I ran my arbitrary values for each of me test runs using this formula:
Arbitrary expression value =10^(-(0.3012*Ct)+11.434)
| Sample |
216.77% |
17.96 |
1057908 |
Ec01 |
|
| Sample |
136.47% |
19.54 |
353632 |
E11 |
|
| Sample |
493.93% |
22.99 |
32316 |
E12 |
|
| Sample |
189.81% |
21.23 |
109528 |
E13 |
|
| Sample |
90.16% |
18.73 |
620189 |
Ec31 |
|
| Sample |
146.45% |
20.63 |
166052 |
Ec02 |
|
| Sample |
145.75% |
20.73 |
154926 |
E21 |
|
| Sample |
178.46% |
22.19 |
56282 |
E22 |
|
| Sample |
N/A |
N/A |
Blank |
||
| Sample |
149.27% |
18.38 |
790576.7 |
E23 |
|
| Sample |
126.74% |
24.08 |
15174.14 |
EC23 |
|
| Sample |
37.36% |
34.98 |
Blank |
November 25th, 2013
Here were my results for qPCR: on
| 11_22_13 |
Ecoli Control 1 Hour 0:
| H1 |
SBGnew |
Sample |
216.77% |
17.96 |
| H2 |
SBGnew |
Sample |
136.47% |
19.54 |
| H3 |
SBGnew |
Sample |
493.93% |
22.99 |
| H4 |
SBGnew |
Sample |
189.81% |
21.23 |
| H5 |
SBGnew |
Sample |
90.16% |
18.73 |
| H6 |
SBGnew |
Sample |
146.45% |
20.63 |
| H7 |
SBGnew |
Sample |
145.75% |
20.73 |
| H8 |
SBGnew |
Sample |
178.46% |
22.19 |
| H10 |
SBGnew |
Sample |
N/A |
N/A |
| 11_19_13 |
Ecoli Treatment 2 Hour 3:
| H1 |
SBGnew |
Sample |
149.27% |
18.38 |
| H2 |
SBGnew |
Sample |
126.74% |
24.08 |
| H3 |
SBGnew |
Sample |
37.36% |
34.98 |
I ran qPCR for the remaining 8 tubes this morning + 1 blank
A master mix was made, combining 500uL of SsoFast EvaGreen Supermix, 20uL of 10x diluted upstream DNMT primer 5' GCAACTGAAGCGTCAGCAAA 3' , 20uL 10x diluted downstream DNMT primer 5'CCGCATAACGCAACTGATGG and 420uL of ultra pure water. 24uL of the master mix was added to 9 white PCR wells. 1uL of the cDNA was placed in 9 reaction wells, 1uL of ultra pure water was added to the blank well. Clean lids were placed on the tubes, then loaded into the PCR machine for the following conditions: 95°C for 10 minutes, 95°C for 15s, 55 °C for 15 s, 72°C for 15 s (+ plate read). The Procedure was repeated from step two then repeated 39 more times. Finally the PCR machine tempered at 95°C for 10s, then processed the melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read).
November 20th, 2013
The 2 tubes + blank we ran were successful. we will prepare to run the remainder of the tubes on Friday the 22nd.
November 19th, 2013
We ran 3 tubes for qPCR for our bacteria as a test for expression.
To begin the PCR process, 5ul of RNA, 0.5 uL of random primers for prokaryotes and 12.25uL of nucleate free H20 were added to a sterile PCR tube for each treatment. The mixture was left to incubate for 5 minutes at 70C in a thermocycler then transferred to ice. A combination of 5ul M-MLV 5X, 1.25uL of dNTPs, 0.5 uL of M-MLV RT and 0.5uL of nuclease free H20 was added to each PCR tube. The mixture was incubated for 60 minutes at 37C then at 70C for 3 minutes on the thermocycler. The sample was spun on a centerfuge then stored on ice at -20C.
qPCR
A master mix was made, combining 50uL of SsoFast EvaGreen Supermix, 2uL of 10x diluted upstream DNMT primer 5' GCAACTGAAGCGTCAGCAAA 3' , 2uL
10x diluted downstream DNMT primer 5'CCGCATAACGCAACTGATGG and 42uL of ultra pure water. 24uL of the master mix was added to 4 white PCR wells. 1uL of the cDNA was placed in 2 reaction wells, 1uL of ultra pure water was added to the blank wells. Clean lids were placed on the tubes, then loaded into the PCR machine for the following conditions: 95°C for 10 minutes, 95°C for 15s, 55 °C for 15 s, 72°C for 15 s (+ plate read). The Procedure was repeated from step two then repeated 39 more times. Finally the PCR machine tempered at 95°C for 10s, then processed the melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read).
November 14th, 2013
The RNA was defrosted then quantified with a nanodrop spectrophotometer to test the concentration of the samples and calculated (RNA)= 40uL/mL x A360 x dilution factor.
EC1
November 12th, 2013
E. coli UV treatment
RNA ISOLATION
500uL of TriReagent was added to Snap cap tubes containing the 4 control and 6 treatment groups of E coli. The bacteria was homogenized using a pestle and vortexed for 5 seconds. An additional 500uL of TriReagent was added then vortexed for 15 seconds. 200uL of chloroform was added then the samples were vortexed vigorously for 30 seconds. The tubes was then left to incubate for 5 minutes. In a refrigerated microfuge, the samples were placed to spin on 14,000g for 15 minutes. The aqueous layer was pippeted of and placed into a sterile snapcap tube. 500 uL of isopropanol was added, then inverted until fully mixed. The RNA was left to incubate at room temperature for 10 minutes, then centerfuged a 14,000 g for 8 minutes. The supernatant was then pipetted off the RNA pellets and 1mL of 75% EtOH was added. The tubes were vortexed briefly then spun in the refrigerated microfuge at 7500 g for 5 minutes. Using a small pipette tip, the remaining supernatant was removed from the RNA pellet. The cap was left open for five minutes to let the pellet dry, then re-suspended in 100uL of 0.1 DEPC-H20. The mixture was then incubated at 55C for 5 minutes in a hot water bath and then placed in -80C for storage .
November 7th, 2013
E. coli UV treatment
The control group was wrapped in tinfoil and two control replicates were taken from the LB medium at time zero. The treatment was placed into a hood and the UV light was turned on. Treatment replicates were sampled at 1,2 and 3 hours from the LB medium, for a total of 6 samples. A final control sample was taken at the 3 hour time point. Each sample was placed into the vortex for 1 minute at 10,000 rpm to pellet, then the supernatant was removed. Pelleted samples were then transferred to storage at -80C for future RNA extraction.
November 6th, 2013
To grow the bacteria, swipes from each plates: LB 10/28/13 SWJ E. coli, and LB/./. 10/28/13 SJW for Vt RE22 were added to two sterilized beakers/bacteria, each containing 50mL of LB broth. The 4 beakers were placed into the incubator for 16 hours at 37C. The stir-plate was set to 75 revolutions/minute. The beakers were transfered to 4C for storage.
November 5th, 2013
Due to the large number of samples we will be processing for heat and UV stress (36) during our experiment, we have decided to perform a comparative response among our two bacteria types: Escherichia coli and Vibrio tubiashii. This will decrease our samples to a more manageable amount (18). We will now subject both Vt and E. coli to UV stress to compare their responses. I will be examining expression of DNMT in Vt and Dam in E. coli, by taking samples from our LB broths injected with the bacterial strains. On the off chance that one bacteria does not respond, we will introduce heat stress as an additional factor.
November 4th, 2013
To assemble the LB Broth, 5g NaCl, 5 g Bactotryptone, and 2.5 g Yeast Extract were added to a sterilized container. The 500mL of D/W was added then placed on a stir plate to mix. 500mL additional D/W was then added to make 1L of broth for E. coli. An additional bottle was made with the previous ingredients and supplimented with with 5g of sodium chloride. The LB broths were labeled and placed in the Autoclave for sterilization.
October 29th, 2013
Lab 5: Project Finalization
- Summary of the lab
- Materials and Methods
- Results
| MR_EscherichiaColi_DAM_U00096.3F |
GCAACTGAAGCGTCAGCAAA |
| MR_EscherichiaColi_DAM_U00096.3R |
CCGCATAACGCAACTGATGG |
the primers ordered for Vt are:
| MR_VibrioTubiashii_DNMT_19109 VITU9109_1F |
TTTGCTCTGGAAAACGCTGC |
| MR_VibrioTubiashii _DNMT_19109 VITU9109_1R |
GATTGCTAAGCGCCTCAACG |
October 22nd, 2013
Lab 4: protein SDS/PAGE and Western blot, analyze conventional PCR (via agarose gel) & qPCR data
- Summary of the lab
- Materials and Methods
The gel was loaded into the box and filled with 1x TAE buffer to cover the wells. The comb was removed then 7uL of ladder was placed in the far left column. 20uL of the PCR sample was loaded into lane #3, then the gel was run for 100V at 1hr. The gel was then visualized on a UV transilluminator.
SDS-PAGE
In a screw-cap, 15ul of previously extracted protein stock was added to 15uL of 2X reducing sample buffer. The sample was mixed then boiled for 5 minutes. The gel box was assembled and the entire sample was loaded into well #3. The electrophoresis box was run at 150V for 45 minutes. The gels were removed and trimmed to prepare for the Western Blotting protocol
WESTERNBREEZE CHROMOGENIC WESTERN BLOT IMMUNODETECTION
Filter paper, nitrocellulose membrane and gel were soaked in Tris-Glycine Transfer Buffer for 15 minutes on a shaker. The blotting sandwich was assembled in the blotting apparatus as anode (+++), filter paper, membrane, gel filter paper, cathode (---). The blot was transferred for 30 minutes at 20V, the gel removed then rinsed with transfer buffer. After placing the membrane in a plastic box, 10mL of blocking solution was added and incubated for 30 minutes on the rotary shaker at 1 rev/sec. The liquid was decanted, rinsed with 20mL of water for 5 minutes then decanted. This was repeated once more then the membrane was incubated in 10mL of primary antibody solution. After decanting the antibody solution, the membrane was rinsed with 20mL antibody wash for 5 minutes, decanted and repeated 3 times. The membrane was incubated in 10mL secondary antibody solution for 30 minutes and decanted. The membrane was washed for 5 minutes in 20mL of antibody wash, decanted and repeated 3 times. 20mL of pure water was added to wash the membrane for 2 minutes, decanted and repeated twice. The membrane was incubated in 5mL of chromogenic substrate until a purple band appeared, between 1-60 minutes after the substrate was added. The membrane was dried on filter paper and exposed to the air.
- Results
Pacific oyster mantle tissue was isolated and qPCR performed. The results for all wells are as follows:
| F9 |
SBGnew |
Sample |
91.43% |
31.26 |
|
| F10 |
SBGnew |
Sample |
150.11% |
34.26 |
|
| F11 |
SBGnew |
Sample |
220.21% |
27.15 |
|
| F12 |
SBGnew |
Sample |
76.40% |
26.5 |
Arbitrary expression value =10^(-(0.3012*Ct)+11.434)
F9 =10^(-(0.3012*31.26)+11.434)= 104.35
F10=10^(-(0.3012*34.26)+11.434)=13.03
F11=10^(-(0.3012*27.15)+11.434)=1804.76
F12=10^(-(0.3012*26.50)+11.434)=2832.70
PCR STRAND LENGTH ELECTROPHORESIS PROCEDURE
The stand length was run in lane #3 from cDNA combined in test well F11. The strand lengths were equally distributed in the 300 base pair and 200 base pair ladder zones.
SDS-PAGE
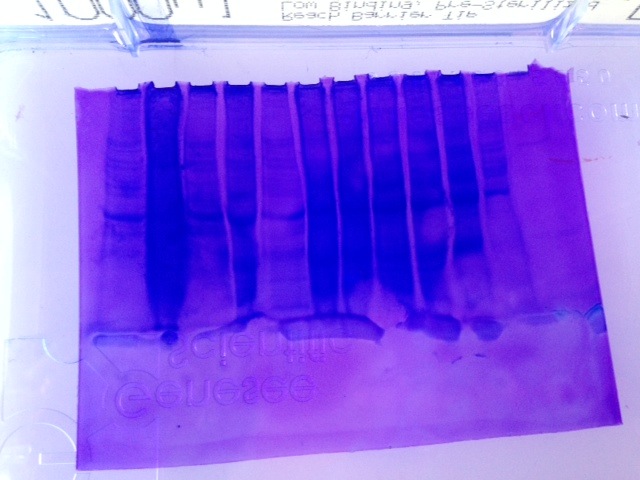
Figure 1. SDS-PAGE gel results. The ladder is the first lane on the left. Lane 3 was run with tissues extracted from pacific oyster mantle and DNMT primer.
Figure 2. Western blot from SDS-PAGE gel (Figure 1). The ladder is the first lane on the right hand side. Lane 3 was run with tissues extracted from pacific oyster mantle, and amplified with DNMT primer. The band indicates the presence of HSP70 proteins.
- Conclusions
REALTIME PCR
The results show that my samples were contaminated. Both the control and test tubes amplified cDNA as a result of cross contamination. It is also possible that the primer unsuccessfully laid down in the test group strands and amplified two regions.PCR STRAND LENGTH ELECTROPHORESIS PROCEDURE
Due to cross contamination demonstrated in the qPCR data, two strands of differing lengths were amplified. This can be seen from the amount of material concentrated in the 200bp and 300bp segments, demonstrated by the ladder results.
SDS-PAGE
The gel demonstrates a band of darker color where dye bound to the protein of interest: HSP70. The Western blot also confirms the presence of the protein transferred from the gel.
- Reflection
Lab #3 October 15th, 2013
- Summary of the lab
Reatime PCR (polymerase chain reaction or qPCR) was performed on cDNA that was reverse transcribed from isolated RNA. An additional protein extraction procedure was completed with Olympia oyster mantle tissue.
- Materials and Methods
qPCR
A master mix was made, combining 62.5uL of SsoFast EvaGreen Supermix, 2.5uL of upstream DNMT primer, 2.5uL downstream DNMT primer and 52.5uL of ultra pure water. 24uL of the master mix was added to 4 white PCR wells. 1uL of the cDNA was placed in 2 reaction wells, 1uL of ultra pure water was added to the negative control wells. Clean lids were placed on the tubes, then loaded into the PCR machine for the following conditions: 95°C for 10 minutes, 95°C for 15s, 55 °C for 15 s, 72°C for 15 s (+ plate read). The Procedure was repeated from step two then repeated 39 more times. Finally the PCR machine tempered at 95°C for 10s, then processed the melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read).
Protein Extraction
A snap tube cap was labeled #43 OlyM, dated 10/15/13 and initialed, then 500uL of CellLytic MT solution was added to the cut piece of frozen tissue. The tissue was homogenized with a sterile disposable pestle then inverted several times. The tube was spun in a refrigerated microfuge for 10 mins at 14,000 x g. A clean tube was labeled "Protein", #43 OlyM, initialed and dated. The supernatant was then transferred to the clean tube then frozen for future analysis.
- Results
- Conclusions
- Reflection
Lab #2 October 8th, 2013
- Summary of the lab
- Materials and Methods
October 1st, 2013
A Snap cap tube containing pacific oyster #43 was initialed, labeled with the date, and placed on ice. 500uL of TriReagent was added and the tissue was homogenized using a pestle and vortexed for 5 seconds. An additional 500uL of TriReagent was added then vortexed for 15 seconds. The homogenized tissue was then placed into a freezer storage for future RNA extraction.
October 8th, 2013
The RNA sample was defrosted and left to incubate at room temperature for 5 minutes. 200uL of chloroform was added then the sample was vortexed vigorously for 30 seconds. The tube was then left to incubate for 5 minutes. In a refrigerated microfuge, the sample was placed to spin on 14,000g for 15 minutes. The aqueous layer was pippeted of and placed into a sterile snapcap tube. 500 uL of isopropanol was added, then inverted until fully mixed. The RNA was left to incubate at room temperature for 10 minutes, then centerfuged a 14,000 g for 8 minutes. The supernatant was then pipetted off the RNA pellet and 1mL of 75% EtOH was added. The tube was vortexed briefly then spun in the refrigerated microfuge at 7500 g for 5 minutes. Using a small pipette tip, the remaining supernatant was removed from the RNA pellet. The cap was left open for five minutes to let the pellet dry, then re-suspended in 100uL of 0.1 DEPC-H20. The mixture was then incubated at 55C for 5 minutes in a hot water bath and then placed in ice. The RNA was quantified with a nanodrop spectrophotometer to test the concentration of the sample and calculated (RNA)= 40uL/mL x A360 x dilution factor. To begin the PCR process, 5ul of RNA, 1 uL of oloigo dT and 4uL of nucleate free H20 were added to a sterile PCR tube. The mixture was left to incubate for 5 minutes at 70C in a thermocycler then transferred to ice. A combination of 5ul M-MLV 5X, 5uL of dNTPs, 1uL of M-MLV RT and 4uL of nuclease free H20 was added to the PCR tube. The mixture was incubated for 60 minutes at 42C then at 70C for 3 minutes on the thermocycler. The sample was spun on a centerfuge then stored on ice at -20C.
- Results
- Conclusions
- Reflection
October 8th, 2013
I found an interesting article that pertains to the genetics of bacterial bioluminescence:
http://www.annualreviews.org/doi/pdf/10.1146/annurev.ge.28.120194.001001
Five lux genes have found to be responsible for the luminescent system in certain bacteria that emit light:
(luxAB) and (lux CDE)
I am interested in Puget Sound water quality and its effects on bioluminescent bacteria that use luciferase reactions to create light. I would like to measure if there is a difference in light reactions from water samples taken in various areas throughout the Puget Sound.
Lab #1 October 1st, 2013
- Summary of the lab
- Materials and Methods
A Snap cap tube containing pacific oyster #43 was initialed, labeled with the date, and placed on ice. 500uL of TriReagent was added and the tissue was homogenized using a pestle and vortexed for 5 seconds. An additional 500uL of TriReagent was added then vortexed for 15 seconds. The homogenized tissue was then placed into a freezer storage for future RNA extraction.
DNA ISOLATION
A snap cap tube containing the Olympia Oyster sample #87 was initialed, labeled with the date and placed on ice. The tissue was homogenized with 0.5mL DNazol and an additional 0.5mL of DNasol was added and mixed. The sample was left to incubate for 5 minutes at room temperature then centerfuged at 10,000 x g for 10 minutes. The supernatant was transferred to a new tube and 0.5mL of 100% ethanol was added. The sample was inverted 8 times, left to rest for one minute at room temperature and then centerfuged at 5000 x g for 5 minutes. The lysate around the DNA was pipetted off then washed with 1mL of 75% ethanol. This step was repeated twice, ensuring all ethanol was removed from the DNA. 300uL of 0.1% DEPC water was added to dissolve the DNA. The DNA was quantified using a Nanodrop spectrophotometer and computer program to assess the DNA concentration (mg/uL), A260/280 and A260/230 ratios. To blank the machine, 2uL of o.1% DEPC-H2O was pipetted onto the nanodrop pedestal and dsDNA was selected from the pulldown menu. A KimWipe was used to clean the surface of the sensor then 2uL of the DNA sample was pipetted onto the pedestal. The measure button was selected and the results appeared after a moment.
- Results
- Conclusions
- Reflection