x TAE buffer (to fully cover wells)
- Remove combs from wells
- Load 7uL 100bp ladder in far left lane
- Load 20 uL of PCR sample into the gel (retain the remaining vol at -20ºC)
- Run gel at ~ 100V for ~ 1hr
- Visualize the gel on the UV transilluminator
*While the gel was running we proceeded with protein extraction below.
* I used the third sample in the tray ( 0-0-DNA-DNA )
Protein Extraction and Analysis Part 2
Materials- micropipettes (1-1000 μL)
- sterile filter pipette tips (1-1000 μL)
- sterile gel loading tips
- 1.5 mL screw cap tubes
- microcentrifuge tube rack
- lab coats
- safety glasses
- gloves
- lab pen
- timers
- heating block with water bath
- tube "floatie" (8 tube capacity)
- glass container for boiling water that can accommodate "floatie"
- protein gel box (SR provided)
- SDS/PAGE gels
- gel loading tips
- trays for staining gels
- power supply
- platform rocker/shaker
- plastic wrap
- 2X SDS reducing sample buffer
- protein ladder marker
- gel running buffer
- light box
- Begin boiling water on hot plate.
- In a fresh, 1.5mL screw cap tube add 15uL of protein stock and 15uL of 2X Reducing Sample Buffer. Return protein stock to the box in the -20C freezer labeled protein samples.
- Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
- Boil sample for 5 mins.
- While sample is boiling, rinse gel wells thoroughly.
- When sample is finished boiling, immediately centrifuge for 1 min
- Slowly load the entire sample (30uL) into the appropriate well using a gel loading tip.
- My sample was in well #10.
- Put lid on gel box and plug electrodes in
- Set voltage to 150V and turn power supply on. Run for 45mins.
- Turn off power supply and disconnect gel box from power supply.
- Remove lid from gel box.
- Disengage the tension wedge and remove gel from gel box.
- Carefully crack open cassette to expose gel.
- Proceed to Western Blotting protocol.
WesternBreeze Chromogenic Western Blot Immunodetection
Materials- Nanopure water
- gel staining tray
- Blocking Solution
- rotary shaker
- Primary Antibody Solution
- Antibody Wash
- Secondary Antibody Solution
- Chromogenic Substrate
- timers
- lab coats
- safety goggles
- gloves
- SDS-PAGE gel
- Tris-Glycine transfer buffer
- filter paper
- nitrocellulose membrane
- semi-dry transfer station
- Soak the filter paper, membrane and gel in Tris-Glycine Transfer Buffer for 15 minutes.
- Assemble the blotting sandwich in the semi-dry blotting appartus:
- anode (+++)
- blot paper
- filter paper
- membrane
- gel
- filter paper
- blot paper
- cathode (---)
- Transfer the blot for 30 minutes at 20V
- Remove the gel from the sandwich and rinse off adhering pieces of gel with transfer buffer.
- Wash membrane 2 times, for 5 minutes each, with 20 mL of pure water.
- Put the membrane in the plastic box and add 10 mL of Blocking Solution. Cover and incubate 30 minutes on a rotary shaker set at 1 revolution/second. -- Following steps by TA within next 24 hours.
- Decant liquid.
- Rinse the membrane with 20 mL of water for 5 minutes, then decant. Repeat.
- Incubate the membrane in 10 mL of Primary Antibody Solution. Decant the solution.
- Rinse the membrane with 20 mL of Antibody Wash for 5 minutes, then decant. Repeat 3 times.
- Incubate the membrane in 10 mL of Secondary Antibody Solution for 30 minutes. Decant.
- Wash the membrane for 5 minutes with 20 mL of Antibody wash, then decant. Repeat 3 times.
- Rinse the membrane with 20 mL of pure water for 2 minutes, then decant. Repeat twice.
- Incubate the membrane in 5 mL of Chromogenic Substrate until a purple band appears. This will occur between 1-60 minutes after adding the Chromogenic Substrate.
- Dry the membrane on a clean piece of filter paper to the open air.
Results
Western BlotThe blot process was not complete in the allotted lab time but was completed the next day during class time.
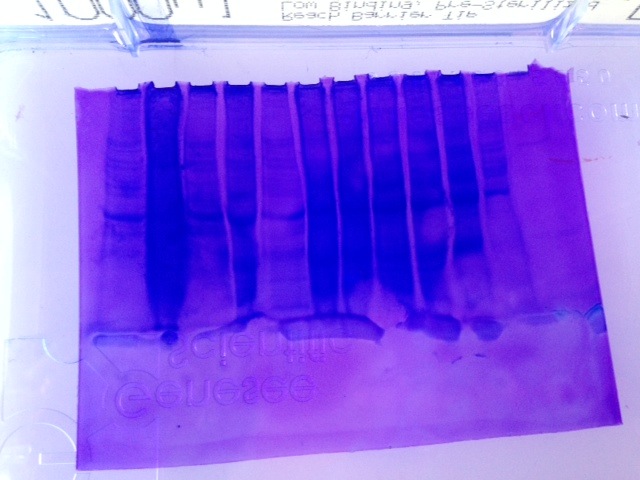
Figure 1. SDS PAGE gel
Figure 2. Western Blot
My sample was in well number 10, our gel/blot was placed backwards-- the ladder is on the far right instead of the far left. My sample is the third from the left, it appears that there was not very much expression of HSP70 when compared to other samples here.
PCR
I expected my product to be about 300 base pairs long. However, my results indicate little to no amplification. There was only a small dot of color approximately 100 base pairs long.
Conclusions
Western BlotRelative to other samples strength in color, I would conclude that there was not enhanced expression of HSP70 in my sample. This may indicate that the oyster from which this sample was taken had little to no cellular stress prior to sampling.
PCR
My results indicate that there was either no amplification of my target gene (HSP70) or I used the wrong sample. qPCR analysis from last week shows that one of my two DNA samples amplified but the other did not. When selecting the DNA sample I thought I was using the sample that amplified but my PCR results might imply that I chose the well that did not. I may have had pipetting errors when starting my PCR last week-- such as leaving the droplet of DNA on the side of the tube prior to thermocycling.
Reflections
I think the purpose of this lab was to analyze the proteins expressed in oyster tissue to become familiar with Western blotting and techniques to see how gene expression translates to the creation of proteins. The procedure in this lab was used to separate proteins and transfer them to blot paper via Western blotting. I wish there was a little bit more information as to what the western blot machine was doing as far as the electrode configuration and what the results will mean.==
==
Lab 3: qPCR, analyze qPCR (via computer data and agarose gel), primer design, prep for experiment, and protein extractions
Lab Summary
The purpose of this lab was to perform qPCR on cDNA, meet up with groups to creat a mock-up experiment, and to extract protein from a new tissue sample. For this lab we used an Opticon thermal cycler with Immomix master mix to create qPCR curves and I used Olympia oyster gill tissue for protein extraction using CelLytic MT Cell Lysis agent to be analyzed later.
Quantitative PCR
Materials- PCR Plates (white); optically clear caps
- 1.5 ml microfuge tubes
- 10.5 uL Nuclease-free water
- filter tips
- Opticon thermal cycler
- kim wipes
- 2x Immomix Master Mix
- 1 uLPrimers (downstream and upstream)
- SYTO-13 Dye
- microfuge tube racks
- ice buckets
- timers
- cDNA samples (student provided)
qPCR PROCEDURE
1. Prepare master mix: enough for 5 total treatments; 2 negative controls, 2 experimental treatments, and one to make up for pipetting errors.
For a 25μl reaction volume:
| Component |
Volume per well |
Final Conc. |
Total Volume Used |
| Master Mix (SsoFast EvaGreen supermix) |
12.5µL |
1x |
62.5 uL |
| *upstream primer, 10μM |
0.5μl |
2.5μM |
2.5 uL |
| *downstream primer, 10μM |
0.5μl |
2.5μM |
2.5 uL |
| Ultra Pure Water |
10.5uL |
NA |
52.5 uL |
- Used HSP70 protein (HSP70-F and HSP70-R)
2. Add mastermix to wells of PCR plate
3. Thaw cDNA samples
4. Add 1uL cDNA template to each reaction
5. Add 1uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. If necessary, spin the strips to collect volume in the bottom of the wells.
8. Ensure the lids are clean and place strips on ice.
9. Load the plate, verify the PCR conditions stated below and start the thermocycler.
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 15 s
5. Read plate
6. Return to step 2 39 more times
7. 95°C for 10s
8. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Making an agarose gel (I didn't do this process, done ahead of time)
Supplies and Equipment:- Micropipettes (1-1000 μl)
- Sterile filter pipette tips (1-1000 μl)
- Tip waste jar
- 1L flask
- agarose
- 1X TAE
- Ethidium bromide
- Microwave
- Gel rigs
- Kimwipes
- Lab coat
- Safety glasses
- gloves
AGAROSE GEL POURING PROCEDURE
- Weigh 2g of agarose and mix with 150mL 1x TAE in a 1L flask
- Microwave solution for ~ 3 minutes. Keep an eye on the solution so that it does not boil over. You want the solution to be clear - no precipitate and no bubbles.
- Cool solution (you should be able to touch the flask for a few seconds), then add 12uL ethidium bromide(EtBr). WARNING: EtBr is a carcinogen be sure to wear gloves and appropriately dispose tip waste.
- Mix thoroughly by swirling, then pour into gel tray.
- Add gel combs. Using a clean pipet tip, pop any bubbles that could get in the way of your PCR product.
- After gel is set, wrap in plastic wrap (label with your initials and date) and place gel in the fridge if not using immediately.
Protein Extraction Part 1
Materials
- micropipettes (1-1000uL)
- sterile filter pipette tips (1-1000uL)
- sterile (RNase free) 1.5mL microcentrifuge tubes
- sterile disposable pestles
- microcentrifuge (refrigerated or in fridge)
- ice buckets
- gloves
- Kim wipes
- lab pens
- safety glasses
- 500 uL CelLytic MT Cell Lysis Reagent (with Protease Inhibitor Cocktail added)
PROTEIN EXTRACTION PROTOCOL
- Label the snap cap tube
- 82 gi Oly JB
- Add 500 ul of CellLytic MT solution to the 1.5mL snap cap tube containing your cut piece of frozen tissue.
- Homogenize the tissue with a sterile disposable pestle.
- Close the tube and invert the tube several times.
- Spin the tube in a refrigerated microfuge for 10 mins at max speed.
- While spinning, label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
- Carefully transfer supernatant (the clear or translucent liquid on top) to labeled tube and store tube on ice.
Results
qPCRBoth negative controls remained a flat line at 0 meaning no contamination occurred. However, only one of my two DNA samples had a fluorescence curve and it appeared to begin rather late along the cycles. I didn't see the final curve once the qPCR was finished but this preliminary look doesn't appear to indicate cellular stress prior to the sample removal-- though nothing solid can be proven just yet.
Protein Extraction
I will be able to see how successful the extraction was next week when we quantify the DNA through Western Blotting. Unfortunately I wasn't able to obtain the initial mass of the sample but I'm not sure what effect this will have on the blot.
Group meeting
My group met up to discuss possibilities for our own experiment. We were all interested in cnematocyst firing mechanisms in nudibranchs but weren't sure how to go about designing an experiment-- especially since nudibrachs don't produce their own cnematocysts, rather they steal the stinging cells from cnidarians. By the end of our discussion it sounded like we settled on a simpler test subject: sea anemones. The next step is a treatment and primer to select. Some ideas included using a predator as a stressor, or exposure to different water qualities, or regeneration of the stinging cells, or doing Western Blot of proteins expressed at different times or with different stresses. We still have some research and meeting to do. There may be some discussion of getting jellyfish tentacles on ice later if we decide to do something cross-species instead.
Conclusions
My qPCR results were not what I expected. I expected both of my cDNA samples to fluoresce. I'm curious to see the entire curve to find out if the sample that hadn't fluoresced when I saw it initially started to amplify. Next I will quantify my protein extracted and work with my experiment group to settle on what exactly our experiment will entail to order primers next week.Reflection
I think the purpose of this lab was to practice common genetic techniques for discovering the responses to environmental stress by using qPCR (to build on last week's cDNA synthesis and PCR) and protein extraction to be expanded upon later. Also we were to get the ball rolling on our personal experiments to get primers ordered by next week. I'm glad for the chance to see what the different primers were doing within the tissue. I've been interested in what the HSP primer will do for the tissue samples I've been working with this last summer when exposed to Ocean Acidification.=
Lab 2: RNA isolation Part 2
October 8, 2013Summary:
The purpose of the lab is to complete and practice RNA extraction and to begin making cDNA for next week.We will also design primers and discuss group project ideas for the rest of the quarter.
We finished this using chloroform and ethanol to separate, precipitate, and wash the sample.
My tissue sample was 48 g (Pacific Oyster gill).
Materials
- micropipettes (1-1000 μL)
- sterile filter pipette tips (1-1000 μL)
- 1.5 mL microcentrifuge tubes
- microcentrifuge tube rack
- lab coats
- safety glasses
- gloves
- lab pen
- timers
- ice buckets (with ice)
- phenol/chloroform waste containers (liquid/solid)
- vortex
- hot water bath
- Nanodrop spectrophotometer
- 200 uL chloroform
- RNase free water
- 500 uL isopropanol
- 1 mL 75% ethanol (EtOH)
- 100 uL 0.1% DEPC treated water
Procedure:
1. Turn on water bath to 55°C.
2. Incubate homogenized tissue sample (from Lab 1-- 48 G JB) tube at room temperature (RT) for 5 mins.
- Also held in hand for a few seconds at a time
4. Vortex for 30s.
- solution became a milky light pink color
6. Spin tube in microcentrifuge in refrigerator for 15 mins. @ max speed: 14000 G
7. Slowly and carefully transfer most of the aqueous phase to a fresh microfuge tube using a pipette. Do NOT transfer ANY of the interphase.
- May have left small amount of aqueous phase in the interphase tube. left there to avoid contamination, disposed of tube at end of lab.
- Had a total of ~ 0.4 mL aqueous solution in new tube
9. Mix by inverting the tube numerous times until the solution appears uniform.
- no lumps, mildly cloudy
11. Spin in refrigerated microfuge at max speed (14000 G) for 8 mins.
- When placing your tube in the microfuge position the tube hinge pointing up, away from the center of the microfuge.
- A small, white pellet (RNA and salts) should be present, if not, don't fret :)
- I had a small pellet pressed up against the side of the bottom of the tube.
13. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube.
- pellet dislodged quickly and settled back to bottom
15. Carefully remove supernatant with micropipette. Make sure not to remove pellet!
- I used the 1000 uL pipette set to 500 uL
17. Using a 10 uL pipette, remove remaining EtOH.
18. Leave tube open and allow pellet to dry at RT for no more than 5mins.
19. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
- vortex if necessary
21. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample (stored by T.A. at -80 degrees C)
Reverse Transcription/ cDNA:
Materials:
- Micropipettes (1-1000 μl)
- Sterile filter pipette tips (1-1000 μl)
- Tip waste jar
- PCR tubes (0.5 ml; thin walled)
- RNA samples (student provided)
- M-MLV reverse transcriptase
- M-MLV 5X reaction buffer
- Oligo dT
- dNTPs
- Nuclease Free water
- thermal cycler
- microfuge tube racks
- PCR tube racks
- ice buckets
- Kimwipes
- Lab coat
- Safety glasses
- gloves
Procedure:
- Mix stock RNA sample by inverting tube several times.
- In a 0.5 ml PCR tube labeled with your initials and “cDNA” combine the following:
- 5 μl of RNA
- 1 μl of oligo dT
- 4 μl of nuclease free H2O
- Incubate the mixture for 5 min at 70°C on the thermocycler then immediately transfer to ice. Briefly centrifuge you tube and the add the following:
- 5 μl of M-MLV 5X Reaction Buffer
- 5 ul of dNTPs
- 1 μl of M-MLV RT
- 4 μl of nuclease free H2O
- Incubate the mixture for 60 min at 42°C and then heat inactivate at 70°C for 3 min on the thermocycler.
- Spin down the sample in a desk top centrifuge.
- Store on ice or at -20°C
Results
I got cDNA and RNA products! Since I didn't do RNA quantification I don't know exactly how much RNA was in my extraction but I should know more about my RNA and cDNA products in later labs.Reflections
I think the purpose of this lab was to finish the RNA isolation from last week and to make cDNA for sequencing. The procedures we used are meant to make stable cDNA to sequence and to isolate RNA using chloroform and alcohol. I think these methods can be used to find products on both sides of RNA in the central dogma-- we can then sequence DNA as well as RNA products/proteins.Class Notes:
How do you make cDNA?- RNA
- nucleotides (x100 allotment)
- Enzyme (reverse transcriptase)
- buffer
-- Much more stable than RNA, can sequence using reverse transcriptase
Discussed possible group research projects-- need to solidify that for next week
- possible ideas could include the HSP chaperone complex or HSP70 expression in geoducks after exposed to high pCO2.
Lab 1: DNA isolation; initiate RNA isolation (Part 1)
October 1, 2013
Pre-class Notes:
DNA -- transcribed --> RNA -- translated --> proteins/ products
ways to visualize each:
| PCR |
gene expression |
Western blot |
| qPCR |
electrophoresis |
|
| electrophoresis |
PCR= Polymerase Chain Reaction; way to amplify target sequence, exponential increase in products.
DNA extraction-- 25-50 mg tissue
RNA extraction-- 50-100 mg tissue
"Oly" = Olympia oyster
"Pac" = Pacific oyster
"M" = mantle
"G" = gill
Summary:
The purpose of this lab is to become familiar with extracting DNA and RNA from Olympia oyster (DNA) and Pacific oyster (RNA) gill tissue by using TriReagent for RNA prep and DNazol for DNA.RNA Extraction Part 1
Materials- micropipettes (1-1000uL)
- sterile filter pipette tips (1-1000uL)
- sterile (RNase free) 1.5mL microcentrifuge tubes
- sterile disposable pestles
- vortex
- ice buckets
- gloves
- lab pens
- safety glasses
- 1000 uL TriReagent
RNA isolation procedure:
Tissue sample: Pacific oyster gill
- Label the snap cap tube--
- 48 G (JB)
- Add 500 uL of TriReagent to the 1.5mL snap cap tube containing tissue. Stored on ice.
- Homogenized the tissue using a disposable pestle and vortexed
- A few small chunks remained
- After the sample homogenized, added additional 500 uL of TriReagent to the tube
- It took some time to get to the second addition of TriReagent-- there's a chance the sample got warm while waiting
- Mixed with pestle and flicking the tube.
- Gave labeled homogenized tissue sample to the TA for storage at -80ºC to be finished next week.
DNA Isolation (DNazol)
Materials- micropipettes (1-1000 µL)
- sterile filter pipette tips (1-1000 µL)
- 1.5 mL microfuge tubes
- microcentrifuge tube rack
- microcentrifuge (room temperature)
- razor blades
- vortexes
- 1 mL DNazol
- 100% ethanol
- 75% ethanol
- 0.1% DEPC water
- kim wipes
- Nanodrop
DNazol Extraction Procedure (Adapted from MRC manual)
Tissue sample: Olympia oyster gill
- Using a sterile pestle, homogenized tissue sample in 0.5 mL of DNazol in a 1.5 mL sterile microfuge tube. Add 0.5 mL more of DNazol and mixed
- Sample incubated for 5 minutes at room temperature.
- Centrifuged sample at 10,000 x g (room temp) for 10 minutes.
- Transferred supernatant to a new, labeled tube
- 74 g (JB)
- Added 0.5 mL of 100 % ethanol to sample.
- Mixed by inverting tube 8 times.
- Incubated at room temperature for 1 minute.
- DNA formed a cloudy precipitate, relatively homogenous
- Cntrifuged for 2 minutes
- small pellet filled the tip of the microcentrifuge tube
- Removed fluid using 1000 uL pipette, leaving DNA in pellet on bottom of microcentrifuge tube
- Sample sat at room temp for 1 minute. Removed the rest of the liquid and disposed it
- Washed DNA with 1 mL of 75% ethanol: Pipeted the ethanol into DNA tube, inverted 6 times, and let sit for 1 minute. Removed the ethanol from the tube using the 1000 uL pipette and repeated.
- Removed with a 1000 uL pipette and 20 uL pipette, disposed.
- Add 300 µL of 0.1% DEPC water to DNA and pipeted up and down multiple times and vortexed to dissolve.
DNA Quantification
- Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm.
- Select "dsDNA" from the pulldown menu
- Click "Blank", to zero the instrument. NOTE: steps 1 and 2 only need to be done once for the whole class.
- Steps 1-3 above were completed when I arrived to use the Nanodrop
- Pipetted 2µL of DNA sample onto the Nanodrop pedestal and lower the arm
- Click "Measure". Record DNA concentration (ng/µL), A260/280 ratio and A260/230 ratio. NOTE: The Nanodrop uses the Beer-Lambert Law
- Raise the arm and wipe off Nanodrop with a KimWipe
- Labeled DNA sample: 78 g JB
- Store sample at -20ºC.
DNA Quantification Results:
| Sample |
8 |
| lambda |
230 |
| Abs |
3.768 |
| A-260 (10 mm path) |
10.818 |
| A-280 (10 mm path) |
6.329 |
| 260/280 |
1.71 |
| 260/230 |
2.87 |
| ng/uL |
540.9 |
| Notes |
No spike on the far left of graph -- means no excess ethanol in sample? |
Reflections:
I believe the purpose of this lab was to practice using a common DNA extraction technique and prepare tissue for RNA extraction to become familiar with initial steps of genetic analysis of tissue samples. As a short-term goal, the procedures performed here were used to measure just how much DNA was extracted from a tissue sample. In the long term, we should be able to measure how much of a product is made from the gene onward as well as from RNA on-- we will be able to see what gets both translated and transcribed once additional analysis is done. ie) through PCR, qPCR, western blot, electrophoresis, and gene expression. So far this procedure has been pretty clear to me; the only part I am little confused about is the final DNA quantification. I'm not sure what the categories of numbers mean or what optimal ranges of numbers would be.Assignment: name a few genes interested in for project and why they're important.
HSP70-- Heat shock protein 70-- important for protein folding and protecting cells from signs of stress
AQP3-- Aquaporin 3-- codes for a water channel on cell membranes (Homeostasis and osmoregulation)
AFGP-- Antifreeze glycoprotein-- to prevent freezing in arctic fish; often carried by blood
LHP (60?)-- Larval haemolymph protein -- for the production of haemolymph