1. Select tissue previously archived in ultra-cold freezer
Selected black abalone digestive gland tissue preserved in Liq N2 and stored @ -80C
sample # 08:13-4 from 2nd (mini) WS resistance study
TL: 61.0 mm
TW: 31.7g
SW: 12.16g
Sample date: 8/14/08
Sump/Tank: Sump 1 (Exp)/2E (SNI)
Notes: Active but not attaching, bloated – histo positive for weird (protist?) bug
SEX: female
Histo Score: FT atrophy: 0.5-1, PE RLO: 2.5, DG RLO: 1.5, Metaplasia: 0.5
2. Start RNA extractions (add Tri-Reagent to samples and re-freeze)
Samples extracted in duplicate in case of error and also for replication
RNA ISOLATION PROTOCOL*important to use extreme sterile technique when handling RNA
1. Turn on heating block to 55C. Also turn on spectrophotometer.
2. Add 500uL of TriReagent *caution: contains phenol* to a 1.5mL snap cap tube. Store on ice.
3. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.
Tissue sample R1 = 63mg
Tissue sample R2 = 50mg
4. Carefully homogenize the tissue using a disposable pestle.
5. Add an additional 500uL of TriReagent to the tube and close the tube.
6. Vortex vigorously for 15s.
----- Stop here for Lab 1 and freeze sample at -80 - Will continue extraction in Lab 2
3. Extract protein from complementary sample
PROTEIN EXTRACTION PROTOCOL*important to use extreme sterile technique when handling proteases
1. Add 0.5mL of CelLytic MT solution to a 1.5mL snap cap tube.
2. Add 25mg of your tissue to the tube.
Tissue sample P1 = 32mg
Tissue sample P2 = 31mg
3. Homogenize the tissue with a disposable pestle.
4. Close the tube and invert the tube several times.
5. Spin the tube in a refrigerated microfuge for 10mins. @ max speed.
6. While spinning, label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
7. Carefully transfer supernatant to labeled tube and store tube on ice.
8. To a fresh tube, add 1.5mL of Bradford reagent. Note: The Bradford assay works best when samples are mixed well. Invert tubes frequently during incubations, and immediately before measuring absorbance to ensure accurate absorbance readings.
*Also add a control tube (1.5ml Bradford and 30uL CelLytic MT solution) to “zero” out absorbance readings and account for any variations due to the CelLytic MT solution.
9. To this same tube, add 30uL of your protein extract.
10. Invert the tube several times and then incubate at RT for 10mins.
11. Mix the tube several times and transfer 1mL to a plastic, disposable cuvette.
12. Measure the absorbance at 595nm and record the value.
P1 = 1.542 OD
P2 = 1.571 OD
(OD = optical density = absorbance)
13. Remove the cuvette from the spectrophotometer. Using a P1000 set to 1mL, carefully pipette the solution in the cuvette up and down a couple of times to mix.
14. Measure the absorbance at 595nm and record the value.
P1 = 1.541 OD
P2 = 1.568 OD
15. Repeat steps 13 and 14.
Did not perform b/c values were almost identical
16. Average the three absorbance values you recorded.
P1 Avg = 1.5415 OD
P2 Avg = 1.5695 OD
Note: Coomassie Blue dye in Bradford reagent absorbs at 595nm and the absorbance can be directly correlated to a specific amount of protein present in the sample when compared to a standard curve (already determined for us).
Standard curve was generated as per Manufacturers Instructions.
Pierce: Coomassie (Bradford) Protein Assay Kit
17. Plug your average absorbance that you just calculated into the following equation y=1011.9x to determine the concentration of protein in your sample. NOTE: It appears all of our protein values were outside of the curve. Need to dilute an aliquot of samples and run again.
18. Write the concentration on your tube (forgot to do this) and place tube in TA ice bucket. Sample stored at -80C.
4. Design / order Primers to use in Lab 3
1/12/09 Ran a tblastn with all 4 genes in est_others [database] against Haliotis (taxid:6452) [organism]
peptidoglycan recognition protein [Argopecten irradians]AAR92030
peptidoglycan recognition protein S1 precursor [Chlamys farreri]AAY53765
nuclear factor interleukin 3 regulated-like protein [Haliotis discus discus]ACJ65688
interleukin 17 [Crassostrea gigas]ABO93467
EST match for PRG EB531075 (Evalue - 6e-21)
1/14/09 Last minute found something I think may be cool
lipopolysaccharide- and beta-1,3-glucan-binding protein [Chlamys
farreri]
AAP82240EST match for LGBP CX726071 (Evalue 2e-80)
1/13/09 Primer3 Output (PRG)
PRIMER PICKING RESULTS FOR gi|184186070|gb|EB531075.1|EB531075 aa344 Haliotis discus Hannailno Liver and Kidney cDNA library Haliotis discus hannai cDNA clone aa344 5', mRNA sequence
No mispriming library specified
Using 1-based sequence positions
OLIGO [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_START|start]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_LEN| len]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_TM| tm]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_GC| gc%]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_ANY| any]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_REPEAT| 3']] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_OLIGO_SEQ|seq]]
LEFT PRIMER 176 20 59.84 60.00 3.00 0.00 GGTCACAGCCTCAACTCTCC
RIGHT PRIMER 441 20 60.97 55.00 4.00 1.00 ACCCCCATCTTACCGATGAC
SEQUENCE SIZE: 681
INCLUDED REGION SIZE: 681
PRODUCT SIZE: 266, PAIR ANY COMPL: 4.00, PAIR 3' COMPL: 0.00
1 CTGGCAGTAAGGTAAAGTTTTGAAGGATCCTGCGTATATCTATCTTGTCATATATATTTT
61 CTGAAAATAAGCAAGGATGAAGACATTTTTCGGATGCCTACTGTTGGTATTAATGGCACC
121 ACGCTCTGAAGCGGCAGATTGCGCATGCGCCACATATGGCTTGCACGTGCGTTCCGGTCA
>>>>>
181 CAGCCTCAACTCTCCTATCATCGGAACCATGACAAACGGTCAATGCGTGACCTTCAAGGG
>>>>>>>>>>>>>>>
241 TGACCGGCAGGTGGCCGATGGCTACACCTGGGCGCACGTCGACTACAACGGAAAGGATGG
301 TTATGCCGCGGTCAACTGGCTCAACATTCATCCATGTGGGGCTCATAATAACGTTCTTCA
361 GCTGAGCGGCTGCCCGCACATCATCACACGTGCTGAATGGGGTGCACGCGCACCTAAATA
421 CGTCATCGGTAAGATGGGGGTCACTCCGAAGTATGTGTTCGTTCACCACGGTGCCACCGC
<<<<<<<<<<<<<<<<<<<<
481 CCCATGTTATAACGAGGCTGCTTGCAAGGCCGAGGTCCTGTCTTATCAGAAGTATCACAT
541 GGACACGCACGGTTGGCCCGACATTGGCTACAGCTTCGTAGTCGGCGAAGACGGTCACGC
601 CTACGAAGCAAGAGGATGGGACACCATCGGCGCCCATACCTACGGCTACAACAGCGTCGG
661 ACTCGGTATCTGCGTTATTGG
KEYS (in order of precedence):
>>>>>> left primer
<<<<<< right primer
ADDITIONAL OLIGOS
[[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_START|start]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_LEN| len]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_TM| tm]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_GC| gc%]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_ANY| any]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_REPEAT| 3']] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_OLIGO_SEQ|seq]]
1 LEFT PRIMER 176 20 59.84 60.00 3.00 0.00 GGTCACAGCCTCAACTCTCC
RIGHT PRIMER 440 19 60.14 57.89 4.00 1.00 CCCCCATCTTACCGATGAC
PRODUCT SIZE: 265, PAIR ANY COMPL: 4.00, PAIR 3' COMPL: 0.00
2 LEFT PRIMER 425 19 60.14 52.63 3.00 1.00 ATCGGTAAGATGGGGGTCA
RIGHT PRIMER 657 20 59.83 55.00 3.00 0.00 ACGCTGTTGTAGCCGTAGGT
PRODUCT SIZE: 233, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 1.00
3 LEFT PRIMER 427 19 59.79 57.89 3.00 1.00 CGGTAAGATGGGGGTCACT
RIGHT PRIMER 657 20 59.83 55.00 3.00 0.00 ACGCTGTTGTAGCCGTAGGT
PRODUCT SIZE: 231, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 1.00
4 LEFT PRIMER 176 20 59.84 60.00 3.00 0.00 GGTCACAGCCTCAACTCTCC
RIGHT PRIMER 308 19 60.28 52.63 2.00 0.00 CGGCATAACCATCCTTTCC
PRODUCT SIZE: 133, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 0.00
Statistics
con too in in no tm tm high high high
sid many tar excl bad GC too too any 3' poly end
ered Ns get reg GC% clamp low high compl compl X stab ok
Left 5161 0 0 0 107 0 1127 3294 124 36 0 85 388
Right 5084 0 0 0 1 0 737 3785 11 16 0 97 437
Pair Stats:
considered 1040, unacceptable product size 1023, high any compl 4, high end compl 2, ok 11
primer3 release 1.1.0
Choose primer set 4 for PRG and submitted order 1/14/09 Primer3 Output (LGBP)
PRIMER PICKING RESULTS FOR gi|82857336|gb|CX726071.1|CX726071 DGT151 Haliotis discus cDNA library (DGT) Haliotis discus cDNA, mRNA sequence
No mispriming library specified
Using 1-based sequence positions
OLIGO [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_START|start]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_LEN| len]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_TM| tm]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_GC| gc%]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_ANY| any]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_REPEAT| 3']] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_OLIGO_SEQ|seq]]
LEFT PRIMER 119 20 59.56 55.00 2.00 0.00 GGCGTCCTCTACATCAAACC
RIGHT PRIMER 354 20 59.53 45.00 4.00 0.00 TGAGCCGATATTTCCACCTT
SEQUENCE SIZE: 632
INCLUDED REGION SIZE: 632
PRODUCT SIZE: 236, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 0.00
1 CGACAATTTCGACACACTGGACTTCAAAGTGTGGGAACACGAGCTGACGGCTGGTGGGGG
61 AGGCAACTGGGAGTTTCAGTTCTACACCAACAACCGCACCAACACCTACGTCCGCGATGG
>>
121 CGTCCTCTACATCAAACCCACGTTGACGGTGGACCAGTTTGGCGAGGCCTTCCTCACATC
>>>>>>>>>>>>>>>>>>
181 CGGAAAACTTGAACTGTGGGGCGCAGGACCGCACGACACCTGTACCGGCAACGCCTTCTA
241 CGGCTGTGAGCGCGTCGGCAACCATCAGTACATCATCAACCCCATTCAGTCGGCCCGACT
301 CCGGAGCTCCAGGGGTCTGAACTTTAAATACGGCAAGGTGGAAATATCGGCTCAACTCCC
<<<<<<<<<<<<<<<<<<<<
361 CAAGGGAGACTGGTTGTGGCCCGCCATATGGATGCTGCCGACGTACACGGAGTATGGCGG
421 TTGGCCGGCGTCCGGCGAGATTGACATCATGGAGAGCAGAGGTAACCGACACTACTACGA
481 CGCGAATGGCCGTTCTGTAGGAGTCGACTCTTACGGCAGTACCATTCACTTCGGCACCGA
541 CTACTTCCACAACGGCTGGTCACGTGCCCACCAGTCCTGGGTCAAAGAGAACGGAACTTA
601 CGGAGATGAGTTTCATACGTACGGAGTCGAGT
KEYS (in order of precedence):
>>>>>> left primer
<<<<<< right primer
ADDITIONAL OLIGOS
[[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_START|start]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_LEN| len]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_TM| tm]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_GC| gc%]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_ANY| any]] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_REPEAT| 3']] [[http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_results_help.cgi#PRIMER_OLIGO_SEQ|seq]]
1 LEFT PRIMER 119 20 59.56 55.00 2.00 0.00 GGCGTCCTCTACATCAAACC
RIGHT PRIMER 355 20 59.53 45.00 4.00 0.00 TTGAGCCGATATTTCCACCT
PRODUCT SIZE: 237, PAIR ANY COMPL: 4.00, PAIR 3' COMPL: 0.00
2 LEFT PRIMER 119 20 59.56 55.00 2.00 0.00 GGCGTCCTCTACATCAAACC
RIGHT PRIMER 353 20 59.53 50.00 4.00 1.00 GAGCCGATATTTCCACCTTG
PRODUCT SIZE: 235, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 1.00
3 LEFT PRIMER 336 20 59.53 45.00 4.00 0.00 AGGTGGAAATATCGGCTCAA
RIGHT PRIMER 489 20 59.37 55.00 4.00 0.00 CCATTCGCGTCGTAGTAGTG
PRODUCT SIZE: 154, PAIR ANY COMPL: 3.00, PAIR 3' COMPL: 1.00
4 LEFT PRIMER 23 20 60.13 45.00 4.00 0.00 TTCAAAGTGTGGGAACACGA
RIGHT PRIMER 292 19 59.90 52.63 2.00 0.00 CGACTGAATGGGGTTGATG
PRODUCT SIZE: 270, PAIR ANY COMPL: 4.00, PAIR 3' COMPL: 1.00
Statistics
con too in in no tm tm high high high
sid many tar excl bad GC too too any 3' poly end
ered Ns get reg GC% clamp low high compl compl X stab ok
Left 4653 0 0 0 28 0 680 3374 129 52 0 87 303
Right 4607 0 0 0 28 0 707 3289 17 10 0 93 463
Pair Stats:
considered 662, unacceptable product size 578, high any compl 65, high end compl 3, ok 16
primer3 release 1.1.0
Chose initial primer set for LGBP and submitted order1/14/09 Lab 2 - Tissue Extraction II
1. re-spec dilution of protein sample
Set up 3 protein dilutions; 1:1, 1:3, 1:5 for both protein samples and re-spec'dP1-1:1 = 1.112 OD
P1-1:3 = 0.729 OD
P1-1:5 = 0.570 OD
1:5 dilution within range of our standard curve (y = 1011.9x)
P2-1:5 = 0.654 OD
P1y = 576.78
P2y = 661.78
Back calculation from 1:5 dilution (6 parts total; multiply # by 6)
P1 = 3,460.70ug/ml = 3.461ug/ul
P2 = 3,970.70ug/ml = 3.971ug/ul
2. Continue with RNA extraction protocol
-----Samples frozen at -80Turn on heating block to 55C.
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube. NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
17. Incubate at RT for 10 mins. Incubated for 15mins
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer (will do lab3)
3. Run SDS-PAGE protein gel
PROTEIN GEL PROTOCOL - See also Manufacturers Protocol / Manual: Precise™ Protein Gels1. Begin boiling water on hot plate.
2. Thaw you protein extract from last week. Mix well by inverting tube several times.
3. In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein sample and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
5. Boil sample for 5 mins.
6. While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly as demonstrated.
7. When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
8. Slowly load your entire sample into the appropriate well using a gel loading tip.
9. Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
10. Turn power supply on and set voltage to 150V. Run for 45mins.
11. Add ~150mL (does not have to be measured - just need enough to cover the gel) of Coomassie Stain to a designated container.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid from gel box.
13. Disengage the tension wedge.
14. Remove gel from gel box.
15. Carefully crack open cassette to expose gel.
16. Trim wells at top of gel.
17. Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
18. Place gel into container with Coomassie Stain.
19. Incubate on shaker/rocker for 5 mins.
20. Carefully pour stain back into original container. Be careful not to dump out gel!
21. Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
22. Add ~250mL (no need to measure) 10% acetic acid to container with gel. Incubate on shaker/rockers for 15mins. Change out buffer and repeat until bands become clearly visible. This may need to incubate O/N. If so, cover container with plastic wrap and leave on shaker/rocker.
Lane ID
1. ladder (10uL)
2. blank
3. Brianna
4. Brianna
5. blank
6. Lisa (30uL = 15uL protein + 15uL 2X Reducing Sample Buffer)
7. Lisa (30uL = 15uL protein + 15uL 2X Reducing Sample Buffer)
8. Blank
9. Mac
10. Mac
11. blank
12. Ladder (10uL)
Figure 1: Brianna/Lisa/Mac FRONT; P1 sample [3.461ug/ul] in duplicate - 15uL protein sample/well = 51.915ug/well
- When cutting the corner on the FRONT GEL we accidentally cut it off at lane 12 instead of lane 1. So the FRONT GEL lane 1 is on the far right. Regardless, my samples are still the middle lanes.
- My bands for both P1 and P2 look alot darker than Brianna (ab gill?) and Mac's (oyster gill?). Perhaps due to the dark green pigment of my sample since using digestive gland (possible inhibitors?). I do notice some dark banding representing Myosin and Phosphoraylase B.
Figure 2: Brianna/Lisa/Mac BACK; P2 sample [3.971ug/ul] in duplicate - 15uL protein sample/well = 59.565ug/well
Invitrogen Ladder

1/21/09 Lab 3 Reverse Transcription and PCR
1. Quantify RNA
-Used nanodrop technology (sweetness!); calibrated machine with H2O, placed 1uL of RNA extract on platform.
R1 = 2,523.2ng/ul
R2 = 1,603.0ng/ul
2. Reverse Transcribe RNA to complementary DNA
REVERSE TRANSCRIPTION PROTOCOL
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
-Add negative control here.
-Because we had enough sample to spare and our [RNA] was within the 25ug range we just used 5ul of RNA (no water was added).
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
*Steps 3 & 4 were skipped by accident…OOPS!
5. Make Master Mix
PER RXN
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
-Made enough for 3 rxns but should have made extra, was short a few ul’s for the negative control
Add 15ul MM to tube with diluted RNA in it (total volume now 20 ul)
Vortex
Spot spin
Incubate at RT for 10 min
Incubate at 37C for 1 hr in thermocycler
Heat inactivate @ 95C for 3 min
Spot spin
Leave on ice or store at –20C
3. Perform PCR
Run each template in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| GoTaq®Green Master Mix, 2X |
25 |
1x |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| DNA template |
1–5μl |
<250ng |
25ul Taq
1ul Forward
1ul Reverse
21 H20
2ul Template
Made enough MM for 9 rxns
Load reactions into thermocycler. Overnight at 4C
1/22/09
Made 1.5% Agarose Gel
Ran out PCR products on agarose gels and photographed
-Used 100bp invitrogen ladder (7ul)
-Added 15ul product/well
-Ran 115V for 60min
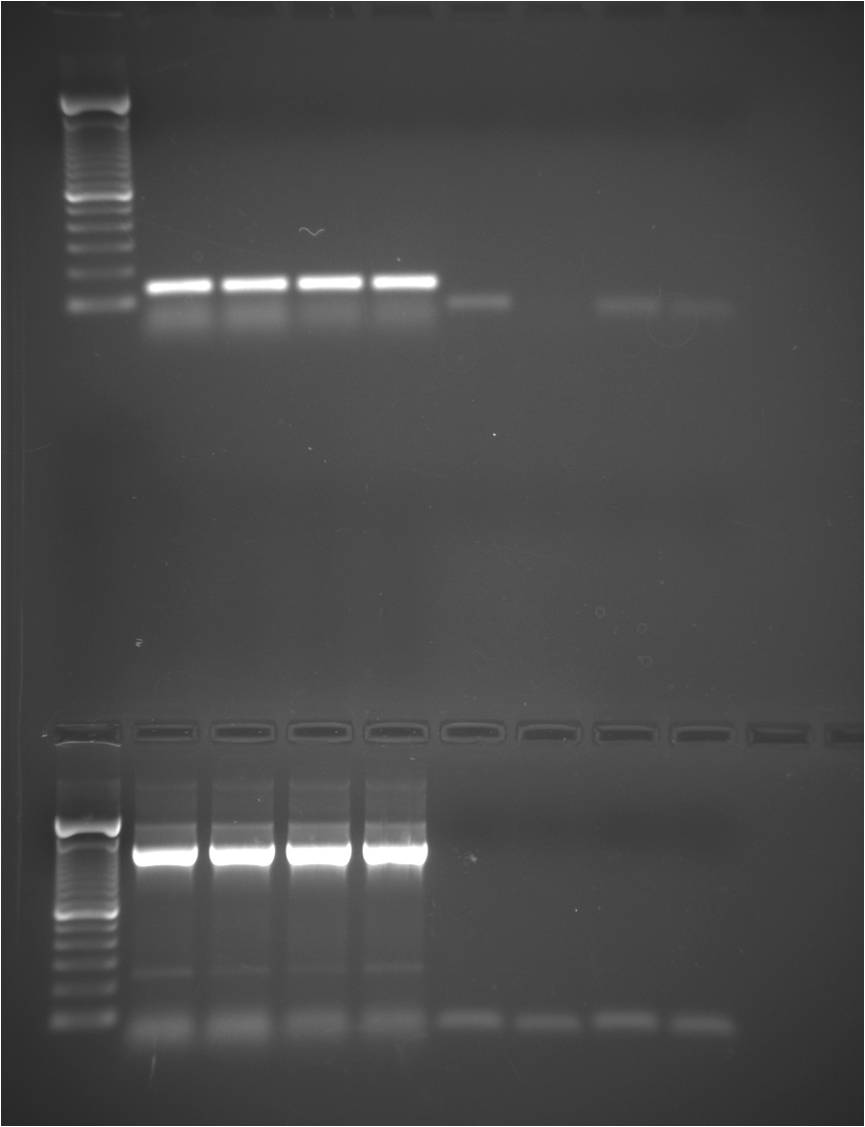
TOP: PRP primers (peptidoglycan recognition protein)
Lane 1: 100bp ladder
Lane 2: P1
Lane 3: P1
Lane 4: P2
Lane 5: P2
Lane 6: reverse transcription negative control
Lane 7: reverse transcription negative control
Lane 8: PCR negative control
Lane 9: PCR negative control
- Product coming up between 100 and 200bp but should be amplifying a 266bp product. Sequence was based off Atlantic bay scallop, may be different size in abalone. Negative controls look clean. Primer dimer issues. However, no dimer in my duplicate reverse transcription negative control. Weird!
- Will excise and sequence my product.
BOTTOM: LGBP primers (lipopolysaccharide- and beta-1,3-glucan-binding protein)
Lane 1: 100bp ladder
Lane 2: L1
Lane 3: L1
Lane 4: L2
Lane 5: L2
Lane 6: reverse transcription negative control
Lane 7: reverse transcription negative control
Lane 8: PCR negative control
Lane 9: PCR negative control
- Super bright big band ~1,000-1,500bp. What could it be? genomic DNA carryover? (solutions: DNAse; design new primers that contain sequence half before and half after introns - check out homologs in sea urchin)
- Faint band coming up between 200-300bp and my product is 236bp so that’s cool. Negative controls look clean. Still primer dimer issues. Might run gel again and excise/clean-up smaller band for sequencing.
Both thermocycler conditions should be optimized for each specific primer set.
1/28/09 Lab4 - Western Transfer - Immunoblots to look for HSP70
1. Run out protein samples as done previously (as in lab 2)
Set up and run by Mac prior to lab2. Transfer proteins from gel to nitrocellulose membrane Western Blotting Protocol
Western Breeze Manufacturer's ProtocolGeneral Guidelines
• Avoid touching the working surface of the membrane, even with gloves.
• Work quickly when changing solutions as membranes dry quickly. If the membrane dries, re-wet the membrane with methanol and rinse with water before proceeding.
• Add solutions to the trays slowly, at the membrane edge, to avoid bubbles forming under the membrane. Decant from the same corner of the dish to ensure complete removal of previous solutions.
1. Cool the transfer buffer to 4°C.
2. Soak the filter paper, membrane and gel in Transfer Buffer for 15 minutes.
3. Assemble the blotting sandwich in a semi-dry blotting apparatus (transfer cell- biorad) as follows:
• Anode (+++)
• 2 thick pieces of Filter paper - roll filter papers from middle to edge to remove bubbles and excess buffer.
• Nitrocellulose Membrane -DO NOT TOUCH!
• Gel - roll gel wet with buffer
• Filter paper - roll again
• Cathode (– – –)
4. Transfer the blot for 30 minutes at 20V.
5. Remove the gel from the sandwich and rinse with transfer buffer.
6. Use a cotton swab to remove any adhering gel from the membrane.
3. Probe membrane with antibody (HSP)
1. Prepare 20 mL of Blocking Solution
Ultra filtered Water 14 ml
Blocker/Diluent (Part A) 4 ml
Blocker/Diluent (Part B) 2 ml
Total Volume 20 ml
2. Place the membrane in 10 ml of the appropriate Blocking Solution in a covered, plastic dish provided in the kit. Incubate for 30 minutes on a rotary shaker set at 1 revolution/sec.
3. Decant the Blocking Solution.
4. Rinse the membrane with 20 ml of water for 5 minutes, then decant. Repeat once.
5. Prepare 10 mL of Primary Antibody Solution (1:3000 dilution)
Blocking Solution 10 ml
HSP 70 antibody 3.3 µl
Total Volume 10 ml
6. Incubate the membrane with 10 ml of Primary Antibody Solution for OVERNIGHT
1/29/09
NEXT DAY
7. Prepare Antibody Wash
Ultra filtered Water 150 ml
Antibody Wash Solution (16X) 10 ml
Total Volume 160 ml
8. Decant Primary Antibody Solution (SAVE- can be re-used)
9. Wash the membrane for 5 minutes with 20 ml of prepared Antibody Wash, then decant. Repeat 3 times.
10. Incubate the membrane in 10 ml of Secondary Antibody Solution for 30 minutes, then decant.
11. Wash the membrane for 5 minutes with 20 ml of Antibody Wash, then decant. Repeat 3 times.
12. Rinse the membrane with 20 ml of water for 2 minutes, then decant. Repeat twice.
13. Incubate the membrane in 5 ml of Chromogenic Substrate until purple bands develop on the membrane. Development is complete in 1 to 60 minutes.
14. Take a digital picture of the membrane
15. Rinse the membrane with 20 ml of water for 2 minutes. Repeat twice
16. Dry the membrane on a clean piece of filter paper to open air
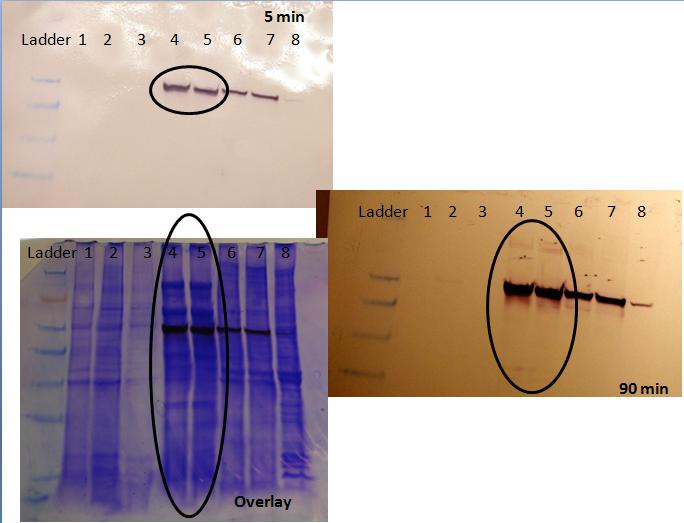
- We took 3 images of our gel: 5 minutes after development, 1.5 hours after development and the "overlay", which is a picture of the membrane overlaid with the coomassie gel (lined up using the markers).
2/4/09 Lab 5 QPCR using Syto-13
- Syto-13 = non specific DNA binding dye fluoresces when bound to any double stranded DNA.
- increase [DNA], increase binding sites, increase fluorescence.
- Rxn specificity determined soley by primers
Each template run in duplicate AND 2 negative controls for each primer set.
1. Prepare master mix
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| Master Mix, 2X (Immomix) |
25µL |
1x |
| Syto-13 dye (50uM) |
2-5µL |
2 - 5µM |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| Ultra Pure Water |
to 48uL |
NA |
Immomix - 25uL
F - 2uL
R - 2uL
Syto - 2uL
H20 - 17uL
2. Add mastermix to wells of a white PCR plate
3. Thaw cDNA samples.
4. Add 2uL cDNA template to each reaction.
5. Add 2uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. If necessary, spin the strips to collect volume in the bottom of the wells.
8. and ensure the lids are clean (wipe with a Kim Wipe) before going into the Opticon.
9. Load the plate, verify the PCR conditions and start the run.
Question of the Day: How can you tell if you have gDNA carryover?
Answer: Run a PCR on your RNA sample. If you get a product, you have gDNA carryover. RNA won't work on PCR bc it's unstable (will denature) and you use DNA polymerase in PCR.
QPCR Results
Row Ct SampleC1 N/A L neg
C2 N/A L neg
C3 30.2 L2
C4 33.01 L2
C5 26.12 L1
C6 25.26 L1
C7 N/A P neg
C8 N/A P neg
C9 27.07 P2
C10 19.64 P2
C11 18.85 P1
C12 19.62 P1
- For both primer sets it looks like sample 2 has the greatest discrepancy among duplicate runs.
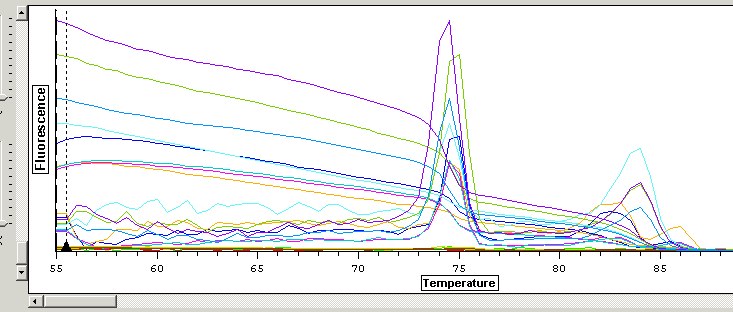
- QPCR Melt curve (dissociation) analysis - amount of fluorescence vs temp - used to differentiate btwn specific anf non specific amplicons based on melting temp. My results show 2 peaks - this is bad news bc it means I am amplifying more than 1 product. Think about increasing annealing temp (increases specificity).
2/11/09 507 project outline.doc
2/12/09 Blk Ab Gill RNA extraction Blk Ab Resist project #2_GILL_RNAlater.xls
2/13/09 Decided to switch sample set and work on Blk Ab Resistance Project #1 using DG samples stored in LN2 (still same project concept)
2/18/09 Gathered ~80 DG samples and moved to Roberts Lab
Google Doc - Abalone Exp1 DG
2/23/09 Extracted RNA from 5 samples using
RNA PowerSoil™ Total RNA Isolation Kit
Note:Sample 08:3-10 @ Step 8 had milky aqueous phase
Step 11 - no visible pellets
Step 12 - used 4mL of SR4 instead of 5mL
2/24/09 RNA Quant on 5 DG samples via Nanodrop (data on google doc spreadsheet)
- DNase Treatment of 5 DG samples
Step 1 Recipe - 1ul DNase, 5ul buffer, 44ul RNA
Step 3 - 5ul DNase inact reagent
2/27/09 Crosson_507ResearchProposal.doc
3/2/09 RT 5 DG samples AMV RTranscriptase - used 5ul DNased RNA
Recipe for 7 rxns
28ul buffer
56ul dntp
7ul AMV RT
7ul dT primer
7ul H2O
total volume/cDNA sample = 20ul
- Received primers for genes of interest (rab7, tollip, b-glucan, MnSD, PGRP, PGRP2, actin)
- DNased 07:12 DG samples (n=19; missing 07-12-19)
Step 1 Recipe - 1ul DNase, 1.5ul buffer, 12.5ul RNA
Step 3 - 2ul DNase inact reagent
total volume/ RNA sample after DNase = 10ul
3/3/09 RT 07:12 samples - used 4ul DNased RNA
Recipe for 22 rxns
88ul buffer
176ul dntp
22ul AMV RT
22ul dT primer
44ul H2O
total volume/cDNA sample = 20ul
3/4/09 Sybr Green QPCR (Sensimix) testing all primer sets (n=7 from Roberts lab + n=1 of PGRP I designed) on 2 samples (06:5-31 SNI Exp Mort and 06:6-43 Carmel Exp Mort) - used only 1ul of template cDNA/rxn to preserve sample
Recipe (per 25ul rxn)
12.5ul sensimix
0.8ul F
0.8ul R
0.5ul Sybr
9.4ul H20
Thermal Profile
95 - 7min
50 cycles of
95 - 10s
60 - 30s
72 - 30
QPCR gene expression data 1st run.ppt
- After looking at the results I will use the following working primer sets to run the rest of my samples for the project: actin, tollip, rab7, and pgrp (lisa)
- Other primer sets should be re-designed/optimized