11/27/12
Pre-Stress Experiment - qPCR and Data Analysis
Summary: During this lab I prepared qPCRs for two different primer sets. The first was for hsp70 and the second was for
glutathione peroxidase. For each of these primer sets I prepared 28 samples along with 2 negative controls per primer set. After the qPCR was run, we normalized gene expression levels and compared the mean expression levels using t-tests.
Materials and Methods:
Micropipettes
sterile filter pipette tips
tip waste jar
qPCR tubes
1.5mL microcentrifuge tubes
cDNA
Primers
Nuclease Free Water
Kimwipes
microfuge tube racks
qPCR tube racks
lab coat
gloves
PCR plates
optically clear caps for PCR
opticon thermal cycler
2X Immomix Master Mix
SYBR Green Dye
spectrophotometer
lab pens
qPCR Protocol (Primer Testing):
1) Prepared Master Mix for qPCR using Table 4, labeled with "MM for qPCR" and my initials
2) We then added Master mix to 6 wells each on two white PCR plates
3) cDNA samples were prepared and then 2u was placed in each well according to labels (1 well for each sample to account for loading/pipetting error)
4) 2uL of PCR water was added to three wells as negative controls to account for potential Master Mix contamination.
5) All caps were secured, and then wiped to ensure clean, clear surfaces.
6) PCR plate was loaded, sample locations were marked on a common spreadsheet and then PCR run under set conditions (Table 2) was started by TA.
7) This entire procedure was completed again for the second set of primers.
Data interpretation
1) qPCR data was downloaded and exported into excel.
2) Treatment by treatment, data was then loaded into PCR Miner and fit together in another excel spreadsheet.
3) The expression of each gene was then calculated using the formula 1/(1+A)^B, where A is the average efficiency of the gene (average per treatment with single primer set) and B is the CT value as given by PCR Miner.
4) Gene expressions were then normalized by using the value calculated in step 3 divided by the normalizing coefficient (calculated for each sample by TA then taken out of spreadsheet on GoPost).
5) Means and standard deviations were then calculated for each treatment, along with two-tailed t-tests to compare across treatments.
Results:
Relative expressions of hsp70 in control, initial heat-stress, and pre-heat stressed experiments is seen in Figure 1. Though we seen a large difference in the mean gene expression values, the only significant difference seen was between control and initial heat-stress treatments in the expressions of hsp70, marked in red, in Table 3. Relative expressions ofglutathione peroxidase across treatments is seen in Figure 2. Though there appears to be an up-regulation of glutathione peroxidase in the pre-heat stress treatments the standard error is large, leaving no significant differences between treatments.
Conclusions:
Unfortunately, we only saw significance in one treatment with a p-value of 0.0495, hsp70 regulation between control and initial heat stressed animals. Even though we see a much greater mean of hsp70 regulation in the Pre-Stress treatment (Figure 1), the range of gene expression levels was from about 600 to 2, leading to extreme error bars and non-significant data. I believe that if we repeated the experiment with more individuals, we could narrow down our mean expression levels for each treatment and give the data a higher likelihood of being significant.
As for levels of glutathione peroxidase between treatments, there was no significant differences between treatments. However, we do see what seems to be an up-regulation of the gene in the pre-stress treatment. In a follow-up experiment this could be explored further to determine if significance could be achieved with a larger sample size.
This data means that the animals are reacting to the heat stress and are apparently responding by higher expression levels when exposed a second time to the heat stress. Even though not all of the differences are significant, we a seeing a trend that should be explored further.
Reflection:
Looking back at this experiment I think that we might have been able to achieve significance in many more of the treatments with a larger sample size. Otherwise, given that all qPCR results seemed to be without contamination and all amplifications appeared successful, I believe that this project overall was a success overall and ought to be pursued further in the future with higher sample sizes.
Figure 1. Hsp70 gene expression as a function of treatment. Positive standard error bars are shown.
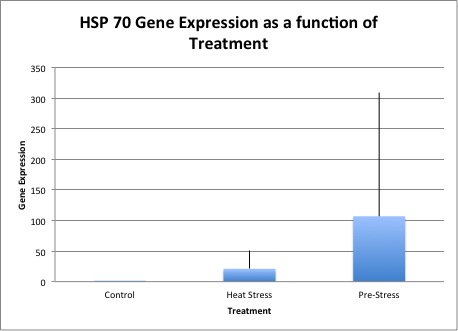
Figure 2. Glutathione peraoxidase as a function of treatments. Standard error bars are shown.
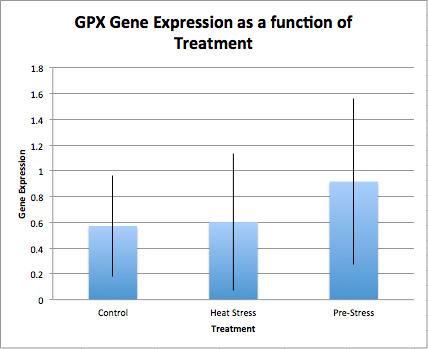
Table 1. Master Mix preparation table for qPCR reactions. Made once for each primer set.
| Reagent |
1 Reaction (uL) |
|
Total in Master Mix (uL) |
| 2X Immomix (Master Mix) |
12.5 |
31 |
387.5 |
| 10uM Forward Primer |
1.25 |
38.75 |
|
| 10uM Reverse Primer |
1.25 |
38.75 |
|
| SYBR Green (50uM) |
1 |
31 |
|
| PCR Water |
7 |
217 |
Table 2. Conditions for qPCR run.
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 30 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Table 3. Means and standard deviations of control, initial heat stress, and pre-heat stressed treatments. Also listed is the p-values by the treatments compared, significant values are highlighted in red.
| average |
standev |
p-value |
||||
| hsp70 |
Control |
0.595338349 |
0.438052559 |
Control/Heat |
0.049459654 |
|
| Heat Stress |
20.98203076 |
30.57204658 |
Control/Pre |
0.112892111 |
||
| Pre-Stress |
106.9144598 |
201.7818607 |
Heat/Pre |
0.224619625 |
||
| average |
standev |
p-value |
||||
| gpx |
Control |
0.571498969 |
0.39197108 |
Control/Heat |
0.887779472 |
|
| Heat Stress |
0.6019879 |
0.532119043 |
Control/Pre |
0.17194842 |
||
| Pre-Stress |
0.916070765 |
0.64404753 |
Heat/Pre |
0.276009918 |
11/20/12
Pre-Stress Experiment - Nanodrop RNA samples and Primer Test
Summary: During this lab I completed my RNA isolation by using a Nanodrop spectrometer to determine contaminant levels and relative RNA concentrations. I then set up a qPCR in order to test my new primer sets for the genes hsp70 and
glutathione peroxidase.
Methods and Materials:
Micropipettes
sterile filter pipette tips
tip waste jar
PCR tubes
1.5mL microcentrifuge tubes
cDNA
Primers
Nuclease Free Water
thermal cycler
Kimwipes
microfuge tube racks
PCR tube racks
ice buckets
lab coat
gloves
PCR plates
optically clear caps for PCR
opticon thermal cycler
2X Immomix Master Mix
SYTO-13 Dye
spectrophotometer
microcentrifuge (in fridge)
lab pens
DI water
RNA Quantification (Nanodrop):
1- Nanodrop pedestal was first wiped down with a kim wipe, and then 2ul of 0.1% DEPC-H2O were pipetted onto the pedestal and the arm was lowered.
2- "Blank" was selected in order to zero the instrument. (These first 2 steps only need to be done once to set for all sampling)
3- 4ul of RNA sample was pipetted onto the nanodrop pedestal and the arm was lowerd.
4- We clicked "Measure" in order to record our RNA concentration (ng/ul). We also recorded the A260/280 and the A260/230 ratios which are calculated by the nanodrop using the Beer-Lambert Law. Numbers were recorded.
5- The arm was lifted and the sample was cleaned off the Nanodrop using a kim wipe.
6- RNA tubes were labeled with tissue type, "RNA," initials, the date, and concentration before storing at -80 degrees C.
qPCR Protocol (Primer Testing):
1) Prepared Master Mix for qPCR using Table 4, labeled with "MM for qPCR" and my initials
2) We then added Master mix to 6 wells each on two white PCR plates
3) cDNA samples were prepared and then 2u was placed in each well according to labels (1 well for each sample to account for loading/pipetting error)
4) 2uL of PCR water was added to three wells as negative controls to account for potential Master Mix contamination.
5) All caps were secured, and then wiped to ensure clean, clear surfaces.
6) PCR plate was loaded, sample locations were marked on a common spreadsheet and then PCR run under set conditions (Table 2) was started by TA.
7) This entire procedure was completed again for the second set of primers.
New Primer Sets:
hsp70:
FW GGCAAATCCAACCGAATCACC
RV TGTCGCCATTTTCCTCGCTT
Glutathione peroxidase:
FW GTCTCCCAAAACAGCCTCCA
RV GAGGTTGGCAAAAGCACAGG
Results:
All Nanodrop results were within the expected ranges.
Both primers amplified well under qPCR conditions.
Conclusions:
The RNA samples that I quantified using the Nanodrop spectrometer all had concentrations or 260/230 and 280/260 ratios within the expected ranges. This means that contaminant levels were low, if there were any, and that there is enough RNA to amplify normally using PCR.
The primers amplified well under this qPCR conditions, showing that they should be successful in amplifying the target regions with cDNA next week.
Reflection:
Everything seemed to go well with this experiment, my concentrations and 260/230 and 280/260 ratios were within normal levels. Also, my primers responded well under PCR conditions, so I do not have any questions about this particular lab.
Table 1. Master Mix preparation table for qPCR reaction.
| Reagent |
1 Reaction (uL |
|
Total in Master Mix (uL) |
| 2X Immomix (Master Mix) |
12.5 |
7 |
87.5 |
| 10uM Forward Primer |
1.25 |
8.75 |
|
| 10uM Reverse Primer |
1.25 |
8.75 |
|
| Syto-13 dye (50uM) |
1 |
7 |
|
| PCR Water |
7 |
49 |
Table 2. Conditions for qPCR run.
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 30 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
11/13/12
Pre-Stress Experiment- RNA Isolation
Summary: During this lab I was able to perform RNA isolations on all of my samples an stored them at -80C. I will then quantify them using the Nanodrop on Monday, before our next Tuesday lab session.
Methods and Materials:
micropipettes
sterile filter pipette tips
sterile (RNase free) 1.5mL microcentrifuge tubes
sterile disposable pestles
vortex
ice buckets
gloves
lab pens
safety glasses
TriReagent
microcentrifuge tube rack
microcentrifuge (room temp)
razor blades
DNazol
75% ethanol
0.1% DEPC water
phenol/chloroform waste containers (liquid/solid)
vortex machine (set to touch)
hot water bath (at 55C)
Nanodrop spectrophotometer
chloroform
RNase free water
chloroform
isopropanol
Samples:
#51-54 were gill samples from the Olympic Oyster Control group
#81-89 were gill samples from the Olympic Oyster Pre-Stressed group
RNA Isolation (Part 1):
1- Microcentrifuge tubes were labeled with initials, date, sample number, and isolation type (RNA). Samples were stored on ice during entire procedure when not in direct use.
2- 500 ul (microliters) of TriReagent was added, and then sample was placed on ice.
3- Tissue was homogenized using sterile disposable pestle. Sample was vortexed and then transferred to a second sterile tube by pipette in order to continue homogenizing.
4- Another 500 ul of TriReagent was added after solution was completely homogenized.
5- Tube was vortexed at high speed for 15 seconds.
RNA Isolation (Part 2):
1- We made sure the hot water bath was turned on and set to 55 degrees C.
2- We confirmed the tissue sample from Lab 2 is homogenized and incubate it at room temperature for 5 minutes.
3- In the fume hood, we quickly added 200uL of chloroform.
4- Making sure the tube was sealed completely, we vortexed the tubes for 30 seconds until it acquired a cloudy appearance.
5- We then incubated the tube at room temperature for 5 minutes.
6-The tube was then spun in the refrigerated centrifuge for 15 minutes at maximum speed.
7- After removing the tube, it was determined that the substances had separated in reverse order, so we vortexed the entire contents and respun the tube in the refrigerator for 10 minutes at maximum speed.
8- Carefully, the aqueous phase of the contents was transferred to a new pre-labeled tube. The tube containing the organic and interphase was disposed of.
9- 500uL of isopropanol was then added to the new tube containing the RNA sample.
10- The tube was then mixed by inversion until the substance no longer appeared viscous.
11- We incubated the tube at room temperature for 10 minutes.
12- Tubes were then spun in the refrigerated microcentrifuge for 8 minutes at max speed.
13- A small white pellet was formed at the bottom of the tube.
14- We then proceeded to remove the supernatant, being carefully not to remove the pellet containing the RNA.
15- 1mL of 75% EtOH was then added to the pellet and the tube was vortexed.
16- Tubes were spun at 7500g for 5 minutes.
17- We again removed the supernatant and then we briefly spun the tubes to pool the last of the supernatant so that we could remove it.
18- Using a smaller pipette for greater removal accuracy, we removed the rest of the supernatant. The tube was left open to sit for no more than 5 minutes so that any remaining supernatant would evaporate.
19- The pellet was then resuspened in 100uL of 0.1%DEPC-H2O and pipetted up and down until the pellet dissolved.
20- The tube was then placed in the hot bath (at 55 degrees C) to incubate for 5 minutes in order to help solubilize the RNA.
21- After removing from heat, we flicked the tubes a few times to mix and then placed the samples in a box to store at -80C until they could be quantified using the Nanodrop the next Monday.
Results:
There were no quantifiable or observable results from this lab session.
Conclusions:
Because there were no measurable results, no concrete conclusions can be drawn from this particular lab session.
Reflection:
Because we have accomplished this procedure in the past, and because there were no measurable results during this section I had no particular questions or confusions from this lab.
11/6/12
Lab 7: Protein SDS/Page and Western Blot, Conventional PCR analysis and & qPCR data
Summary: The purpose of this lab was to use the previously isolated proteins from the experimental animals and analyze the relative protein expressions using a protein SDS/PAGE and a western blot. Using the example animals, the ones upon which DNA, RNA, and cDNA isolations and amplifications were performed, we analyzed PCR products through gel electrophoresis and qPCR data through computer graph outputs.
Combined Materials:
Micropipettes
sterile filter pipette tips
tip waste jar
1L flask
agarose gel
1X TAE
Ethidium bromide
Microwave
Gel rigs
Kimwipes
lab coat safety glasses
gloves
sterile gel loading tips
1.5mL screw cap tubes
microcentrifuge tube rack
lab pen
timers
heating block with water bath
tube floatie (8-capacity)
glass container for water boiling and to accommodate "floatie"
protein gel box
SDS/PAGE gels
gel loading tips
trays for gel staining
platform rocker
plastic wrap
2X SDS reducing sample buffer
protein ladder marker
gel running buffer
light box
digital camera
Nanopure water
Blocking Solution
rotary shaker
Primary Antibody Solution
Antibody Wash
Secondary Antibody Solution
Chromogenic Substrate
Tris-Glycine transfer buffer
filter paper
nitrocellulose membrane
semi-dry transfer solution
Protein Ladder - SeaBlue Plus 2 (Inviteogen)
DNA Ladder - HyperLadder II (Bioline)
Methods:
Agarose gels were pre-made
Electrophoresis Procedure:
1) Secured gel in gel box, making sure DNA was be run from negative (black wire) to positive (red wire) and filled with 1X TAE buffer until all wells were fully covered.
2) Removed combs from wells.
3) Loaded 7uL 100bp ladder (HyperLadder II from Bioline) in far left lane.
4) Loaded 20uL of PCR samples into gels, stored remaining PCR products at -20C.
5) Ran gel at ~100V for 45 minutes then ramped up voltage to 115V for another 15 minutes.
6) Gels were visualized by means of the UV transilluminator.
SDS-PAGE Procedure:
1) Water was set on hot plates to begin boiling.
2) Using a fresh 1.5mL screw cap tube, we mixed 15uL of protein stock and 15uL of 2X Reducing Sample Buffer.
3) Tubes were mixed by flicking and then centrifuged for ~10s to pool liquid at base of tube.
4) Boiled samples for 5 minutes. (Prepared/rinsed gels while waiting).
5) Immediately centrifuged samples for 1 minute to pool liquid.
6) Loaded entire sample into well using gel loading tip.
7) Placed lib on gel box, placed electrodes and hooked into power supply.
8) Began running gel at 150V for 35 minutes.
9) Disconnected power supply and turned off box. We then carefully removed the lid from the gel box.
10) Before removing gel, we had to disengage the tension wedge, then removed the gel.
11) Then we carefully cracked open the cassette in order to expose the inner gel.
12) Gels were marked (by trimming) off the upper left corner so that gel orientation could be determined at a glance.
Western Blot Protocol:
1) Filter paper was soaked for 15 minutes, along with the membrane and gel in the Tris-Glycine Transfer Buffer.
2) Blotting Sandwich was then assembled in the following order. Please note that those things first on the list will actually form the base of the sandwich.
Anode (+++)
2 pieces of filter paper
membrane
gel
2 pieces of filter paper
cathode (---)
3) Blot was then allowed to transfer onto the membrane for 30 minutes at 20V.
4) Gel was then carefully removed and placed in a dye solution on the rocker.
5) Membrane was then carefully washed twice using 20mL of pure water for 5 minutes each.
6) 10mL of Blocking Solution was then added to a plastic box where the membrane was allowed to soak (covered) overnight on the rotary shaker at 1rev/sec.
7) The next day, liquid was decanted and membrane was again rinsed for 5 minutes. Rinsing and decanting procedure was then repeated.
8) Membrane was incubated in 10mL Primary Antibody Solution then decanted.
9) Membrane was rinsed using 20mL of Antibody Wash for 5 minutes then this step seas repeated for a total of 4 times.
10) For 30 minutes membrane was incubated in the Secondary Antibody wash for 30 minutes and decanted.
11) Membrane was rinsed using 20mL of Antibody Wash for 5 minutes then this step seas repeated for a total of 4 times.
12) Membrane was then rinsed using 20mL of pure water for 2 minutes then decanted. This step was then repeated twice.
13) Membrane was incubated in 5mL of Chromogenic Substrate until purple bands began to appear (usually between 1-60 minutes after exposure).
14) Finally, the membrane was dried on filter paper by extended exposure to open air.
Results:
Electrophoresis gel- My samples appeared to have no binding with the hsp70 primers I chose. If anything is present (beyond detection of the human eye) it is possible that there was a small amount of primer dimer or that the fragments were amplified at very low densities (Figure 1).
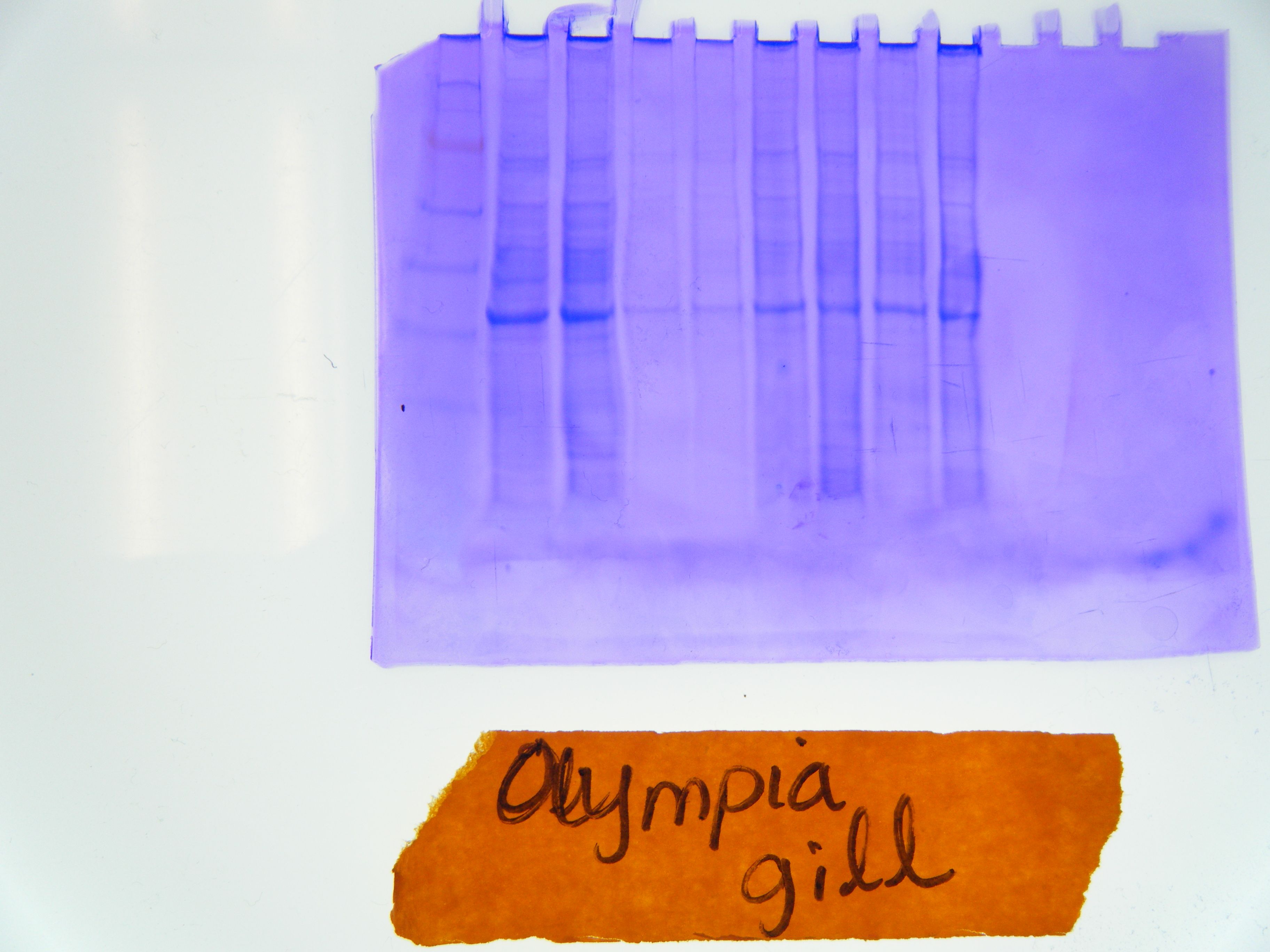
Western Blot- Many gel slots have a variety of proteins throughout the gel, ladder is easily visible (Figure 2).
Protein Membrane Results- Protein ladder is easily visible whereas only one or two lanes seem to have amplified proteins right around the length of hsp70 (Figure 3).
qPCR- Unfortunately, no DNA seemed to be bind to the hsp70 primers used in this initial experiment. There were a few small peaks we observed on the qPCR graphs which were most likely due to primer dimer. This will lead to a primer redesign.
Conclusions:
The conclusions were reached were that hsp70 is a gene that is commonly turned on in relation to general stress, and that relative levels of protein may be a better indication of protein affects then present or absent data such as gels might provide. In this case, we actually saw surprising results in that the strongest hsp70 protein bar was in a control animal, one that supposedly experienced little to no stress throughout the experiment.
With the Western Blot results we saw that though many proteins were expressed, hsp70, a general stress protein was not a reliable indicator of stressed state in this experiment (unless the animals were not feeling any stress at all in reaction to the heat).
qPCR results indicated to me that there was an error with my primers in that they were not binding to any present DNA. What little results that we saw were likely the effects of primer dimer.
Reflection:
As I have already spoken of, I think I was under the impression that genes in the genome were specific to a singular purpose or response in an organism. I wasn't thinking that they may have multiple purposes or that their purposes might overlap. I think I need to do a lot more research, redesign my primers, and look for a second gene to support my study. I believe this lab made me realize something truly fundamental about the function of genes, and something that I will apply to future laboratory settings and studies I may find myself in.
Figures:
Figure 1. A view of the results of the Olympia Electrophoresis gel under UV light so as to highlight the bans. The 4 slots above the lower left hand ladder are where any results from my PCR reaction would have appeared.
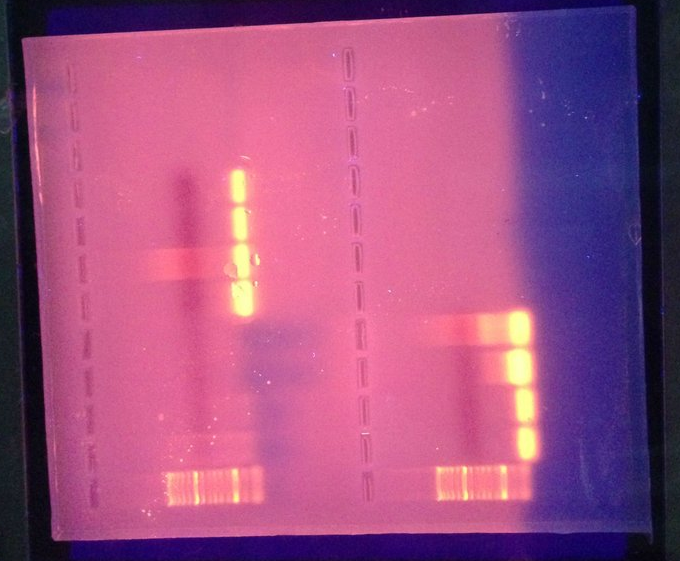
Figure 2. A view of the Olympia oyster Western Blot gel. Protein ladder can bee seen on the far left.
Figure 3. The membrane to which our Olympia Western Blots were transferred after protein binding. A few lines are visible near the size bin of hsp70.
10/30/12
Lab 6: cPCR, qPCR, and Protein Isolation
Summary: The purpose of this lab was to set up and perform conventional PCR, quantitative PCR, and the protein analysis. At the completion of this lab, we will have amplified our particular DNA region surrounding the gene of interest, determined our original concentration of DNA based on calculations from qPCR, and back calculated the protein concentration in the tissue.
Combined Materials:
Micropipettes
sterile filter pipette tips
tip waste jar
PCR tubes
1.5mL microcentrifuge tubes
cDNA
dNTPs
2x Apex Red (Master Mix)
Primers
Nuclease Free Water
thermal cycler
Kimwipes
microfuge tube racks
PCR tube racks
ice buckets
lab coat
gloves
PCR plates
optically clear caps for PCR
opticon thermal cycler
2X Immomix Master Mix
SYTO-13 Dye
cDNA samples
sterile microcentrifuge tubes 1.5 mL
sterile 2mL screw cap microcentrifuge tubes
sterile disposible pestles
spectrophotometer
microcentrifuge (in fridge)
lab pens
safety glasses
CelLytic MT Cell Lysis Reagent (with Protease Inhibitor Cocktail added)
Coomassie Protein Assay Reagent
DI water
PCR Protocol:
1) We created Master Mix using amounts described in Table 2 and labeled the tube with MM and my initials.
2) Then we pipetted 23 uL of Master mix into each of my 0.5L PCR tubes that were pre-labeled with sample names and initials (note that negative controls are also labeled).
3) Sample Names were as follows:
Gill 81
Gill 82
Negative 1
Negative 2
4) 2uL of DNA template was added to the appropriate tubes, and 2uL of PCR water was added to each negative control to help determine if the Master Mix was contaminated.
5) Tubes were then loaded into the thermocycler after determining all caps were sealed.
6) Tubes were then cycled according to Table 3, and then stored at -20C.
qPCR Protocol:
1) Prepared Master Mix for qPCR using Table 4, labeled with "MM for qPCR" and my initials
2) We then added Master mix to 6 wells on a white PCR plate
3) cDNA samples were prepared and then 2u was placed in each well according to labels (2 wells for each sample to account for loading/pipetting error)
4) 2uL of PCR water was added to both negative control wells to account for potential Master Mix contamination.
5) All caps were secured, and then wiped to ensure clean, clear surfaces.
6) PCR plate was loaded, sample locations were marked on a common spreadsheet and then PCR run under set conditions (Table 5) was started by TA.
Protein Extraction Protocol:
1) Recorded Tissue weights.
2) Labeled snap-cap tube according to sample number and initials.
3) Added 500uL of CellLytic MT solution to each tube containing sample tissues.
4) Homogenized tissue using sterile pestle.
5) We then mixed the tube by inversion several times.
6) Tubes were then centrifuged for 10 minutes in a refrigerated centrifuge.
7) Fresh tubes were labeled and supernatant was transferred to clean, labeled tubes.
Protein Quantification:
1) New 2mL screw-cap tubes were labeled.
2) 15mL of protein sample was pipetted into new tube along with 15uL of DI water, then mixed by pipetting.
3) 1.5mL of Bradford reagent was pipetted into each tube, then mixed well by piping up and down.
4) Tubes were mixed via inversion and then allowed to sit at room temperature for 10 minutes.
5) Protein samples were transferred to clean disposable curvettes, and abosorbance was measured through the samples at 595nm.
6) To confirm measurements, curvettes were removed, wiped down, mixed and remeasured. Both measurements were averaged to get a final reading.
7) Protein concentrations were then back-calculated according to the formula y(concentration)=996.52x - 43.64.
Results:
Results of PCR and qPCR will be interpreted during the next lab. Protein concentrations were determined to be 731.653 for g81, and 837.782 for g82. Absorbance for g81 was 0.778 and was 0.8845 for g82.
Conclusions:
According to these results my protein concentrations were interesting because there was a difference in over 100 between the two oysters. This implies that there are large fluctuations in the individual protein expression and concentrations of these oysters.
Reflection:
I believe I understood everything that occurred in this lab well enough and after redoing my calculations I believe I reached the correct concentration conclusions.
Table 2. Master Mix for PCR reaction.
| Reagent |
1 Reaction (uL |
|
Total in Master Mix (uL) |
| 5X Apex Red |
12.5 |
5 |
62.5 |
| 10uM Forward Primer |
10 |
50 |
|
| 10uM Reverse Primer |
10 |
50 |
|
| 10uM dNTP Mix |
2 |
10 |
|
| PCR Water |
8.5 |
42.5 |
Table 3. Time table followed by the thermocycler to complete PCR.
| Step |
Temperature |
Time |
Cycles |
| Denaturation |
95C |
5 min |
1 |
| Denaturation |
95C |
30 sec |
40 |
| Annealing |
55C |
30 sec |
|
| Extension |
72C |
90 sec |
|
| Final extension |
72C |
3 min |
1 |
| Hold |
4C |
∞ |
1 |
Table 4. Master Mix preparation table for qPCR reaction.
| Reagent |
1 Reaction (uL |
|
Total in Master Mix (uL) |
| 2X Immomix (Master Mix) |
12.5 |
7 |
87.5 |
| 10uM Forward Primer |
1.25 |
8.75 |
|
| 10uM Reverse Primer |
1.25 |
8.75 |
|
| Syto-13 dye (50uM) |
1 |
7 |
|
| PCR Water |
7 |
49 |
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 30 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
10/23/12
Lab 5: Experiment, Oyster Dissection and Primer Dilution
Summary: During this lab session and the day before experiments were set up and performed. These experimental designs for Olympia Oysters are explained in Table 1 at the end of this lab entry. Primers were then diluted in preparation for PCR in the next lab.
Materials and Methods:
microcentrifuge tubes
gloves
oyster shucker
Kimwipes
10% bleach
Dilute ethanol
dissecting scissors
forceps
forward and reverse primers
PCR water
Dissection:
1- After experiments were performed as described in Table 1, oysters were removed from water and placed in a tray for dissection.
2- Each oyster was measured, then shucked according to protocol explained in the previous write up, and then placed in a numbered weight boat.
3- Using dissection equipment sterilized first in bleach, then ethanol, the oysters were dissected to remove sample tissues of the gills and mantle.
4- These tissues were placed in microcentrifuge tubes pre-labeled with tissue type and sample number corresponding with the weight boat number. Dissection equipment was sterilized each time between oysters.
5- Tissues were placed on dry ice and stored at -80C for future use.
Primer Dilution:
1- Primers were diluted by adding 283uL of PCR water to the forward primer bottle, and then 384uL of PCR water to the reverse primer bottle.
2- 10uL of each primer was taken out and placed in a corresponding microcentrifuge tube labeled FW or RV working sample.
3- 90uL of the PCR water was added to each of these tubes, leaving us with a 10uM working stock of each primer.
Results:
Only one oyster from the Heat-stress sample was determined to be dead after experimentation, and so was removed from sampling.
Conclusions:
It seems as though the lowering of the heat-stress temperature from 40C to 35C prevented a significant amount of death in the heat-stressed oysters.
Reflection:
Because this lab was mainly dissection, I feel as though I understood everything. I enjoyed our oyster anatomy lessons, which included learning to sex oysters.
Table 1. Olympic oyster experimental design. Includes samples to be used in comparison with Pre-Stress experiment.
| Olympia Oyster Table |
Experiment Summary |
Experiment Day 0 |
Experiment Day 1 |
Tissue Sample Numbers |
| Control |
Oysters were kept in a control tank during the entire experiment without additional stress |
Oysters were placed in control tank for holding |
Oysters remained in control tank until dissection |
51-60 |
| Heat-Stress |
Oysters were heat stressed to determine gene expression |
Oysters were placed in control tank for holding |
Oysters were placed in heated seawater at 35 C for 1 hour |
61-70 |
| Pre-Stress |
Oysters were pre-stressed before the experiment to determine if heat tolerance is acquired |
Oysters were pre-stressed at 40 C for 1 hour, then returned to control tank |
Oysters were placed in heated seawater at 35 C for 1 hour |
81-89 |
10/16/12
Lab 4: Reverse Transcription and primer design; Prep for Experiment
Summary:
The purpose of this lab was to learn techniques for reverse transcription of RNA and cDNA. We also learned dissection techniques for our experimental animals, and began to prepare our set-ups for our experiments. Finally, we chose genes and designed primers for our experiments for the following week.
Materials and Methods:
Reverse Transcription Materials:
micropipettes (1-1000uL)
Sterile Filter pipettes tips (1-1000uL)
tip waste jar
PCR tubes (0.2 mL, thin walled)
RNA samples (student provided)
M-MLV reverse transcriptase
M-MLV 5X reaction buffer
Oligo dT
dNTPs
Nuclease Free water
thermal cycler
microfuge tube racks
PCR tube racks
ice buckets
Kimwipes
lab coat
safety glasses
gloves
Oyster Dissection Materials:
protective gloves
oyster schuckers
lemon juice
Reverse Transcription Methods:
1- Stock RNA was mixed by inverting the tube several times after it had thawed.
2- Using a 0.2mL PCR tube (labeled with cDNA and initials) we mixed:
- 5uL of RNA mixture
- 1uL of oligo dT (primer)
- 4uL of nuclease free H2O
3- Tubes were then incubated at 70 degrees C for 5 mins in the thermocycler and transferred to ice immediately.
4- Tubes were mixed by inversion several times and then the following was added:
- 5uL of M-MLV 5X Reaction Buffer
- 5uL of dNTPs
- 1uL of M-MLV RT
- 4uL of nuclease free H2O
5- The mixture was then incubated for 60 minutes at 42 degrees C and heat inactivate at 70 degrees C for 3 minutes on the themocycler.
6- Samples were then spun down using a centrifuge and stored at -20 degrees C.
Experimental Preparation:
1- Labeled 1 net bag set to contain 10 Olympia Oysters for pre-heat stress, then labeled and set up air pumps in control tanks to be filled and stocked at a later date.
Dissection:
1- After placing oyster in gloved left hand, we inserted the shucker into the back of the oyster, as near to the hinge as possible.
2- Once the shucker broke through the shell, we twisted the shucker to pop open the shell while simultaneously slicing through the adductor muscle.
3- We then removed the top half of the shell, made sure the oyster was cut clean off the other half of the shell, and ate them (sometimes with lemon juice).
Primer Design:
1- We searched through the annotated sequences of our species of interest to find our specific gene.
2- Then we copied the sequence and pasted it into the primer BLAST website.
3- Making sure the following parameters were met, we collected the forward and reverse primers that best fit, and ordered them for use in our experiment.
The primer parameters (put into BLAST) were:
- Amplifying 80-200bl
- Melting temps at ~60C (within 3C)
- Set the database to "nr"
- primers within 18-22 bases in length
- limited search to Phylum Mollusca
- chose primers based also on a lower self-complementary score
Results:
There were no measurable results during the reverse transcription procedure.
Forward Primer:
CCGAATCACCATCACCAACG
Reverse Primer:
GTCGCCATTTTCCTCGCTTG
Conclusions:
These primers were chosen based primarily on their compliance with all of the parameters required. These primers were what I had expected, and are designed to selectively bind to the hsp70 gene in Olympia Oysters. Based on these results, next week we will be performing our experiment, extracting tissue, and using the primers to amplifying the cDNA of our gene of choice so we can discover on a molecular level how the animal responds to it's given stress.
Reflection:
I think the purpose of this lab was to familiarize us with the final step in transitioning our samples to cDNA so that we may measure their initial concentrations using the primers we chose and a method known as qPCR, which we will be performing during our next lab session.
The only thing about this lab I did not understand was if the primers we were designing we for the example samples we ave been looking at, or for the experimental samples we will collect once we are done.
10/9/12
Lab 3: RNA Isolation (Part 2)
Summary:
The purpose of this lab is to familiarize us with the entire procedure involved in RNA isolation, from the start in Lab 2 until our Nanodrop measurements at the end of Lab 3. In this way, the lab was meant to better prepare us for our own DNA/RNA sampling and qualification after the completion of our class experiment. By learning the procedures, and potentially learning from mistakes we made during these procedures, we should be able to enhance our future results by troubleshooting problem areas before they occur.
Materials and Methods:
micropipettes (1-1000uL)
sterile pipette tips (1-1000uL)
1.5 mL microcentrifuge tubes
microcentrifuge tube rack
lab coats
safety glasses
gloves
lab pen
timers
ice buckets
phenol/chloroform waste containers (liquid/solid)
vortex machine (set to touch)
hot water bath (at 55C)
Nanodrop spectrophotometer
chloroform
RNase free water
chloroform
isopropanol
75% ethanol
0.1% DEPC treated water
Sample Tissue:
RNA- #232 Pacific Oyster (male gonad tissue)
RNA Extraction:
1- We made sure the hot water bath was turned on and set to 55 degrees C.
2- We confirmed the tissue sample from Lab 2 is homogenized and incubate it at room temperature for 5 minutes.
3- In the fume hood, we quickly added 200uL of chloroform.
4- Making sure the tube was sealed completely, we vortexed the tubes for 30 seconds until it acquired a cloudy appearance.
5- We then incubated the tube at room temperature for 5 minutes.
6-The tube was then spun in the refrigerated centrifuge for 15 minutes at maximum speed.
7- After removing the tube, it was determined that the substances had separated in reverse order, so we vortexed the entire contents and respun the tube in the refrigerator for 10 minutes at maximum speed.
8- Carefully, the aqueous phase of the contents was transferred to a new pre-labeled tube. The tube containing the organic and interphase was disposed of.
9- 500uL of isopropanol was then added to the new tube containing the RNA sample.
10- The tube was then mixed by inversion until the substance no longer appeared viscous.
11- We incubated the tube at room temperature for 10 minutes.
12- Tubes were then spun in the refrigerated microcentrifuge for 8 minutes at max speed.
13- A small white pellet was formed at the bottom of the tube.
14- We then proceeded to remove the supernatant, being carefully not to remove the pellet containing the RNA.
15- 1mL of 75% EtOH was then added to the pellet and the tube was vortexed.
16- Tubes were spun at 7500g for 5 minutes.
17- We again removed the supernatant and then we briefly spun the tubes to pool the last of the supernatant so that we could remove it.
18- Using a smaller pipette for greater removal accuracy, we removed the rest of the supernatant. The tube was left open to sit for no more than 5 minutes so that any remaining supernatant would evaporate.
19- The pellet was then resuspened in 100uL of 0.1%DEPC-H2O and pipetted up and down until the pellet dissolved.
20- The tube was then placed in the hot bath (at 55 degrees C) to incubate for 5 minutes in order to help solubilize the RNA.
21- After removing from heat, we flicked the tubes a few times to mix and then placed the samples on ice to take to the Nanodrop spectrophotometer as our stock RNA samples.
RNA Quantification:
1- Nanodrop pedestal was first wiped down with a kim wipe, and then 2ul of 0.1% DEPC-H2O were pipetted onto the pedestal and the arm was lowered.
2- "Blank" was selected in order to zero the instrument. (These first 2 steps only need to be done once to set for all sampling)
3- 4ul of RNA sample was pipetted onto the nanodrop pedestal and the arm was lowerd.
4- We clicked "Measure" in order to record our RNA concentration (ng/ul). We also recorded the A260/280 and the A260/230 ratios which are calculated by the nanodrop using the Beer-Lambert Law. Numbers were recorded.
5- The arm was lifted and the sample was cleaned off the Nanodrop using a kim wipe.
6- RNA tubes were labeled with tissue type, "RNA," initials, the date, and concentration before storing at -80 degrees C.
Results:
The results obtained from this RNA isolation were as follows
Concentration: 601.7 ng/uL
A260/280: 1.95
A260/230: 1.56
Conclusions:
My results of this procedure were more in line with what I expected then the results of my DNA isolation were. This concentration of RNA, though much higher than the DNA, is what I expected. This is mainly because the RNA sample was of male gonad tissue, which not only contains a large amount of DNA, but is one that should be constantly replicating and so producing more RNA. My A260/280 ratio was expected, as normal ranges are within 1.8-2.0. My A260/230 was better than my previous isolation with DNA, but still indicates ethanol contamination, as normal levels are seen between 2.0-2.2. Based on these results, in the future i will continue to work on removing ethanol contamination. I believe our next step will be to select primers to translate and amplify certain parts of the RNA so that we can determine original concentrations of the RNA, and therefore the genes that are being amplified according to environmental stresses.
Reflection:
I believe that the purpose of this lab was to give us hands on experience with RNA isolations so that we may perform them on our own when we are analyzing our own samples for epigenetic responses. The procedures outlined in this lab are used to measure the concentration of RNA in our sample tissue as well as the amount of contamination on both ends of the scale. These methods might be used in the future to compare relative RNA concentrations to DNA concentrations and by isolating certain genes, we should be able to quantify their responses to environmental stimuli. Having completed the last procedure, very little was unclear or confusing to me during this lab.
Extra Assignment:
1) Do oysters of either species better respond to heat stresses if they have been previously exposed to the stress?
2) The gene I would like to design primers for is HSP70.
10/2/12
Lab 2: DNA isolation, Initiate RNA isolation
Summary:
The purpose of this lab was to experience preparing individual samples for DNA and RNA isolations, so that we may troubleshoot in future sample preparations. The technique used to perform this extraction was DNazol adapted from the MRC manual. DNA samples were isolated from the gills of a pacific oyster, whereas the RNA isolation was prepared from the gonad tissue of a male olympia oyster because of an insufficient amount of tissue in the original sample.
Materials and Methods
micropipettes
sterile filter pipette tips
sterile (RNase free) 1.5mL microcentrifuge tubes
sterile disposable pestles
vortex
ice buckets
gloves
lab pens
safety glasses
TriReagent
microcentrifuge tube rack
microcentrifuge (room temp)
razor blades
DNazol
100% ethanol
75% ethanol
0.1% DEPC water
kim wipes
nanodrop machine
Samples:
RNA- #232 Pacific Oyster (male gonad tissue)
DNA- #19 Olympia Oyster (gill tissue)
RNA Isolation (Initiation):
1- Microcentrifuge tubes were labeled with initials, date, sample number, and isolation type (RNA). Samples were stored on ice during entire procedure when not in direct use.
2- 500 ul (microliters) of TriReagent was added, and then sample was placed on ice.
3- Tissue was homogenized using sterile disposable pestle. Sample was vortexed and then transferred to a second sterile tube by pipette in order to continue homogenizing.
4- Another 500 ul of TriReagent was added after solution was completely homogenized.
5- Tube was vortexed at high speed for 15 seconds.
6- The homogenized tissue samples were then placed in a freezer at -80 degrees C to store.
DNA Extraction:
1-Tissue was homogenized in 0.5mL of DNazol in 1.5mL microcentrifuge tube. After homogenization another 0.5mL of DNazol was added and mixed into the solution.
2-Samples were incubated at room temp for 5 minutes, the centrifuged at 10,000xg at room temp for 10 minutes.
3- Supernatant (the liquid, not cell debris) was transferred to a new tube labeled with initials, date, sample number, and isolation type (DNA).
4- 0.5 mL of100% ethanol was added to the 1.5mL microcentrifuge tube. Sample was mixed by inverting 8 times.
5- Sample sat at room temperature for 1 minute.
6- After a cloudy precipitate formed, we centrifuged for 5 minutes at 5000xg, and then removed the lysate.
7- DNA was washed with 1 mL of 75% ethanol, inverted 6 times, then let sit for 1 minute. Ethanol was removed and this step was repeated.
8- Excess ethanol was removed with a smaller pipette (at a later time, I discovered I did not remove enough ethanol at this point).
9- 300ul of 0.1% DEPC water was added to the DNA and solution was piped up and down to dissolve completely.
10- Sample containing DNA was brought to the Nanodrop for quantification.
DNA Quantification:
1- Nanodrop pedestal was first wiped down with a kim wipe, and then 2ul of 0.1% DEPC-H2O were pipetted onto the pedestal and the arm was lowered.
2- "dsDNA" was selected from the pulldown menu
3- "Blank" was selected in order to zero the instrument. (These first 2 steps only need to be done once to set for all sampling)
4- 4ul of DNA sample was pipetted onto the nanodrop pedestal and the arm was lowerd.
5- We clicked "Measure" in order to record our DNA concentration (ng/ul). We also recorded the A260/280 and the A260/230 ratios which are calculated by the nanodrop using the Beer-Lambert Law. Numbers were recorded.
6- The arm was lifted and the sample was cleaned off the Nanodrop using a kim wipe.
7- DNA tubes were labeled with tissue type, initials, the date, and concentration before storing at -20 degrees C.
Results:
The only results obtained from this lab were from the DNA quantification.
Concentration: 144.5 ng/uL
A260/280: 1.89
A260/230: 0.25
Conclusions:
For the most parts, my results were expected. A concentration of 144.4ng/uL is considered normal for this type of tissue (gills). The A260/280 ratio is considered normal as long as it is within the range of 1.7-1.9, which my results of 1.89 are. The A260/230 ratio, however, is abnormal (outside the normal range of 2-2.2 for this ratio), and in this case is the result of ethanol contamination, as stated earlier in the procedures. Next, we will probably compare the results of concentration from our DNA to the RNA and protein concentrations to determine which genes are more active than others. This will benefit our future experiments by allowing us to determine which genes are activated according to environmental stresses that we subject them to.
Reflection:
I believe that the purpose of this lab was to familiarize us with another method of DNA extraction and quantification that we will use for our projects in the future dealing with the effects of environmental stresses (other than extraction vis PCR, which is my previous experience).
I think that the procedures performed in lab are universal procedures for measuring the concentration of DNA, and determining where, if anywhere, on the spectrophotometer scale contaminates may be polluting the solution.
These methods might be used for measuring the concentration of DNA in various tissues of the body (ie. gills versus gonads). Also, when we finish the RNA isolation, they might be used to assess the relative activity of certain genes tied to environmental stresses.
One of the situations in the lab that was unclear to me, was how to effectively remove all of the excess ethanol without damaging or removing part of the pellet of DNA. Is it possible to remove the ethanol with another wash of something that will remove the excess ethanol without dissolving the DNA pellet? I wish there was more information on the procedure and what the benefits of using this procedure for extraction and isolation in comparison to others such as PCR. Is it better to use this one because it is similar protocol for both RNA and DNA isolation?
9/26/12
Setting up the lab notebook with my first entry.