Summary
In this lab, we performed qPCR on plate four and discussed our previous results.
Materials and Methods
Plate four was loaded with Elongation Factor, HSP 70B, and Superoxide Dismutase.
The plate was arranged as follows:
HSP 70B
Column 1: Control
Column 2: Heat
Column 3: Mechanical
Column 4: M->H
Column 5: C. Virginica
Column 6: NTC
SOD
Column 7: Control
Column 8: NTC
Results
The HSP 70B amplified in the mechanical treatment group. This provided us with the opportunity to compare the unstressed control group's expression of HSP 70B to the mechanically stressed treatment. Neither of the Superoxide Dismutase columns amplified.
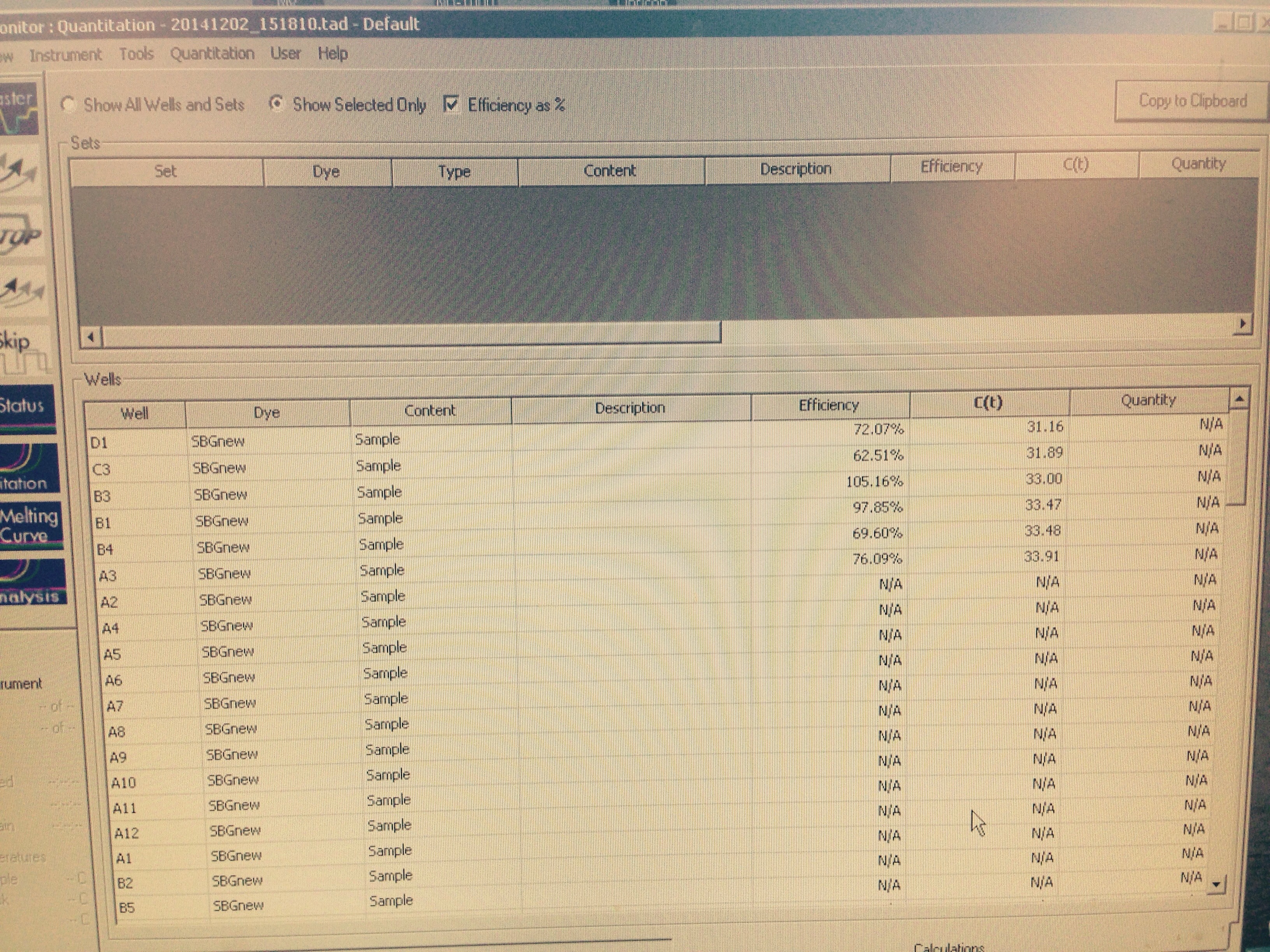 |
| The arbitrary expression of Elongation Factor compared to HSP 70B |
Conclusion
Plate four did give us some good insight into the nonspecific response of C. gigas to mechanical stress. There are many things we would do differently if given the chance to continue these experiments, but the end of the quarter means that this line of inquiry will have to rest here.
11/25/14 Lab 8
Summary
In this lab, we performed qPCR on the previously obtained oyster RNA.
Materials and Methods
Sophie's HSP 70B and HIF 1a and Sean's Superoxide Dismutase were the primers used in this lab. Elongation factor was also measured as a control.
qPCR reactions were set up using the following proportions:
SSOFast: 100uL
Forward primer: 4uL
Reverse primer: 4uL
H2O: 84uL
cDNA (or H2O for control): 1uL
Superoxide Dismutase and Elongation Factor were used in equal parts in the first plate. HSP and HIF were put into plate two, but it unfortunately spilled and was unrecoverable.
Results
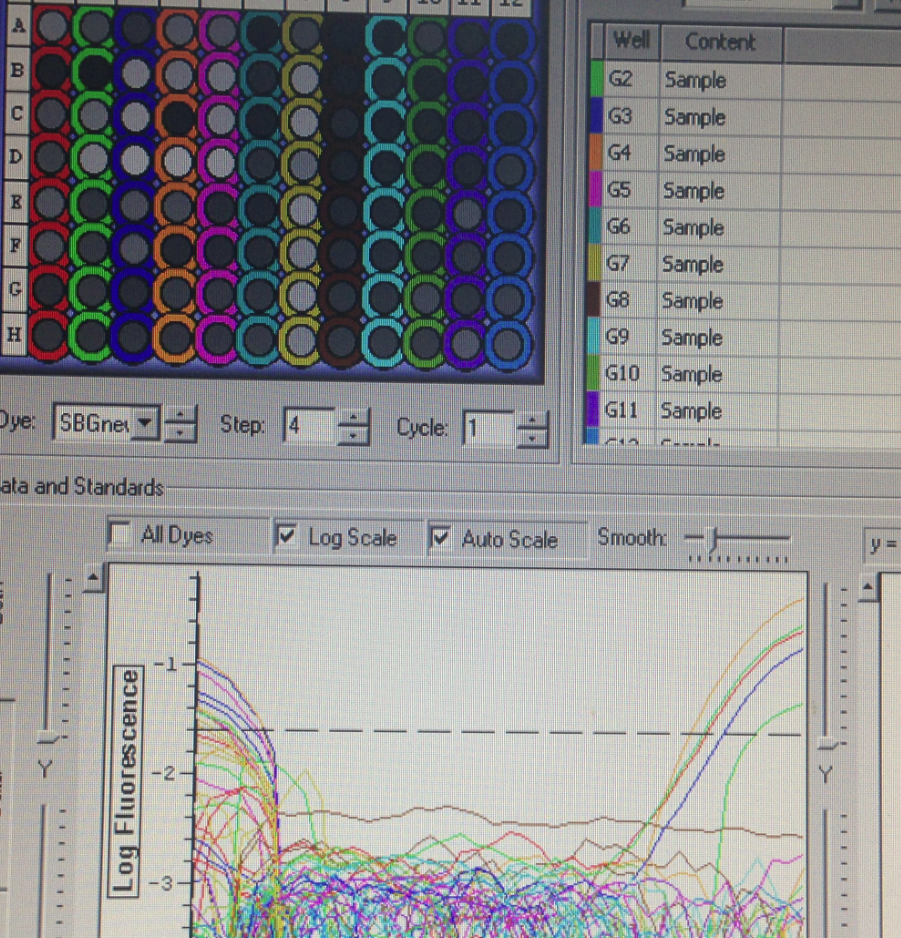
Plate one amplification
Conclusion
These results were not as expected, because most of the wells did not amplify. This could occur because the RNA degraded after being kept out of the freezer for too long, or perhaps due to errors in extraction. We were therefore unable to collect data that could have come from the lack of amplification of Superoxide Dismutase, because we are unable to tell whether it was due to our error or the true lack of expression.
The purpose of this lab was to collect qPCR data on the stress responses of oysters. The procedure was pretty clear, as it was similar to previous labs, but our execution was disorganized. However, I don't need any further clarification on the lab activities.
11/18/14 Lab 7
Summary
In this lab, we homogenized Crassostrea gigas tissue, and then extracted RNA in order to prepare for reverse transcription.
Materials and Methods
24 hours before the lab, 40 oysters were separated into five treatments of eight oysters each. The description of the groups is as follows:
-Control group
-Heated at 30 degrees C for two hours
-Placed on a plate shaker for ten minutes
-Heat at 30 degrees C for two hours then placed on a plate shaker for ten minutes
-Placed on a plate shaker for ten minutes then heated at 30 degrees C for two hours
The oysters were first shucked, then gill tissue was removed from each specimen. Each tissue sample was placed in 500 uL of TRI reagent and homogenized using a disposable pestle. 200 uL of Chloroform was then added to each sample and was mixed again by inverting and briefly vortex in the tubes. Each tube was labeled and spun in the refrigerated centrifuge on high for 15 minutes. The supernatant was then removed from each tube and placed in a corresponding tube with 500 uL isopropyl alcohol. The tubes were then spun in the refrigerated centrifuge at max speed for eight minutes. The supernatant was then removed from the tube and disposed of. The pellet at the bottom of the tube was dried to allow for the evaporation of excess alcohol, and then 100 uL of Nanopure H2O was added. The sample was then stored at -80 degrees C in the Roberts lab.
Results
All 40 samples were successfully prepared for reverse transcription.
Conclusion
The procedures from this lab seem to have been carried out well, as the samples seem to be prepared for reverse transcription. However, it is impossible to know if the chemical aspects were carried out correctly until the samples are quantified with the Nanodrop Spectrophotometer.
I did not need any clarification about the processes in this lab, because they were very similar to the steps taken in lab two and are the typical procedures associated with gene expression.
11/04/14 Lab 6
Summary
In this lab, we received our primers and performed qPCR on our oyster tissue samples for the species C. gigas and C. virginica.
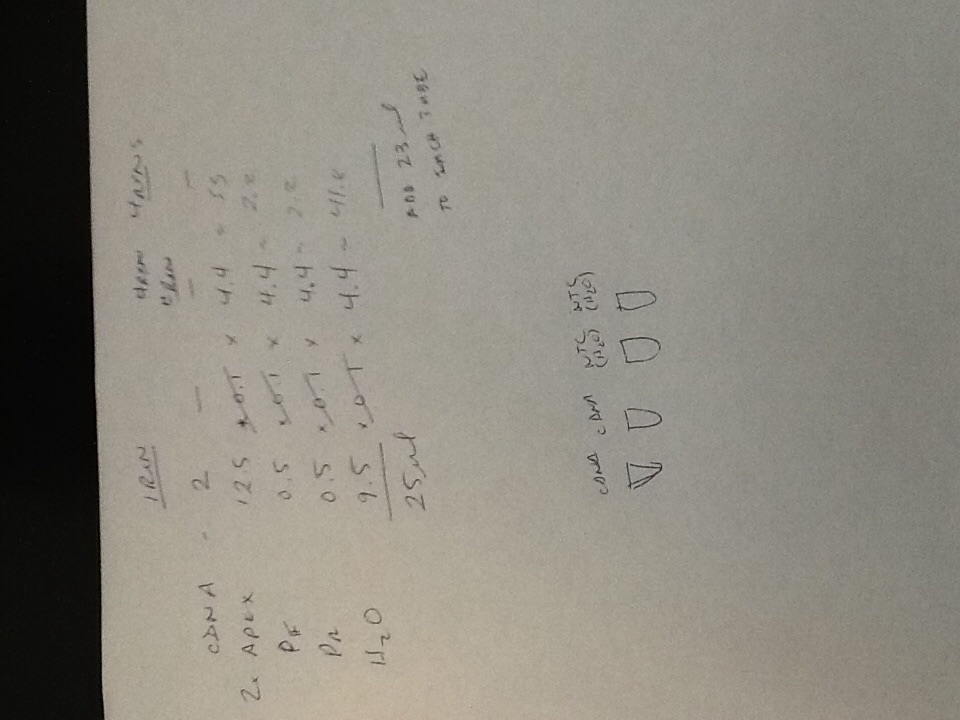 |
| The first pair of primers is for species C. gigas, the second pair for C. virginica. |
Materials and Methods
Nanopure water was added to each primer at a 1:10 primer to water ratio to create the concentrated stock solution. That solution was then diluted again at a 1:10 ratio to create the working stock solution. The working solution was therefore one part primer to 100 parts Nanopure water.
Each species had its cDNA diluted in five different cDNA to Nanopure water ratios to make a template: 1:0, 1:10, 1:100, 1:1000, 1:10000.
The qPCR master was as follows:
12.5 uL SsoFast EvaGreen Supermix
0.5 uL 10x Upstream Primer for Elongation Protein
0.5 uL 10x Downstream Primer for Elongation Protein
10.5 uL Nanopure water
The ratio was multiplied by eight to create enough solution for each of my reactions.
Results
The high efficiency values indicates that the tissue samples may have been contaminated with Ethanol, etc. Gel electrophoresis would indicate whether or not the correct product is being amplified. The C(t) values of about 37 indicate that if only the correct substance is being amplified, the HSP 70 and CYP 450 proteins were indeed present.
Reflection
I still have difficulty interpreting qPCR results. I also found designing a primer by myself to be a challenge, and I wasn't sure if I was overlooking any important aspects. In future labs we will be stressing our oysters to measure their stress protein/hormone production in the face of heat and mechanical stressors.
10/21/14 Lab 4
Summary
In this lab, we used the protein samples from the previous week to perform SDS-PAGE before using Western blotting to identify HSP 70 proteins from a tissue sample. We also determined whether our amplification of cDNA in the previous lab was successful using agarose gel electrophoresis.
Materials and Methods
Electrophoresis
Gel was put in the gel box and filled with 1x TAE buffer. 20 uL of the previously obtained PCR sample and 2 uL 10x DNA loading dye were added. 7 uL 100 bp ladder was added to the leftmost lane And the gel ran at 100V for one hour.
SDS-PAGE
15 uL of protein stock was combined with 15 uL 2x Reducing Sample Buffer. The sample mixed by flicking and then centrifuged for 10 seconds. The solution was then boiled for 5 minutes and then centrifuged again for one minute. Sample was then loaded into cell five and run at 150V for 35 minutes.
Western Blot
Membrane and gel were soaked in Tris-Glycine Transfer Buffer for 15 minutes. The filter paper was not soaked. The blotting materials were assembled in the order of:
- Anode (+++)
- filter paper
- membrane
- gel
- filter paper
- cathode (---)
The blot was then run at 20V for 30 minutes. The Invitrogen WesternBreeze Chromogenic Western Blot Immunodetection Kit was used. The membrane was rinsed in 10mL of blocking solution and incubated for 30 minutes on a rotary shaker set at 1 revolution/second. The liquid was then decanted. The membrane was then rinsed with 20 mL of Nanopure water for five minutes. The liquid was then decanted and rinsed again in 20 mL of Nanopure water for five minutes. The membrane was then incubated in 10 mL Primary Antibody Solution for an hour, then the liquid was then decanted. The membrane was then rinsed with 20mL of Antibody Wash for five minutes, then the liquid was decanted. This was repeated three times. 10 mL of Secondary Antibody Solution was added, and the membrane incubated in it for 30 minutes. The solution was decanted, and then washed with 20 mL of Antibody Wash for five minutes. This was repeated three times. The membrane was then rinsed with 20 mL of Nanopure water for two minutes, then decanted. This was repeated twice, then the membrane incubated in 5 mL of Chromogenic Substrate for 60 minutes. The membrane was then dried in the open air.
Results
10/14/14 Lab 3
Summary
In this lab, the RNA previously extracted is mixed with Master Mix 1 to create cDNA. Quantitative PCR was then performed to amplify the DNA. We also made an agarose gel with agarose, EtBr, and 1x TAE in the microwave, which was stored for future use. Additionally, we used CellLytic MT to extract proteins from a tissue sample.
Materials and Methods
Reverse Transcription
Master Mix 1 was created with 98 uL nuclease free H2O and 4 uL Oligo dT. 5 uL of the previously extracted RNA was added to a clean test tube and combined with 0.5 uL of primers, 12.75 uL of Master Mix 1, and 12.25 uL of nuclease free H2O. The mixture was then incubated for five minutes at 70 degrees C and then transferred to ice. Master Mix 2 was created by combining 40uL M-MLV 5X reaction buffer, 10 uL dNTP, 4 uL M-MLV reverse transcriptase, and 4 uL nuclease free H2O. 7.25 uL of Master Mix 2 was added to the reaction and centifuged. The solution was then incubated at 42 degrees C for 60 minute, spun down in the centrifuge, and stored on ice.
Q-PCR
Master Mix 3 was created with 12.5 uL Sso Fast and EvaGreen supermix, 0.5 uL of 10 uM upstream primer, 0.5 uL of 10 uM downstream primer, and 10.5 uL Nanopure H2O. The solution was added to the wells of a PCR plate. 1 uL of the thawed cDNA was added to the PCR well B9. Nanopure H2O was added to the control wells and the PCR run was started.
Making an Agarose Gel
1g agarose and 75 mL 1x TAE were microwaved for three minutes. The solution was cooled and 6 uL of EtBr was adde and mixed, and the solution was poured into the gel tray. Gel combs were added and the gel was put in the refrigerator for future use.
Protein Extraction
The tissue sample was transferred to a tube labeled "KH 10/14" and weighed. The weight recorded was 0.010g for sample HB 26. 500 uL of CellLytic MT was added. The solution was then homogenized using a disposable pestle and inverted several times. The tube was then spun in a refrigerated microfuge at max speed for 10 minutes. The supernatant was transferred to a tube marked "Protein, KH, Oyster tissue, 10/14" and stored on ice. The protein was not quantified.
Results
Quantitative PCR Results
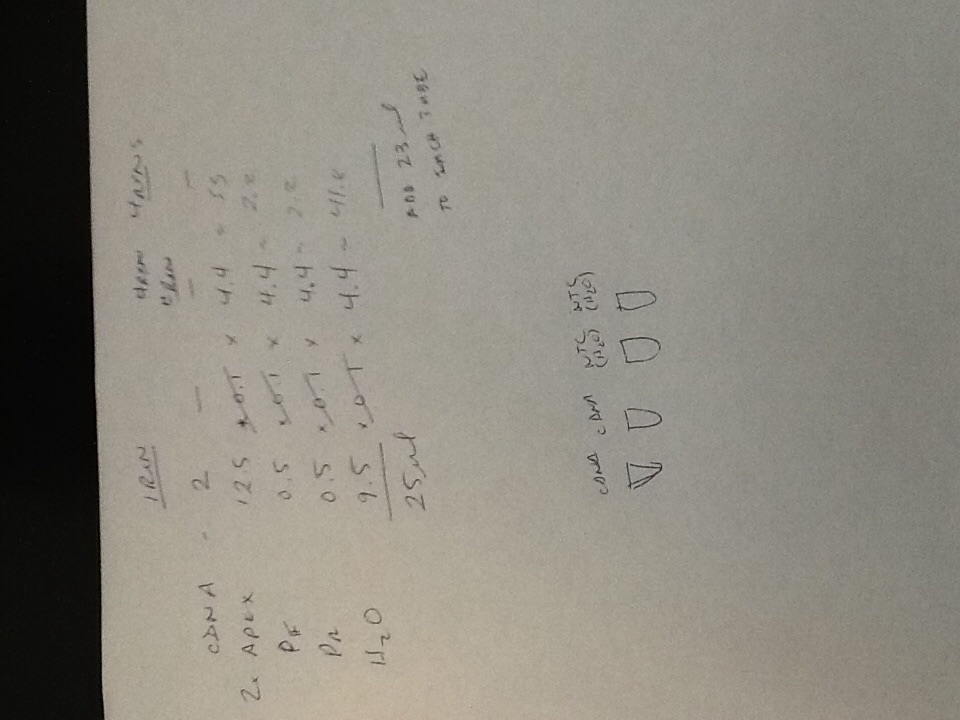
The plate ended up getting shifted over three rows and down one row, so my results can be found in well D5.
Conclusion
The relatively low Ct value of 15.10 was surprising because I thought my inability to distinguish the RNA pellet in the previous experiment would have resulted in less successful RNA extraction. However, the efficacy value of 2.44 indicates over 100% amplification efficacy, which indicates contamination of the sample, likely with EtOH.
Based on the success of the past procedures, next week we will be able to perform agarose gel electrophoresis to determine if the correct substance (cDNA) was amplified during PCR.
Reflection
Just as in previous labs, the processes done in this experiment had the goal of preparing the sample for the analysis of gene expression. PCR was used to measure the number of double stranded sequences were in my solution. Ideally, these would be cDNA, if not contaminated. These are typical procedures to determine gene expression once RNA has been extracted.
When performing the lab, I was a bit confused about which part of the procedure we were on, and whether or not it was part of the PCR measurement we were doing on 10/14. Again, I wish there had been more of an explanation of what the significance of the results was. However, through independent research I was able to find some clear explanations.
10/7/14 Lab 2
Summary
At different steps, I added chloroform, ethanol, and isopropanol to my homogenized oyster gill tissue sample. I used a refrigerated microfuge to separate the RNA into a pellet at the bottom of my vial, to be stored for later testing.
Materials and Methods
The homogenized tissue sample from 9/30/14 was incubated at room temperature (RT) for five minutes. 200 uL of chloroform were added to the sample and the solution was vortexed until it became milky. It was incubated again at RT for five minutes before it was spun in the refrigerated microfuge at max speed for 15 minutes. The aquaphase was then transferred to another vial and mixed with 500 uL of isopropanol. It was then incubated at RT for 10 minutes and then spun in the microfuge at max speed for eight minutes. The supernatent was then removed, but the pellet was not visible, so it was uncertain how much of the pellet and supernatant were being removed or kept. 1 mL of Nanopure H2O was added in error, then removed and replaced with 75% EtOH and the solution was spun in the microfuge at 7500 g for five minutes. Supernatant was removed and the pellet was dried at RT for 5 minutes.The sample was then suspended in 100 uL of Nanopure H2O and the RNA was quantified using Nanodrop Spectrophotometer. The sample was then labeled "KH RNA 10/7/14 Oyster gills" and stored at -80C for future use.
Results
The RNA concentration was 44.2 ng/uL. The A260/280 ratio was 2.07, and the A260/230 ratio was 3.20.
Conclusion
I did not have many expectations about my data, due to my unfamiliarity with the procedures. However, after receiving my results I found out that my A260/230 ratio is somewhat high, which is what I would have expected given the knowledge that it is a determination of the degree of purity of my RNA sample and the fact that my EtOH removal was probably incomplete. Since my RNA extraction was successful, next week I will be using the enzyme reverse transcriptase to transcribe RNA to DNA, and amplify it using PCR.
The purpose of this lab was to extract RNA from the tissue sample. The only facet of our procedure that was used to measure was the Nanodrop Spectrophotometer, which measured the purity of the RNA sample. These are the standard procedures used for RNA extraction and gene expression. The procedure was unclear as to exactly how important it is to remove all of the supernatant at the risk of accidentally removing some of the RNA, which made it hard to interact with my RNA pellet which was difficult to discern. I wish that we had more information on the significance of our Nanodrop Spectrophotometer results and what that indicates about our success in the lab.
9/30/14 Lab 1
Summary
In this lab, we sampled oyster tissue and used both TRI reagent and a pestle to break down the material. This was done so we could prepare the tissue for RNA removal.
Materials and Methods
Using tweezers and smalls scissors, I removed 82 mg of tissue from an oyster's gills. It was then placed in a vial, and I used an automatic pipettor to add 0.5 mL of TRI regent. I then crushed the tissue using a disposable pestle. Another 0.5 mL of TRI reagent was added, and the mixture was vortexed to further homogenize it. The solution was then labeled "Hartke Oyster gills" and placed on ice to store for a week.
Results
The tissue did not get nearly as homogenous as I expected. The pestle seemed to be only marginally effective at crushing the tissue, as it kept sliding around the bottom of the vial.
Conclusion
I expected that the methods used would be more effective than they ultimately were. The pieces of tissue in my vial were about 4 mm across, so it might have actually been more effective if I had used the scissors to cut them into smaller pieces. This is a surprise, because I wouldn't have expected such a crude instrument to outperform our standard lab methods.
Next, I will extract the RNA from the tissue sample.