Summary
I created 10 uMol primer stock for all three primer sets (F+R) and ran qPCR on 2 of my cDNA samples to see if the primers worked
Materials
-PCR plates (white with caps)
-1.5 mL microfuge tubes (RNAse free)
-Nuclease Free water
-pipettes + tips
-Opticonthermal cycler
-2x Immomix Master Mix
-50 uM Syto-13 dye
-microfuge tube racks
-ice bucket
-cDNA samples from
-3 sets of primers (Strongylocentrotus pupuratus HSP gp96 F+R, Strongylocentrotus pupuratus CRH F+R, and Odontaster validus HSP70 F+R)
Procedure
Creating 10 uM primer stock
1) Each concentration of primer was multiplied by 10 individually and then that amount of water was added directly into each primer tube
O. validus HSP 70 forward- 30.3 nm*10 = 303 uL of water added
O. validus HSP 70 reverse-28.2 nm*10 = 282 uL of water added
S. pupuratus HSP gp96 forward-25.8 nm*10 = 258 uL of water added
S. pupuratus HSP gp96 reverse-36.4 nm*10 = 364 uL of water added
S. pupuratus CRH forward-22.4 nm*10 = 224 uL of water added
S. pupuratus CRH reverse-30.7 nm*10 = 307 uL of water added
2) 10 uL of that mixture was pipetted into a new 1.5 mL tube labeled with my initials and its specific primer and 90 uL of water was added to each to create the 10 uM stock
Testing primers with qPCR
1) Enough master mix was made for 7 reactions (4 samples, 2 negative controls, and 1 for pipetting error). The master mix consisted of 175 uL of Immomix, 14 uL of Syto-13 dye, 3.5 uL upstream primer, 3.5 uL downstream primer, and 140 uL of PCR water
3 separate master mixes were made, one for each primer set
2) On the PCR plate each of three columns was for its own specific primer set (1 column for HSP gp96, 1 for CRH and another for HSP 70)
3) 48 uL of master mix (specific to each column) was added to each well
4) 2 uL of PCR water was added into each of the first two wells of all three columns (the negative controls)
5) 2 uL of cDNA from 2 samples (P1, P2, C1, C2) were added to the remaining wells in each column
Plate Layout
| |
P1 (HSP gp96) |
P2 (CRH) |
P3 (HSP70) |
| 1 |
Negative control |
Negative control |
Negative control |
| 2 |
Negative control |
Negative control |
Negative control |
| 3 |
cDNA from P1 |
cDNA from P1 |
cDNA from P1 |
| 4 |
cDNA from P2 |
cDNA from P2 |
cDNA from P2 |
| 5 |
cDNA from C1 |
cDNA from C1 |
cDNA from C1 |
| 6 |
cDNA from C2 |
cDNA from C2 |
cDNA from C2 |
| 7 |
X |
X |
X |
| 8 |
X |
X |
X |
Analyze qPCR results to see if all primers worked and run the rest of the samples.
11/25/09
Summary
I finished specing my DNased RNA in order to determine the RNA concentrations of each sample and I made cDNA from my DNased samples.
Materials
-1.5 mL snap cap tubes
-ice bucket
-gloves
-refrigerated microfuge
-pipettes + tips
-spec tubes
-tube holder
-thermocycler
-0.1% DEPC-H20
-PCR water
-M-MLV RT Buffer
-2.5 mM mixed dNTPs
-M-MLV RTranscriptase
-Oligo dT primer
Procedure
Spec and Normalizing RNA
1) The DNased RNA samples were quantified using nanodrop (See results for concentrations and absorbancy ratios in Table 1 below)
2) To obtain DNased RNA samples with a concentration between 20-25 ng/ ul only P1 & C2 at 39.6 ng/ uL and 34.9 ng/ uL respectively were diluted with 0.1% DEPC-H20. Using the equation: C1*V1 = C2*V2, 7.43 uL of water was added to P1 and 6.405 uL was added to C2.
Making cDNA
1) 8 PCR tubes were labeled as follows: KS P1 cDNA, KS P2 cDNA, KS P3 cDNA, KS P4 cDNA, KS C1 cDNA, KS C2 cDNA, KS C3 cDNA, KS C4 cDNA and 5 ul of the corresponding RNA sample was added to each tube
2) The tubes were incubated in the thermocycler at 75 degrees celsius for 5 minutes and then put on ice for 5 minutes
3) Enough Master Mix was made for 9 reactions with 36 uL RT buffer, 72 uL dNTPs, 9 uL Oligo dT Primer, 9 uL RNase free water, and 9 uL RTranscriptase
4) 15 uL of Master Mix was added to each tube (which already included 5 uL of RNA for a total of 20 uL)
5) The tubes were vortexed and placed in the thermocycler to incubate the tubes at room temperature for 10 minutes, then at 37 degrees celsius for an hour and then heat inactivate at 95 degrees celsius for 3 minutes
6) The cDNA was stored at -20 degrees celsius for future use
Calculations
P1: C1*V1 = C2*V2, (39.6 ng/uL)(?) = (20 ng/ uL)(15 uL)
(20*15)/ 39.6 = 7.575, 15uL-7.575 = 7.43 uL of water to be added
C2: (34.9 ng/ uL)(?) = (20 ng/ uL)(15 uL)
(20*15)/ 34.9 = 8.59, 15-8.59 = 6.40 uL of water to be added
Results
Table 1. Results of RNA quanitification for DNased samples including the concentration of RNA in each sample and the absorbancy ratios for each
Sample ID |
RNA Concentration (ng/ul) |
A260/280 |
A260/230 |
| Predator 1 (P1) |
39.6 |
1.78 |
0.76 |
| Predator 2 (P2) |
22.6 |
1.70 |
0.52 |
| Predator 3 (P3) |
19.3 |
1.69 |
0.53 |
| Predator 4 (P4) |
25.6 |
1.98 |
0.78 |
| Control 1 (C1) |
22.2 |
1.72 |
0.56 |
| Control 2 (C2) |
34.9 |
1.85 |
0.82 |
| Control 3 (C3) |
13.3 |
1.78 |
0.41 |
| Control 4 (C4) |
23.1 |
1.80 |
0.69 |
After DNasing the samples all the concentration values for the new RNA increased significantly while the absorbancy values for A260/280 and A260/230 ratios decreased.
Next Steps
I will be making 10 uMol primer stock for all three primers (F+R) and testing to see if the primers worked with qPCR
11/17/09
Summary
I began DNase of my 8 RNA samples to check for carryover.
Materials
-0.5 mL snap cap tubes
-ice bucket
-gloves
-20 uL of DNase buffer (2.5 uL per tube)
-8 uL of Turbo DNase (1 uL per tube)
-20 uL of Inactivation Reagent (2.5 uL per tube)
-refrigerated microfuge
-pipettes + tips
-spec tubes
-tube holder
-water bath with tube holder
Procedure
Start DNase of RNA samples
1) Thawed RNA samples from -80 freezer and pipetted 20.5 uL of each RNA sample into a fresh tube (8 tubes total) labeled as follows: KS C1, KS C2, KS C3, KS C4, KS P1, KS P2, KS P3, KS P4
2) 2.5 uL of DNase buffer and 1 uL of Turbo DNase was added to each of those tubes (total volume of 24 uL in each tube)
3) The tubes were incubated in the thermocycler at 37 degrees celsius for 30 minutes
4) 2.5 uL of inactivation reagent was added was added to each tube and the tubes were incubated at room temperature for 2 minutes, mixing occasionally
5) The tubes were spun at 10,000 rcf for 1.5 minutes and the supernatant from each tube was transfered into a corresponding spec tube with an identical label
Next Steps
I didn't finish DNasing my samples and will need to spec the samples and then normalize with 0.1% DEPC H20 before I can make cDNA.
11/12/09
Summary
I made stock RNA and quantified my RNA
Materials
-8, 1.5 mL snap cap tubes
-ice bucket
-gloves
-chloroform (and waste containers)
-isopropanol
-ethanol
-water bath (with floating tube holders)
-tube holder
-RNase free water
-pipettes + tips
-refrigerated microfuge
-Kim Wipes
-0.1% DEPC-H20
Procedure
RNA Extraction
1. Brittle star tissue that was stored in TriReagent at -80 degrees celsius was thawed at room temperature
2. Quickly added 200 uL of chloroform to each tube in the hood and vortexed it for 30s until the solution was milky looking
3. Sample was left at room temperature for 5 minutes and then spun in the refrigerated microfuge at full speed for 15 minutes
4. 4 new 1.5 mL tubes were labeled as follows: KS C1, KS C2, KS C3, KS C4, KS P1, KS P2, KS P3, KS P4 with RNA written on each lid and the the aqueous phase (the top clear liquid in the tube) was pipetted from each tube into its corresponding new tube. The remaining organic and interphase liquid was disposed of in the liquid waste in the hood
5. 500 uL of isopropanol was added to each new tube containing only the aqueous phase (with RNA) and the tube was inverted to mix until the liquid appeared uniform
6. Tubes were incubated at room temperature for 10 minutes and then spun in the refrigerated microfuge for 8 minutes at full speed
7. The supernatant was removed (all the liquid excluding the white pellets at the bottom of the tubes)
8. 1 mL of 75% ethanol was added to the pellets and the tubes were vortexed to dislodge the pellets
9. The tubes were spun in the microfuge for 5 minutes at 7500g and the remaining supernatant was removed. The microfuge and small pipette tips were used to try and remove as much of the residual ethanol as possible.
10. Pellets were dried up to 5 minutes and redissolved in 100 uL of 0.1% DEPC-H20
11. The stock RNA tubes were placed in a water bath set at 55 degrees celsius for 5 minutes and then into the -80 degrees celsius freezer
Observations
Small dark pellets of RNA were only visible in the C2, P1, and P4 tubes after isopropanol was added and the tubes were spun in the microfuge. It was difficult to pipette as much ethanol as possible off of the other samples because I couldn't tell where the RNA was, as a result some residual ethanol was probably left in the samples.
RNA Quantification
1) The Nanodrop was zeroed with 2uL of 0.1% DEPC-H20
2) 2 uL of each RNA sample was placed on the Nanodrop one at a timeand the following values were recorded: the A260 absorbance, the RNA concentration (in ng/L), the ratio of A260/280, and the ratio of A260/320
3) The Nanodrop pedestal was wiped with a Kim Wipe between measurements for each sample
Results
Table 1. Results of RNA quanitification including the concentration of RNA in each sample and the absorbancy ratios for each
Sample ID |
RNA Concentration (ng/ul) |
A260/280 |
A260/230 |
| Predator 1 (P1) |
43.69 |
1.75 |
1.13 |
| Predator 2 (P2) |
22.82 |
1.70 |
0.72 |
| Predator 3 (P3) |
20.73 |
1.60 |
0.95 |
| Predator 4 (P4) |
28.16 |
1.63 |
1.12 |
| Control 1 (C1) |
30.29 |
1.61 |
1.20 |
| Control 2 (C2) |
41.13 |
1.68 |
1.59 |
| Control 3 (C3) |
13.20 |
1.61 |
0.65 |
| Control 4 (C4) |
28.13 |
1.66 |
1.43 |
Conclusions
The tubes in which I saw an obvious pellet of RNA after the isopropanol was added and the samples were vortexed (C2, P1, and P4) not surprisingly had some of the highest concentrations of RNA in the sample. All the values for absorbancy ratios of A260/A280 fell below the range of 1.8-2.0 for clean RNA which means that stock RNA is contaminated with ethanol or phenol. This is further confirmed with the A260/A230 absorbancy ratio values, none of which fall between 1.5-2.0 (with the exception of the C2 sample). Most likely the samples are contaminted with ethanol because it was very difficult to pipette all the ethanol from the sample as I was afraid that I would suck up the remaining RNA when I couldn't see any visible pellet at the bottom of most of the tubes. I tried to get as much out as possible, but the results indicate that residual ethanol was left.
Next Steps
-
Create 10 mmol stock for all F+R primers and make cDNA from all 8 stock RNA samples
Primer Information
Primer Name, Sequence, #bp, GC%, Melting Temp, Organism, Gene Gene Acc., # Size
KS_strongylocentrotus_CRH_F, TCGCGGGTTCCGAACAACGG, 20, 65, 59.98, S. pupuratus, CRH, XM_001195663.1, 214
KS_strongylocentrotus_CRH R, TAGCGCTCACGACGTCCCCA, 20, 65, 59.97, S. pupuratus, CRH, XM_001195663.1, 214
KS_strongylocentrotus_HSPgp96_F, TCCAGGCCGAGGTCAACCGT, 20, 65, 59.83, S. pupuratus, HSP gp96, NM_214643.2, 815
KS_strongylocentrotus_HSPgp96_R, ATCCCGAGGCGCTCTGGTCC, 20, 70, 60.39, S. purpuratus, HSP gp96, NM_214643.2, 815
KS_validus_HSP70_F, ACGGCGGCAAGCCCAAAGTT, 20, 60, 60.11, Odontaster validus, HSP 70, AM408048.1, 118
KS_validus_HSP70_R, CCGAGGAAGGCCTCTGCCGT, 20, 70, 60.6, Odontaster validus, HSP 70, AM408048.1, 118
11/10/09
Lab 6-Experiment Set-Up for Project
Summary
I set up my experiment to test the stress response for Ophiura leutkani (brittle star) to the introduction of a sea star predator (Pycnopodia helianthoides).
Materials
-8, 1.5 mL snap cap tubes
-8 mL of TriReagent
-ethanol
-8 razor blades
-forceps
-2 tanks with filtration
-tube holder
-bucket with ice
-gloves
-pipette + tips
-lab pen
-Kim wipes
-dissecting tray
-8 disposable pestles
Procedure
1) 8, 1.5 mL tubes were labeled as follows: KS C1, KS C2, KS C3, KS C4 (the control tubes), KS P1, KS P2, KS P3, KS P4 (the treatment tubes).
2) 500 uL of TriReagent was added to each tube and then they were put on ice.
3) 10 brittle stars (approximately 10 mm disk diameter) were selected.
4) 5 brittle stars were placed into the filtered control tank and another 5 into the treatment tank.
5) The predator was added (approximately 2.9 mm disk diameter) to the treatment tank and left to induce stress response in the brittle stars for one hour.
6) After an hour the brittle stars were removed one by one and approximately 50-100 mg of tissue was removed using a razor blade from the interambulacral space on the ventral side of each animal. A fresh razor blade was used to take tissue from each individual and the forceps and dissecting tray were cleaned with ethanol between each dissection. The tissue was chopped up as much as possible to break down the hard calcareous endoskeleton.
7) The tissues taken from the animals in the control tank were put into the "C" tubes and the tissues from the treatment tank were put into the "P" tubes.
8) The tissues were homogenized in their tubes using a disposable pestle and then an addition 500 uL of TriReagent was added to each tube.
9) The tissues were placed in the -80 degrees celsius freezer for future use.
Observations
1) When the Pycnopodia was added to the treatment tank the brittle stars had a very clear, strong reaction to its presence and moved around the tank. The individuals in the control tank were non reactive as we would expect. When I returned to take tissues the predator had crawled up the side of the tank and the brittle stars were stationary down below.
2) I had to sample control brittle stars from another tank because very "leggy" individuals were needed for another student's experiment. This should not effect the results of my experiment as all these individuals were in tanks inside of a larger container and receiving the same filtered water.
Table 1. Recorded disk diameters of brittle stars used in experiment
| Control 1 (C1) |
Control 2 (C2) |
Control 3 (C3) |
Control 4 (C4) |
Predator 1 (P1) |
Predator 2 (P2) |
Predator 3 (P3) |
Predator 4 (P4) |
|
| Brittle Star Size |
13.0 |
9.0 |
10.0 |
10.0 |
10.0 |
12.0 |
12.0 |
10.0 |
1) Continue RNA extraction protocol to make stock RNA.
Project Timeline
11/10: Run experiment and store tissue samples in TriReagent at -80
11/12: RNA extraction and quantification, determine primers
11/17: Create primer stock and make cDNA (if primers are in, if not, set aside lab time later in the week-
11/24: run qPCR
12/1 & 12/8: Analyze results and work on presentation and extended abstract
11/3/09
Lab 5-Quantitative PCR
I was absent in lab this week due to illness so I didn't load my samples myself. Thanks Rachel!
Summary
1) ran qPCR on cDNA from Crassostrea gigas gill tissue
Procedures
1) Master mix was prepared for 7 reactions (2 controls, 2 RNA reactions, 2 cDNA reactions, and 1 extra for pipette error). The following concentrations went into the master mix for each reaction.
-25 uL 2X immomix
-2 uL Syto-13 dye (50 uM)
-0.5 uL upstream primer (10 uM)
-0.5 uL downstream primer (10 uM)
-20 uL Ultra Pure Water
2) 48 uL of master mix was added into 6 tubes. 2 uL of cDNA was added to two of the tubes, 2 uL of RNA was added to another two tubes and 2 uL of ultra pure water was added to the last two tubes. After being capped the tubes were loaded onto the PCR plate
Thermal Profile for qPCR
1. Incubate at 95 degrees celsius for 10 minutes
2. Incubate at 95 degrees celsius for 15 seconds
3. Incubate at 55 degrees celsius for 15 seconds
4. Plate read
5. Incubate at 72 degrees celsius for 30 seconds
6. Plate read
7. Repeat steps 2-6, 39 times
8. Incubate at 95 degrees celsius for 1 minute
9. Incubate at 55 degrees celsius for 1 second
10. Melting curve from 55 degrees celsius to 95 degrees celsius, read every 0.5 seconds and hold for 30 seconds
11. Incubate at 21 degrees celsius for 10 minutes
END
Results
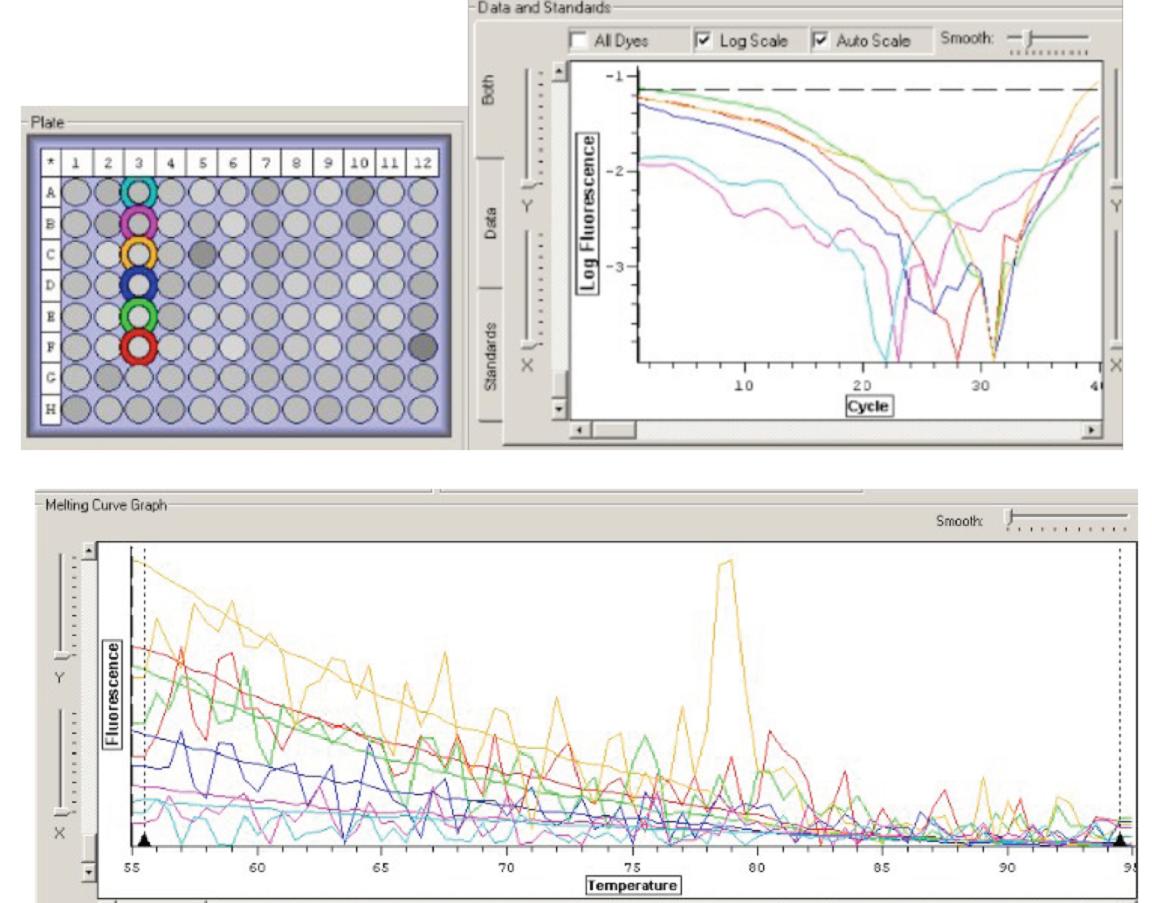
Figure 1. qPCR results for cDNA from oyster gill. The first two wells in the PCR plate are the negative controls, the next two are RNA and the last two are cDNA.
Conclusion
My results clearly indicate that nothing was amplified (including my negative control samples, which you would expect), which could mean that the superoxide dismutase gene wasn't expressed. Next week we will set up our experiments. -
10/27/09
Lab 4-Western Transfer + Immunoblots
Summary
1) Run PCR on gel from lab 3
2) Run Western Blot
3) Discuss research projects
Procedures
Running PCR products on gel
1) filled gel box with 1x TAE buffer to cover the gel and removed the combs
2) 7 uL of 1000bp ladder was added to the far left lane
3) 25 uL of PCR sample were added to the gel wells in the following order (sampe 1, sample 2, control 1, control 2)
4) After all the samples were loaded the gel was run at 100 V for one hour and later visualized
*My samples were loaded into gel 2
Western Blot
1) The filter paper, membrane, and gel were soaked in cooled transfer buffer for 15 minutes
2) The blotting aparatus was assembled as follows: (from the bottom up) anode (+), filter paper, nitrocellulose membrane, gel, filter paper, cathode (-) making sure to push bubbles out after each layer was laid down
3) Blot was run at 20V for 30 minutes
4) The gel was removed, rinsed with buffer and gel was carefully removed from the membrane
5) Blocking solution was prepared with 14 ml of filetered water, 4 ml of blocker (part A), and 2 ml of blocker (part B)
6) The membrane was put into 10 ml of blocking solution and placed on a shaker (1 rev/sec) for 30 minutes
7) The blocking solution was poured off and the membrane was rinsed with 20 ml of water twice
8) Primary Antibody Solution (1:3000 dilution) was prepared as follows: 10 ml of blocking solution and 3.3 ul of HSP 70 antibody
9) The membrane was placed in 10 ml of Antibody Solution overnight
10) The membrane was rinsed with 20 ml of Antibody Wash 3 times
11) It was then set in 10 ml of Secondary Antibody Solution for 30 minutes and washed in 20 ml of Antibody Wash for 5 minutes
12) The membrane was washed in 20 ml of water for two minutes and repeated
13) It then sat in 5 ml of Chromogenic Substrate until bands appeared and was rinsed in 20 ml of water
14) Finally, the membrane was dried on filter paper
Results
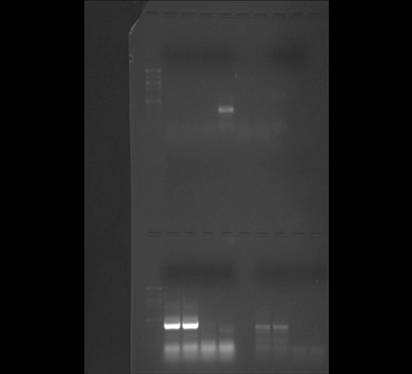
Figure 1. PCR Gel #2 lanes (left-right, top): 7uL ladder, CB1, CB2, CBP1, CBP2, KS1, KS2, KS3, KS4
(bottom): 7uL ladder, SRB, SRC, SRD, SRE, blank, LH1, LH2, LH3, LH4
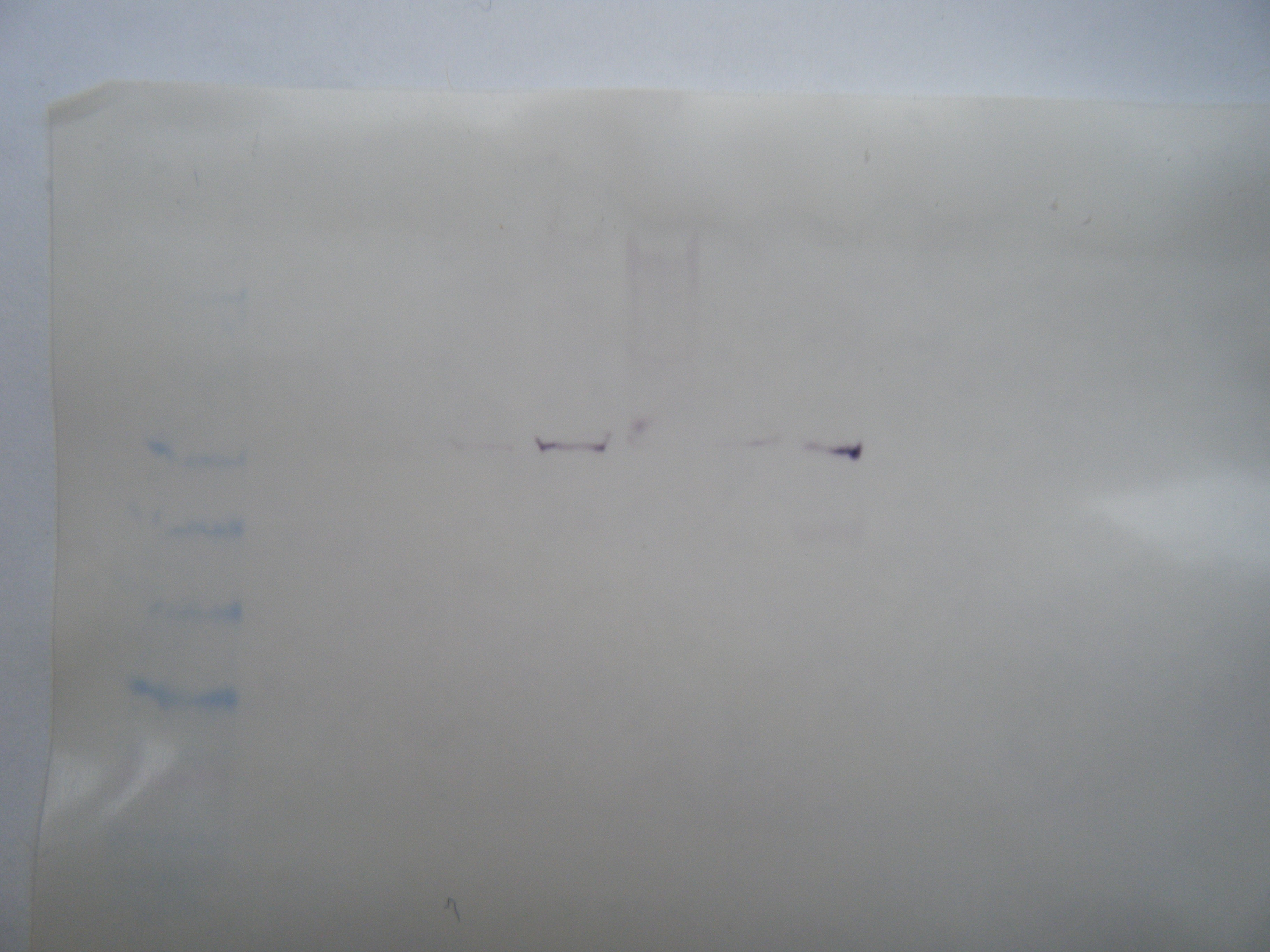
Figure 2. Western blot membrane (HSP 70) with lanes loaded as follows (left to right):
1) SeeBlue ladder
2) Vibrio tubiashii (15 uL)
3) Vibrio tubiashii (+ oyster) (15 uL)
4) Sepia eye (9.5 uL)
5) Salmon brain (15 uL)
6) Salmon brain (15 uL)
7) Herring heart (6.9 uL)
8) Herring brain (10.4 uL)
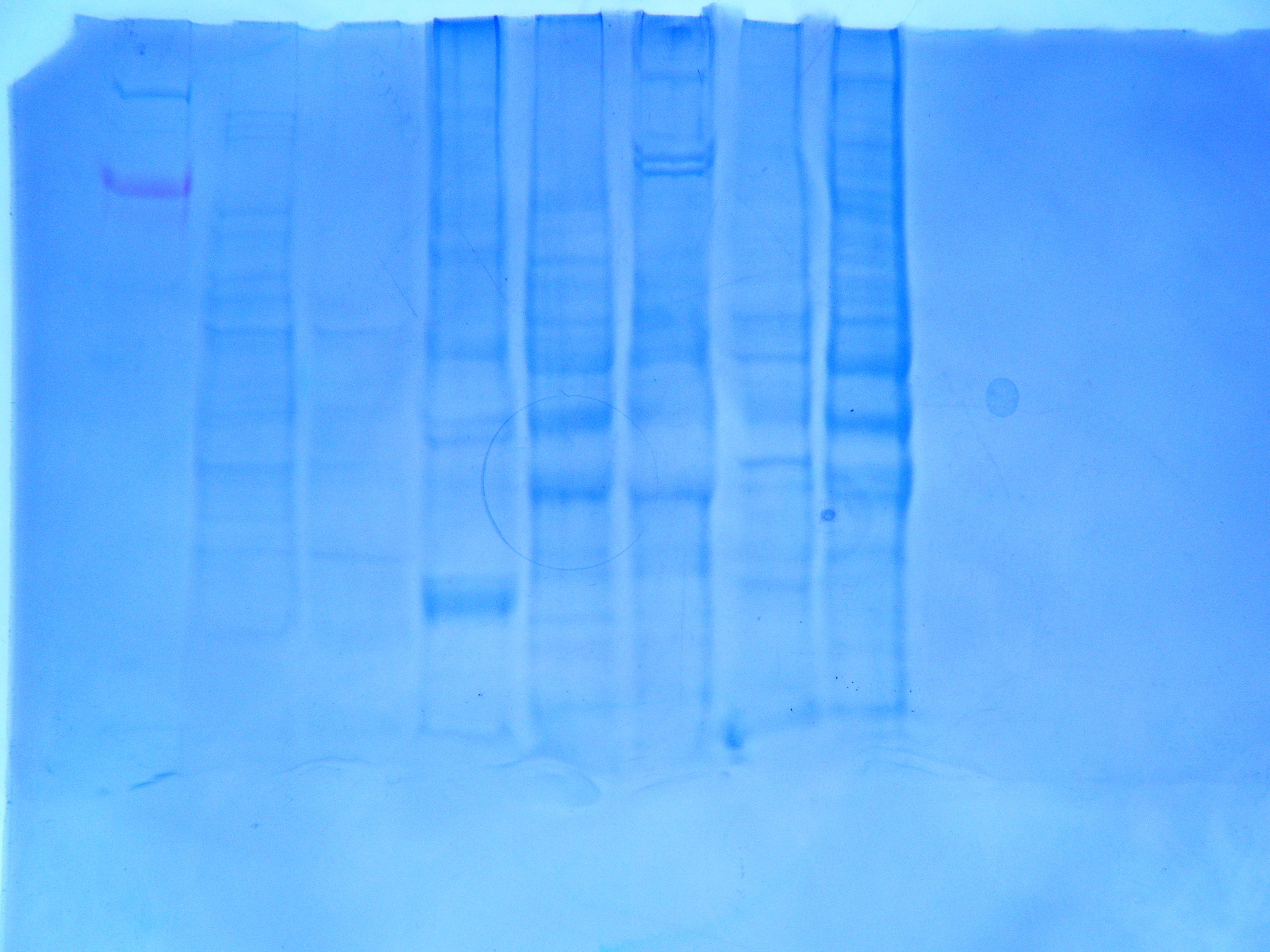
Figure 3. Western blot gel (HSP 70) with lanes loaded as in Figure 2
Conclusions
I wasn't able to copy the PCR image with arrows into my notebook, but looking at the original image I can see that something went wrong. I loaded the sample into my first two wells, however there are no bands (it's possible that they are just very light and I can't see them well or that I mixed up my samples and controls when I was loading them). There appears to be a band above 500 bp in the last lane (which if this is one of my controls, it clearly indicates that I either mixed up my samples or there was some contamination between samples). The primers should have amplified a section of 199 bp and no band shows up in this region on the gel (See Figure 1). It is not apparent that the HSP 70 antibodies binded to my protein if you look at figure 2, the bands are very faint. However, you can see several bands in Figure 3, which is not what you would expect from non-heat shocked protein (-
10/20/09
Lab 3-Reverse Transcription & PCR-
Summary
1) Quantified RNA with Nanodrop
2) Created 10 mmol stock for forward and reverse primers for Crassostrea gigas SOD
3) Created cDNA from stock RNA sample (of Crassostrea gigas oyster)
4) Prepared samples for PCR and set up gel boxes
Procedures
RNA Quantification
1) The Nanodrop was zeroed with 2uL of 0.1% DEPC-H20
2) 2 uL of the oyster gill RNA sample was placed on the Nanodrop and the following values were recorded: the A260 absorbance, the RNA concentration (in ng/L), the ratio of A260/280, and the ratio of A260/320.
3) The Nanodrop pedestal was wiped with a Kim Wipe
Reverse Transcription
1) 5 uL of the stock RNA sample (from Crassostrea gigas gill) and 5 uL of PCR water were pipetted into a fresh tube and the solution was incubated for 5 minutes at 75 degrees celsius
2) The tube was placed on ice for 5 minutes
3) A master mix consisting of 4 uL 5x Buffer (AMV RT Buffer), 8 uL of dNTPs, 1 uL of AMV RTranscriptase, 1 uL of Oligo dT Primer, and 1 uL RNase free water was added to the tube and the tube was spun
4) The tube sat at room temperature for 10 minutes and then was incubated at 37 degrees celsius for 1 hour and then at 95 degrees celsius for 3 minutes
5) The tube was spun and stored at -20 degrees celsius
PCR
Crassostrea gigas superoxide dismutase forward primer sequence: 5'-GAG AGG CAC GTG GGG GAC CT-3'
Crassostrea gigas superoxide dismutase reverse primer sequence: 5'-ATC GTC CGC CAG CGT TTC CG -3'
10umol stock of forward primers was created by adding 313 uL of nuclease free water to 31.3 nm of primer (which makes 100 umol stock) and then pipetting 10 uL of this 100 umol stock into a tube with 90 uL of water. 10 umol stock of reverse primers was created by adding 408 uL of nuclease free water to 40.8 nm of primer (which makes 100 umol stock) and then pipetting 10 uL of this 100 umol stock into a tube with 90 umol of water.
1) The master mix was prepared by combining 90 uL of water, 125 uL of Promega's GoTaq Green Master Mix (2X 25), 12.5 uL of reverse primer and 12.5 uL of forward primer which makes enough for 5 reactions (50 uL total).
2) 4 tubes were prepared (two with 2 uL of cDNA & 48 uL of master mix (these are the samples) and two more with 2 uL of water and 48 uL of master mix (these are the negative controls)).
Thermal profile:
95 degrees celsius for 10 minutes
40 cycles:
-95 degrees celsius for 30 seconds
-55 degrees celsius for 30 seconds
-72 degrees celsius for 90 seconds
72 degrees celsius for 3 minutes
4 degrees celsius for forever
Agarose Gel Preparation
1) 2g of agrose was mixed with 150 mL of 1x TAE in a flask and microwaved for a few minutes
2) The agrose was cooled slightly and 12 uL of ethidium bromide was added
3) The solution was swirled to mix, poured into the gel tray and the combs were added
4) Gel was left to set and stored in a fridge
*Note: Our gel solution boiled over in the microwave so we added more buffer and agrose until it approximated the appropriate volume. We also neglected to make sure all the bubbles were out of the gel before it set.
Results
Nanodrop measurements:
-A260: 5.942
-RNA concentration (ng/uL): 237.7
-A260/280: 1.86
-A260/230: 1.79
Conclusions
A range of 1.8-2.0 for the A260/A280 ratio and a 1.5-2.0 ratio for the A260/A230 ratio indicates a clean stock RNA sample. Since my values for A260/280 and A260/230 were 1.86 and 1.79 respectively they fall within the range that indicates my stock RNA is likely clean without residual ethanol or salt content. PCR is likely to be more successful without these byproducts when the gel is run. We will be running out PCR products on the agrose gel during next week's lab.
10/13/09
Lab 2-Tissue Extraction II
Summary
1) Continued RNA extraction of Crassostrea gigas gill (rather than cuttlefish eye) to produce stock RNA sample
2) Ran SDS-PAGE on protein samples from lab 1 (using cuttlefish eye protein) and visualized gels
Procedures (continued from Lab 1)
RNA Extraction
1. Sample tissue in TriReagent from Lab 1 that was stored at -80 degrees celsius was thawed at room temperature
2. Quickly added 200 uL of chloroform to sample in the hood and vortexed it for 30s until the solution was milky looking
3. Sample was left at room temperature for 5 minutes and then spun in the refrigerated microfuge at full speed for 15 minutes
4. Only the aqueous phase (the top clear liquid in the tube) was pipetted into a fresh tube. The remaining organic and interphase liquid was disposed of in the liquid waste in the hood
5. 500 uL of isopropanol was added to the new tube containing only the aqueous phase (with RNA) and the tube was inverted to mix until the liquid appears uniform
6. Tube was incubated at room temperature for 10 minutes and then spun in the refrigerated microfuge for 8 minutes at full speed
7. The supernatant was removed (all the liquid excluding the white pellet at the bottom of the tube)
8. 1 mL of 75% ethanol was added to the pellet and the tube was vortexed to dislodge the pellet
9. The tube was spun in the microfuge for 5 minutes at 7500g and the remaining supernatant was removed. The microfuge and small pipette tips were used to try and remove as much of the residual ethanol as possible.
10. Pellet was dried up to 5 minutes and redissolved in 100 uL of 0.1% DEPC-H20
11. RNA tube was placed in a heat bath at 55 degrees celsius for 5 minutes and then into the -80 degrees celsius freezer
Originally the RNA extraction procedure was done using cuttlefish eye tissue, however, I accidentally threw away the aqueous phase containing the RNA (Step 4) during this lab. As a result, I started this lab over using Crassostrea gigas (Pacific oyster) gill tissue and this stock RNA will be used in future labs.
SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
1. Added 15uL of the cuttlefish eye protein extract (from Lab 1) and 15uL of 2X Reducing Sample Buffer to a 1.5 mL screw cap tube
2. Sample was mixed, quickly centrifuged, then boiled for 5 mintues and centrifuged again for 1 minute
3. All samples were pipetted into gel wells and the gel was run at 150V for 25 minutes
4. Gel was removed; placed into a container with 150 mL of Coomassive Stain and then on the rocker for 5 minutes
5. The excess stain was removed and the gel was rinsed with 10% acetic acid
6. The gel was left in acetic acid solution overnight and removed
2 gels were run (Figure 1 & Figure 2)
Results

Figure 1. SDS PAGE, 4-20% Tris-HEPES gel results (Group 2): Gel lanes 1-8 correspond to the following protein concentrations loaded into each well (left to right): SeaBlue 2 ladder, herring heart (26.17 ug), salmon brain (10.31 ug), hard clam foot (32.89 ug), sea scallop (24.2 ug), barnacle (17.31 ug), gigas (4.57 ug), oyster gill (18.855 ug).
The 18.855 ug of oyster gill protein loaded into well 8 should actually be labeled as cuttlefish eye protein

Figure 2. SDS PAGE, 4-20% Tris-HEPES gel results (Group 1): Gel lanes 1-9 correspond to the following protein concentrations loaded into each well (left to right): SeaBlue 2 ladder, vt oyster (3.214 ug), heat shock barnacle (23.43 ug), blank, 9.165 ug, heat shock barnacle (77.3496 ug), salmon brain (9.562 ug), herring brain (16.2 ug), 9.57 ug.

Figure 3. Approximate molecular weights (kDa) of the NuPAGE MES protein gel (which is closest to the 4-20% Tris-HEPES gel we used). Position of bands and their weights were compared to the gels that we ran (Figure 1 & Figure 2). Smaller (lower molecular weight) proteins move further down the gel, quicker than larger (higher molecular weight) proteins.
1257.7917 ug/mL (amount of protein sample-see lab 1)*.001=1.2577 ug/uL*15uL=18.855 ug (the concentration of protein loaded into the gel well).
Conclusions
18.855 ug of cuttlefish eye protein (Figure 1, Lane 8) was loaded into the gel. Comparing the lane to the ladder there are two distinct bands at 98 & 49. It's interesting to note that darkest and the lightest lanes have the highest and lowest loaded protein concentrations of the gel. We did not have the time to quantify our RNA samples using Nanodrop during this lab so we will continue on 10/20/09.
10/6/09
Lab 1-Tissue Extraction I
Summary
1) Began RNA extraction with TriReagent and stored sample at -80 degrees celsius.
2) Extracted protein from cuttlefish eye tissue and measured the average absorbancy at 595 nm with a spectrophotometer. Sample was stored at -20 degrees celsius.
Procedures
RNA Extraction
1. 500 uL of TriReagent was added to 1.5 mL tube
2. A small piece of tissue (~50-100 mg of cuttlefish eye) was added to the tube and macerated with a pestle
3. Additional 500 uL of TriReagent was added
4. Sample vortexed for 15 seconds and stored at -80 degrees celsius for future use
Protein Extraction
1. Added 0.5mL of CelLytic MT solution to a 1.5 mL tube with a piece of tissue (~25 mg) and macerated with a pestle
2. Tube was inverted for a few seconds to mix the contents
3. Tube was placed in refrigerated microfuge and spun at the maximum speed for 10 minutes
4. The supernatant material was transferred to a new tube labeled "Protein" and put on ice
5. 15uL of supernantant and 15 uL of DI water were put into a new tube labeled "sample" and inverted to mix
6. 30 uL of DI water were put into a "blank" tube
7. 1.5 mL of Bradford reagent was added to each tube
8. Both tubes were inverted to mix and sat at room temperature for 10 minutes
9. The "blank" tube was inverted and 1 mL of the fluid was transferred to a cuvette. The absorbance at 595 nm was measured in the spectrophotometer
10. The "sample" tube was inverted to mix and 1mL of the solution was transferred to a cuvette. The absorbance at 595 nm was measured
11. The "sample" cuvette was removed and the fluid was mixed by pipetting the solution up and down several times (set to 1mL)
12. The absorbance at 595 nm was measured again and the two values were averaged
13. The protein concentration of the sample was calculated using the standard curve
14. The protein sample was stored at -20 degrees celsius
Results
-The equation for the standard curve used to calculate the protein concentration from the sample is Y= 1011.9x
-The value of x is the average of the two absorbancy values at 595 nm from the sample cuvette.
-The calculated total protein concentration of the sample at 1257.7917 ug/mL is a higher concentration value than we would expect for an absorbancy at 0.6215 based on the curve.
Calculations
(0.615+0.628)/2 = 0.6215 is the average absorbancy of the sample
1011.9*(0.6215)x2 = 1257.7917 ug/ml is the calculated total protein concentration of the sample
The average absorbancy value was multiplied by two in order to account for the 1:2 dilution of sample to DI water.
Conclusions
-The calculated total protein concentration of the sample at 1257.7917 ug/mL** is a higher concentration value than we would expect for an absorbancy at 0.6215 based on the curve.
-Continue RNA extraction and quantification and set up protein gel for SDS-PAGE on 10/13.