Summary: Hydrated new primers. Run qPCR primers for testing, then on olympia oyster samples.
Materials and Methods:
Primer Hydration:
- Find the primer length (nm) and add the appropriate amount of uL H2O
- To make working stock, mix 10uL of primer with 90uL of H2O (used for qPCR)
qPCR:
- Create a master mix with the following components per reaction:
- Sensimix- 12.5uL
- SYBR - 1uL
- upstream primer - 1.25uL
- downstream primer - 1.25uL
- Ultra Pure Water - 7uL
- Mastermix was then combined with either 2uL of cDNA samples or 2uL of pure water (blanks) into white qPCR wells
- Wells were spun to collect volume at bottom of wells then run under PCR conditions
Results:
Primers:
Cytochrome c oxidase:
Forward: 34.9nm
Reverse: 40.1nm
ATP Synthase:
Forward: 38.6nm
Reverse: 29.2nm
raw qPCR results can be found here under "Michael_1128_cytc"
Analyzed results:
below are the results taking the mean of all normalized gene expression values and graphing them.
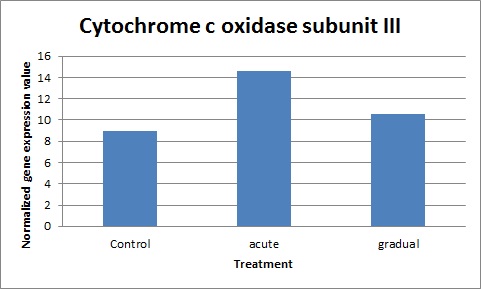
p-values are:
| cont vs. acute |
cont vs. grad |
acute vs grad |
| 0.321373501 |
0.747123 |
0.58748 |
Conclusions:
While p-values are higher than the .05 needed for significance, it does seem that acute and gradual treatments did cause the cytochrome c oxidase gene to be expressed more. What is interesting and different from expected is that the acute treatment seemed much higher than the gradual. The graph does not account for any outstanding values. Will probably add a standard deviation at some point to the graph. The results are a little inconclusive, so possibly testing with ATP synthase gene will clear it up.
Reflections:
Purpose of this lab was to finally get results on gene expression and quantify them. Procedures during this lab were used to measure gene expression and get a value for each sample. These methods can be used for measuring the gene expression of an organism and comparing the measurement against a control. Instructions were pretty clear and no other questions.
11/19/12, 11/20/12
Summary: Finished isolation of RNA from gill tissues, spectro-analyzed two samples using nanodrop, and designed new primers.
Materials and Methods:
RNA Extraction: for samples 55-57, 61,62
- Homogenized gill tissue with added tri-reagent (500uL)
- Added more tri-reagent (500uL) then used a centrifuge for 15 seconds
- add 200uL of chloroform to room temp RNA sample from Lab 2 and vortex for 30s
- after 5 mins at room temp, refrigerated microfuge for 15 mins.
- Take the aqueous phase (top layer) and added 500 uL of propanol to new solution.
- After mixing and sitting at room temp, another refrigerated microfuge for 8 mins.
- After removing supernatant, added 75% EtOH (1mL) to pellet and votexed
- Spun at 7500g for 5 mins. then removed supernantant
- Tubes spun 15s then pipetted removing remaining EtOH
- Tubes left open for max 5 mins, and added 100uL of 0.1%DEPC-H2O
- Tubes incubated at 55C for 5 mins then stored on ice
- Tubes labeled with initials MT, sample number, date 11/19, and RNA
- Pipette 2uL 0.1%DEPC-H2O and click "Blank"
- Pipetted 2uL for spectro analysis in samples 56, 79 to check for purity; lowered arm and clicked "Measure"
Results:
Spectro-analysis:
| Sample # |
Concentration |
260/280 ratio |
260/230 ratio |
| 56 |
679.4 |
1.98 |
.97 |
| 79 |
700.1 |
1.94 |
1.91 |
ATP synthase beta subunit
FW: TACCACTCGACCACTAGCCA
RV: AATGCAGCAAGGAAAGCGTG
Cytochrome c oxidase subunit 3:
FW: GCATAGAAGACTACGCCACTCT
RV: CACAACCATAGCCGCAATCAC
Conclusions:
Spectro-analysis results were generally what was expected. The normal ranges are: 260/280=1.8-2.0 and 260/230=1.5-2.0. The in-range A260/280 ratios indicate the samples were clean of proteins. Sample #56 result of a low 260/230 ratio indicating carryover of phenol, ethanol or high salt in the sample. Since the samples were all gill tissues, there is a possibility that salt concentrations could be higher and thus the cause for the low A260/230 ratio. As seen with sample #79, the ratios were in range, and can assume the rest of the samples were clean and pure, with the possibility of high salt concentration. Since RNA has been isolated, as a class they were put into wells in preparation for cDNA. Since primers have been designed, next will be to re-hydrate them and create a master mix.
Reflections:
Purpose and procedures of the first part of the lab was like last week in extracting RNA and testing the purity. This can then be used to make cDNA which is better to use for PCR than genomic DNA since it will not include extras like introns. The primers were designed to give another gene to test for metabolic rate. The cytochrome c oxidase is a redesign since the previous one had a little too much primer dimer. While the previous cyt-c oxidase primer may work, the redesign is just for insurance. The primers will be used for future PCR runs. Procedures were pretty clear and no issues.
11/13/12
Summary: Isolated RNA from gill tissues from half of assigned Olympia oysters
Materials and Methods:
- extracted and weighed tissues
- Homogenized gill tissue with added tri-reagent (500uL)
- Added more tri-reagent (500uL) then used a centrifuge for 15 seconds
- add 200uL of chloroform to room temp RNA sample from Lab 2 and vortex for 30s
- after 5 mins at room temp, refrigerated microfuge for 15 mins.
- Take the aqueous phase (top layer) and added 500 uL of propanol to new solution.
- After mixing and sitting at room temp, another refrigerated microfuge for 8 mins.
- After removing supernatant, added 75% EtOH (1mL) to pellet and votexed
- Tubes stored at -80 C
- Tubes labeled with initials MT, sample number, date 11/13, and RNA
| Sample Number |
Weight in g |
| 55 |
|
| 56 |
.062 |
| 57 |
|
| 61 |
.045 |
| 62 |
.004 |
| 63 |
|
| 64 |
|
| 76 |
Jingle shell (no data) |
| 77 |
|
| 78 |
.030 |
| 79 |
.062 |
| 80 |
Black= Olympia Control
Blue = Olympia 35 C
Purple = Olympia Gradual Temp
Conclusions:
Only half of the samples were done, and due to time I was only able to stop after washing with alcohol. What will be done next is to finish washing samples with water, then repeat process with other half of the samples (55-57, 61-62). After finishing with RNA extraction, use spectro-analysis to analyze for purity.
Reflection:
Purpose of this lab was to extract RNA in the first step for measuring and studying gene expression. Since procedures have been performed before, nothing was unclear.
11/6/12 Lab 7
Summary: Conventional PCR and qPCR gel electrophoresis and analysis of the previous labs results. Using the western blot technique to analyze qPCR results from SDS -PAGE.
Materials and Methods:
Agarose gel: (pre-done)
- Mix 2g agarose with 150mL 1x TAE in a 1L flask
- Microwave for 3 mins, then let it cool
- add 12uL ethidium bromide and mix
- after pouring gel into a tray, add gel combs (pop any bubbles) then put in fridge
- Place gel in box, fill with 1x TAE buffer, then remove combs from the wells
- load 7uL of 100bp latter in far left lane
- 20uL of PCR sample into the gel
- gels loaded into lanes 6-9
- run at 100V for 1hr, using UV transilluminator view results
- Mix 15uL of Protien stock and 15uL of 2x reducing sample buffer together in a 1.5mL screw cap tube
- After centrifuging for 10s, boil samples for 5 mins.
- Centrifuge for 1 min, and slowly load sample into wells
- Lanes 6,7
- Plug electrodes into receptacles and put the lid on, set voltage to 150V and run for 45 mins
- After turning power off, and removing lid, remove gel and cut off top part of lane one for reference
- Soak filter paper, membrane and gel in Tris-Glycine Transfer buffer for 15 mins
- Assemble the blotting sandwich in semi-dry blotting apparatus rolling each piece after placing them on:
- Anode (+++) at bottom
- filter paper (2 pieces)
- membrane
- gel
- filter paper (2 pieces)
- cathode(---) at top
- Transfer at 20V for 30 mins
- Removing gel from the sanwich, rinse off the adhering pieces with transfer buffer
- Wash membrane with 20mL pure water for 5 mins each
- Membrane placed in plastic box and add 10mL of blocking solution
- (Next steps were done by Mackenzie)
- Membrane was placed in 10mL of Blocking Solution in a covered dish, incubated for 30 mins on rotary shaker set at 1 rev/sec, then decant
- Rinse membrane with 20ml of water for 5 min, decant and repeat once
- Incubate membrane with 10 mL of primary Antibody solution overnight, then decant
- Membrane was washed for 5 mins with 20 mL of Antibody wash then decant and repeated 3 times
- Membrane was then incubated in 10 mL of Secondary Antibody Solution for 30 mins then decanted
- 5 mins with 20mL of Antibody Wash, then decanted and repeated 3 times
- 20ml of Water for 2 mins, decant and repeated twice
- Incubate membrane in 5mL of Chromogenic Substrate until purple bands develop on the membrane
- Rinse with 20 mL of water for 2 mins, repeated twice
- membrane dried on clean piece of filter paper to open air, by a stream of slight warm air or under an infrared lamp
Results:
Conventional PCR:
Results can be found here, the 2nd image in the album with the top 4 on the left having marks ~150 bp or lower. The lanes were: 6=cDNA, 7=gDNA, 8=Blank, 9=Blank
The SDS-protein gel can be viewed here
The Western Blot results could be found here, but there was no band produced in my lanes (6, 7) for samples # 71, 72
Conclusions:
The conventional PCR results were not exactly what was expected since the blank lanes came up with bindings, but the expected product length of my designed cytochrome oxidase primer was supposed to be 122 bp. Due to the marks around that area for my DNA samples, it was relatively successful and the marks for the blank samples could be due to primer dimer. In the western blot, it was expected that HSP 70 should have been shown, and thus expressed, but none of my results bound to the 70 area which would have been positive for expression (only one of the Olympia oysters showed signs of expression in the last lane(8)). This could possibly mean that HSP 70 was not expressed in the Olympia oysters with gradual temperature.
Reflections:
This lab's purpose was to practice the techniques used to visualize data. Since the the procedures and techniques were used to determine/analyze gene expression, it will be very useful for when we try to analyze our own data. The studies of these results can be used to determine if a certain gene is being expressed or not. Procedure seemed pretty clear.
10/30/12 Lab 6
Summary: Using the designed primers, performed conventional PCR and qPCR. With samples of oysters, isolated protein.
Materials and Methods:
Conventional PCR (25uL):
- Thaw cDNA sample
- Create a master mix for the 4 reactions. (make enough for 5 to allow for pipetting errors)
- Each reaction consists of Apex Red 12.5uL, 10uM Forward Primer 1uL, 10uM reverse Primer 1uL, and 2 uL of templet. all into 0.5mL PCR tube.
- Samples then underwent thermocycling
- Thaw cDNA sample
- Created master mix for 6 reactions.
- Reactions consist of Immomix 12.5uL, Syto-13 dye 1uL, forward primer 1.25uL, reverse primer 1.25uL, ultra pure water 7uL, and 2uL of template/ water.
- Samples then underwent varying temperature conditions
- Obtain and note weight of tissue given (tubes labeled with initials and date)
- Homogenize tissue with 500uL CellLytic MT in 1.5mL tube and then spin in refrigerated microfuge for 10 mins at max speed
- Transfer supernatant to fresh tube
- New tube labeled "protein MT 10/30" diluted sample by combining 15uL protein sample with 15uL DI water
- Transfer 1000uL of sample to a cuvette
- Using a spectrophotometer, find the absorbance at 595nm
- Concentration was used by equation given y=996.52x - 43.64
Results:
No results yet from PCR and qPCR.
Protein Isolation and Extraction results:
| Sample # |
Weight in g |
Absorbance |
Concentration |
| 71 |
.024 |
.553 |
507.4245 |
| 72 |
.020 |
.574 |
528.351 |
Conclusions:
The results from the PCR samples will be up for analysis next week. The protein extraction was successful and will be used in SDS-PAGE and western blotting next week as well.
Reflection:
Purpose of this lab was to practice making PCR solutions (conventional and qPCR), and also to practice extracting and analyzing protein samples. Procedures are used in part to measure the expression of a specific gene or isolating protein from a part of an organism. These methods can be used for virtually any experiment looking to analyze gene expression. It was slightly unclear what concentration amount we were looking for in the protein isolation, but everything else seemed pretty clear.
10/23/12 Lab 5
Summary: Dissected oyster tissues from experiments and re-hydrated primers into a stock and working stock solution.
Materials and Methods:
Oyster tissue dissection:
- Tubes were made and labeled for each oyster and tissue sample (i.e. 1g or 1m for oyster #1 gill tissue or oyster #1 mantle tissue)
- Oysters for all experiments were first measured for length and width, then shucked and dissected for gill and mantle tissue.
- Numbers were assigned based on the experimental stress they went under. #1-50 were pacific oysters: 1-15=Control, 16-30=Cyclodextrin, 31-40=35 degree shock, 41-50= 45 degree shock; #51-89 were olympia oysters: 51-60=Control, 61-70= 35 degree acute shock, 71-80=gradual temp, 81-89=pre-stress
- Find weight of primers and add water to dillute: Stock solution = primer weight *10uL nuclease-free H20 (forward = 295uL of H20). Working solution = 10uL of stock solution + 90uL H2O
Gradual temp experiment:
- Obtain Olympia Oysters
- At starting temp, increase temperatures 3-4 degrees every 2 hours. Max = 35 C
Measurements for all oysters can be found here
Gradual temp experiment:
| Time |
Temp in C |
| 9AM |
12 |
| 11AM |
21 |
| 1PM |
24 |
| 3PM |
28 |
| 9AM |
28 |
| 11AM |
30 |
| 2PM |
35 |
Conclusions:
The temperature increase from 9-11AM was more than we anticipated (adjustment to the equipment), but it should not effect the data too much since the temperature was lower and the total temperature rise was over 24 hrs. It was expected that the rise from 30-35 would be like the others, but the heater was not strong enough so it took longer to increase the temperature in that interval. Next, since we have collected tissue samples, we can now start isolating DNA and RNA.
The primers are now set so we can practice with PCR
Reflections:
The purpose of this lab was to begin the process of collecting data for the experiments, and to prepare for the PCR lab. Any experiment would require the procedures we performed to get tissue. Not necessarily shucking and opening up oysters, but to get data there needs to be a time at the beginning where tissue needs to be dissected and extracted. It also helped to work in an assembly line to make the process go a little quicker. Procedures were pretty clear and there were no other issues.
10/16/12 Lab 4
Summary: Using the Isolated RNA from lab 3, reverse transcribed to make cDNA. The other part of lab was getting prepared for experiments the following week, which included designing primers.
Materials and Methods:
Reverse Transcription
- In 0.2mL PCR tube mix 5 uL RNA, 1 uL oligo dT, and 4uL nuclease free H20
- incubate at 70 C for 5 mins using termocycler, then transferred to ice
- After centrifuging, added 5uL m-MLV 5x Reaction buffer, 5uL DNTPs, 1uL M-MLV RT, and 4 uL of nuclease free H20
- mixture incubated at 42 C for 60 min, then heat inactivated at 70 C for 3 min
- Centrifuged and stored on ice at -20 C
Primer Design
- Find gene on NCBI website
- design primers with length 18-22, 80-200 bp, and melting temp 60 C +- 3 (also database to be "nr")
- obtain the forward and reverse sequences
Results:
No data measured in reverse transcription.
Primers for Ostreola conchaphila, cytochrome c oxidase:
Forward: TTCCACTCGGCTTTGTCTCC
Reverse: TGAAGCGGAGCCTTACTTCAA
Conclusions:
With the cDNA, along with the primers we designed, perform PCR.
Reflection:
Purpose of the lab was to acclimate ourselves with converting RNA to cDNA and used to the anatomy of an oyster. The cDNA is now more stable than the RNA and can be used in PCR. No problems occurred during lab, and everything seemed pretty clear.
Potential question to be addressed from class experiment:
How does metabolic rate change in an oyster due to gradual and sudden temperature stress?
Gene: cytochrome c oxidase
10/9/12 Lab 3
Summary: Isolation of RNA from the gill of a pacific oyster. Use a Nanodrop spectrophotometer to quanitify the RNA.
Materials and Methods:
- add 200uL of chloroform to room temp RNA sample from Lab 2 and vortex for 30s
- after 5 mins at room temp, refrigerated microfuge for 15 mins.
- Take the aqueous phase (top layer) and added 500 uL of propanol to new solution.
- After mixing and sitting at room temp, another refrigerated microfuge for 8 mins.
- After removing supernatant, added 75% EtOH (1mL) to pellet and votexed
- Spin at 7500g for 5 min, then removed supernatant and spun tube again for 15 sec.
- Removed remaining EtOH, and left tube open for ~ 4 mins
- add 100uL 0.1% DEPC-H2O and dissolved pellet
- Incubated tube for 5 mins at 55C then used 2uL for spectro-analysis (rest went to storage)
Results:
RNA Concentration: 790.6 ng/uL
A260/280 ratio: 2.00
A260:230 ratio: 1.94
Total RNA Concentration = 790.6 ng/uL * 100 uL = 79060 ng
Conclusions:
The ratios for my results were exactly in line with the expected ratios of clean DNA. The A260/280 of 2.00 is at the top of the range of expected 1.8-2.0, but i can assume it to be clean from proteins. The A260/230 ratio value of 1.94 falls between the expected 1.5 - 2.0, so my sample should also be clean of ethanol, salts or phenols (that extra ~4 mins to remove remaining EtOH might have helped). Next, we will try to take the RNA and transcribe it to cDNA.
Reflections:
The purpose of the lab, like last week was to get us used to the lab techniques of isolating RNA. These procedures can help extract and isolate RNA and then be used to measure the gene expression from a specific tissue. These methods might be used for studying the stress effects on various organisms. Specifically, we will be examining the effects on pacific and olympia oysters. There were no problems during the lab, and everything seemed pretty clear.
10/2/12 Lab 2
Summary: RNA extraction and isolate DNA from the gill of a pacific oyster. After isolating DNA, use a NanoDrop spectrophotometer to analyze the quality.
Materials and Methods:
RNA extraction:
- Homogenized .043g of gill tissue with added tri-reagent (500uL)
- Added more tri-reagent (500uL) then used a centrifuge for 15 seconds
- Stored sample at -80 C for next week
- Tube labeled with initials MT, date 10/2, and an R
- Homogenized .061g of gill tissue with DNazol (1mL total)
- Centrifuge at 10,000 g for 10 mins.
- Obtain the Supernaut (top liquid layer) and add 100% ethanol (.5mL)
- After mixing, isolate the DNA precipitate (centrifuged at 2,000 g for 1 min) and wash with 75% ethanol (1mL)
- Removed the ethanol and dissolve precipitate with .1% DEPC water (150uL)
- Used 2uL for spectro-analysis then stored rest of sample at -20 C “MT, 10/2, DNA”
Observed tissue weight:
RNA: .043g
DNA: .061g
DNA Spec:
Abs. 5.430
260/280: 1.92
260/230: 1.13
DNA concentration (Ng/uL) : 306.3
Total DNA concentration = 306.3 Ng/uL * 150 uL = 45945 Ng
Conclusions:
The DNA A260/280 value of 1.92 is slightly higher than the 1.7-1.9 value expected. This means that the DNA is not as pure as intended. Since protiens generally absorb light at 280nm, I might have needed to dissolve the sample more, or there might have been other contaminants with the sample. There was also a large spike before the 230nm value. This probably indicates that there was excess ethanol still present. If the expected ratio for A260/230 is that of RNA (1.5-2.0), then my value is lower than expected. This would confirm that there is a decent amount of ethanol left over from washing the DNA. The 260/230 ratio could also be lower due to more salts in my sample. Next, since the RNA was put on ice, we will probably try to isolate it and run another spectro-analysis to determine the quality.
Reflection:
The purpose of this lab was to get us acclimated to a type of DNA and RNA extraction. This is probably a common lab technique that we will probably have to use in the future with our projects. The spectrophotometer was used to measure the purity of DNA and to see what kinds of impurities are with our sample; such as phenols, salts, and proteins. These methods could be used for gene sequencing, or anything that needs the use of an isolated DNA (potentially in combination with other lab techniques like PCR). It could have been helpful to know of a better way of extracting ethanol away from our DNA than by just pipetting the excess out, since I believe I still had excess. It would have also been nice to know if the A260/230 ratio range should have been the same as expected from RNA (1.5-2.0) or something different.