Lab 9: Salmon project
This week helped some students set up their experiments and retrieve their data from BioRad. We all worked to get our data averaged, fold over min, graphed and then we ran an one way ANOVA which determined that one of my 4 categories was statistically different than another (<.001). Then we ran a Tukey test that took each category and determined the significance between all of the categories. My LPS control was significantly different, but none of the others were. My plate layout from last week was:
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
|
| A |
49 |
49 |
50 |
50 |
51 |
51 |
52 |
52 |
1 |
| B |
53 |
53 |
54 |
54 |
55 |
55 |
56 |
56 |
2 |
| C |
57 |
57 |
58 |
58 |
59 |
59 |
60 |
60 |
3 |
| D |
61 |
61 |
62 |
62 |
63 |
63 |
64 |
64 |
4 |
| E |
65 |
65 |
66 |
66 |
67 |
67 |
68 |
68 |
|
| F |
69 |
69 |
70 |
70 |
71 |
71 |
72 |
72 |
|
| G |
73 |
73 |
74 |
74 |
75 |
75 |
76 |
76 |
|
| H |
77 |
77 |
78 |
78 |
79 |
79 |
80 |
80 |
November 29-December 3, 2010
Lab 8: Salmon project
Monday Chris and I finished reverse transcription on the lost box samples. Tuesday, half the group ran test qPCR on primers of all of our 13 primers. The other half hydrated our primers and prepared the remaining samples for qPCR. On Wednesday we looked at our results from the qPCR and 8 of the primers worked. On Thursday, I ran prepared my samples (#49-80) with my SAA primers for PCR. I made a master mix with half the reaction volume 25uL instead of 50uL. As seen below:
| Component |
Volume |
Final Conc. |
New Volume (x70) |
| 2x (Immomix) |
25uL |
1x |
1750 |
| Syto-13 dye (50uM) |
2uL |
2uM |
140 |
| upstream primer, 10uM |
2.5uL |
2.5uM |
175 |
| downstream primer, 10uM |
2.5uL |
2.5uM |
175 |
| Ultra pure water |
16uL |
NA |
1120 |
November 22-26, 2010
Snow days/Thanksgiving holiday!
November 15-19, 2010
Lab 7: Salmon project
On Monday, we quantified RNA on the nanodrop and made an excel file for the data. On Tuesday, students normalized all of the samples before lab started. Then we ran a qPCR to determine if there was any DNA carryover on all of the samples. On Wednesday, we learned how to analyze our results from the qPCR and determined which samples we needed to DNase (12 total). On Thursday, the 12 samples were DNased and half of the samples we ran reverse transcription on. On Friday, we ran reverse transcription on the 12 DNased samples and the remaining samples. Next week our primers will be in, where we will test our primers and then begin.
November 9, 2010
Lab 6: Independent project: Salmon fry exposed to pesticide cocktail and LPS (bacterial stimulant) and Poly: IC (viral stimulant) to determine the immune or stress responses
Summary:
Our group went over to the computer lab to find peer reviewed papers conducted on similar species or animals exposed to environmental contaminants and then a second stressor. We also turned to NCBI to find primers on specific genes that would best express immune response, stress response, or normalizers/internal controls. Then we decided how to proceed and laid out next steps for the remainder of the week.
Materials and Methods:
Verbal description of the experiment done by Chris Gru's student. We learned that salmon fry were exposed to pesticide cocktail of 12 pesticides found in urban waterways and of course controls who were not, then injected with secondary stressors either LPS or Poly: IC. We then searched electronic journals on the UW library for papers that might have genes of interest that might be expressed due to similar stressors. Once the genes were found, we used NCBI website, to find the sequence of interest, then we ran a BLAST to see if the gene is conserved or has been tested in a species closely related to our salmon Oncorhynchus kisutch. We then copied the gene via FASTA text and pasted it into Primer 3 Plus, where we then chose our primers and posted them on our wikipage (spreadsheet). Further research was done once everyone posted their primers to see how which pathways these genes belonged to and started to process how these might be expressed in this experiment (increase production or decrease). We also worked on a very simplified plan for this week, and an idea of Monday and Tuesday of next week. Our goal was to order our primers by Friday or Monday at the latest, so we can get started as soon as they come in.
Results:
The primers that I found were the following:
| Pforeman |
tshawytscha_Cyp1A_F |
AATTCATCAACGACGGCAAG |
20 |
45 |
61 |
|||||
| Pforeman |
tshawytscha_Cyp1A_R |
CCATGACGGAGGTAAGCTGT |
20 |
55 |
60.1 |
|||||
| Pforeman |
nerka_VTG_F |
CTCAGGGAGTGTGCAAGACC |
20 |
60 |
60.9 |
|||||
| Pforeman |
nerka_VTG_R |
CTCTGCCGGCACTCTACAC |
19 |
63.2 |
59.6 |
|||||
| Pforeman |
tshawytscha_VTG_F |
CATCCATTTGACCAAGAGCA |
20 |
45 |
59.6 |
|||||
| Pforeman |
tshawytscha_VTG_R |
CGGGCTTCATGAGGTAGTTG |
20 |
55 |
60.6 |
|||||
| Pforeman |
mykiss_SAA_F |
TAAAGACATGTGGCGTGCAT |
20 |
45 |
60.1 |
|||||
| Pforeman |
mykiss_SAA_R |
TTACGTCCCCAGTGGTTAGC |
20 |
55 |
60 |
|||||
| Pforeman |
mykiss_IL1B1_F |
CTGAAGCCAGACCTGTAGCC |
20 |
60 |
60 |
|||||
| Pforeman |
mykiss_IL1B1_R |
GAAAAGGAAACGCACCATGT |
20 |
45 |
60 |
Raida, M.K. and K. Buchmann. 2009. Innate immune response in rainbow trout (Oncorhynchus mykiss) against primary and secondary infections with Yersinia ruckeri O1. Developmental and Comparative Immunology 33: 35-
45.
Veldhoen, N., M.G. Ikonomou, C. Dubetz, N. MacPherson, T. Sampson, B.C. Kelly, and C.C. Helbing. 2010. Gene expression profiling and environmental contaminant assessment of migrating Pacific salmon in the Fraser River watershed of British Columbia. Aquatic Toxicology 97: 212-225.
Conclusions:
We decided to order some primers on Friday and those we didn't get approved we would order on Monday. We are meeting on Monday to measure how much RNA we have in the samples with the nanodrop and some folks will run qPCR on samples to determine if they are contaminated. Our limiting factors for this experiment is the volume of samples, so we were advised on Friday to not DNAase the samples.
Reflections:
I will have to say that our group really needs to pull together and possibly set consistent meetings to keep us on track and divvy up the work. I have spent time looking up each of the genes and tried to understand their function, but would love to get some feedback if we are on the right track. For immune response, we have MHC, XBP1, TNF(a), SAA, and IL 1B1, IL 12B, IL 6. For stress response we have chosen Ptgs2b, Cyp 1A, CRH, and SOD. We also have VTG to see if there is any reproductive response, but I am not sure we have primers for normalizers, like GAPDH, B-actin, or cell death. So we might want to talk as a group on Monday if what we have chosen will be enough in those categories or if we should keep looking.
November 2, 2010
Lab 5: Epigenetics (continued) and Quantitative PCR
Summary:
Last week we conducted two of the three components of methylated cytosine dot blot procedures. We diluted our DNA samples and bound those to a Dot Blotting nylon membrane. This week we developed the dot blot to measure DNA methylation by using the Western Breeze kit and a 5 meC antibody for chromogenic immunodetection. Then we conducted a Quantitative PCR (polymerase chain reaction) with the primers we chose from lab 1 (Oncorhynchus nerka VTG).
Materials and Methods:
We rehydrated our primers with 10mL of blocking solution (14mL of ultra filtered water, 4 mL Blocker/Dilutent part A, and 2 mL of Blocker/Dilutent part B) that was prepared last week. Then covered and incubated for 30 minutes while on the rotary shaker (dial 2; 1 revolution/sec). Then we decanted the solution, rinsed with 20mL of water for 5 minutes, decanted and repeated another time. Next we prepared 10mL of primary antibody solution (1:5000 dilution; 10mL of Blocking solution and 2uL of 5-MeC antibody), incubated the membrane in this solution for hour. Then the primary antibody was poured off, washed in 20 mL of TBS-T for five minutes. This was then repeated two more times (3 total). Then the membrane was incubated in 10mL of secondary antibody solution for 30 minutes, decanted, and then washed for two minutes with 20 mL of TBS-T. Decanted and repeated two more times, totally three washes. The membrane was rinsed with 20mL of water for two minutes, conducted a total of three times. Next we incubated the membrane in 5 mL of Chromogenic Substrate until color development. After about 45 minutes, the membrane was rinsed with 20 ml of water for two minutes, then decanted (repeated two more times), dried on a clean piece of filter paper and photographed.
For the QPCR, a master mix was prepared in a snap cap and labeled (enough for 6 reactions and 1 for good luck). As seen in the table below:
| Component |
Volume |
New volume (vol x 7) |
Final Concentration |
| Master Mix, 2X (Immomix) |
25 uL |
175 uL |
1x |
| Syto-13 dye (50uM) |
2 uL |
14 uL |
2 uM |
| Upstream primer, 10uM |
2.5 uL |
17.5 uL |
2.5 uM |
| Downstream primer, 10uM |
2.5 uL |
17.5 uL |
2.5 uM |
| Ultra pure water |
16 uL |
112 uL |
NA |
Results:
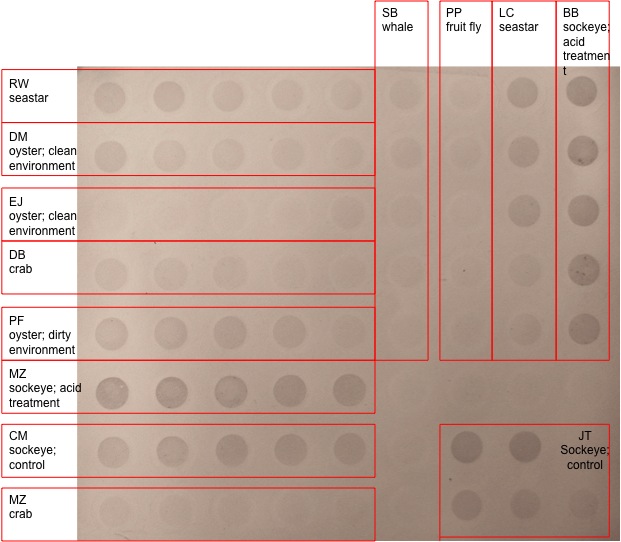
According to the photo below, the chromogenic immunodetection for my samples (PF oyster 2; dirty environment) are seen in the 5th row down on the left. There was visible precipitate in all 5 of the dots; however the first four look quite similar and the last dot is less dark. In comparison to the oyster from the clean environments, there was definitely a difference in relative abundance of protein.
In regards to qPCR, my results can be seen in the graph below:
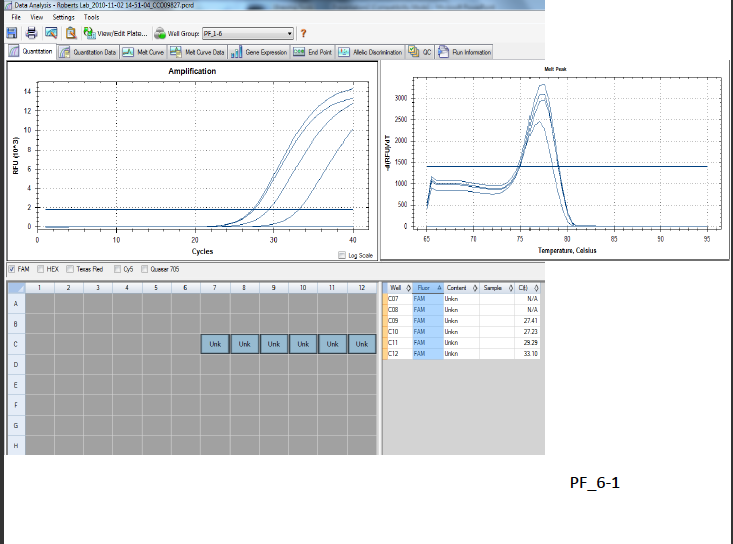
Hypothetically, in the graph on the left my RNA and negative controls should have been flat lines (RFUs of 0). My negative control was a flat line, with a RFU of 0 (N/A amplification), but my RNA sample indicated amplification of 27.23 and 27.41 RFU and my cDNA was 29.29 and 33.1. In the graph on the right, both my RNA and cDNA had a similar peak when the temperature was between 77 and 77.5. When the RFU/dT was 2400, the T was 77; 2900 the T was 77.5; 3100 the T was 77.5; 3200 the T was 77.5.
Conclusions:
The cytosine methylation measured using dot blot and visualized with immunodetection methods revealed that the oyster in the dirty environment expressed more protein than the oyster from a clean environment as far as precipitate color is concerned. However the color concentration or density bands in the first four dots were very consistent possibly indicating that the samples might be over developed, or have a high signal to noise ratio. It is only different in the lowest concentration dot on the far right. However, this dot is darker than all the other colors in the oyster from the clean environment.
In regards to the qPCR, it appears that there was operator error in my RNA sample, because I saw amplification. In lab two when I extracted RNA from sockeye salmon juv. B we were supposed to be very careful in extracting the aqueous phase and two times I accidentally lifted the interphase with my pipette. Because I created my cDNA from that single stranded RNA this didn’t work out. In the future, I would add a step before I reverse transcribe my RNA into cDNA; I would conduct a DNA digestion which would prevent any DNA carryover and avoid contamination. By treating the RNA with DNA digestion methods before I synthesize my cDNA I hope to see different amplifications (only cDNA).
Reflection:
It is hard to evaluate qualitative measures like the dot blot when the species are from clean vs dirty environments. How do I know if the methylated cytosine expression is due to the cleanliness of the environment and not from another factor? If epigenetics is a structural change “on top of” the a gene of concern how do people know “which” gene to look on top of? Couldn’t it be missed or misunderstood? Wow, I’m not quite sure I fully understand how to interpret this from an ecological perspective or even on a physiological perspective; I feel like I need to know the stimulus and then complete pathway before hand.
Lab 4
October 26, 2010
Summary:
We ran our two (25uL) PCR samples and two blanks on the agrose gel and used electrophoresis and 5uL of 100bp ladder to determine amplification of our cDNA. Then for our epigenetics we measured cytosine methylation using dot blot and chromogenetic immunodetection methods.
Materials and Methods:
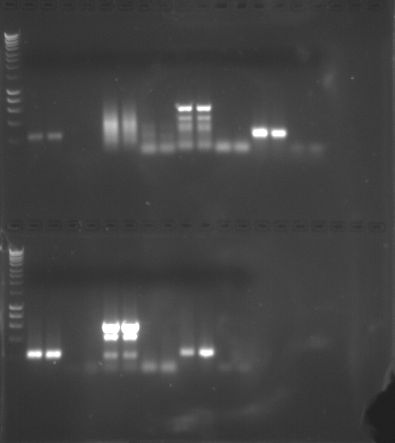
Agrose gel was placed in gel box and covered with 1x TAE buffer and combs were carefully removed. A 5uL 100bp ladder was added to the top and bottom far left column of both gels. Then I loaded 25uL of my two PCR samples into columns 6 and 7, then placed my two blanks/controls in columns 8 and 9. The remaining concentrations were stored in -20C. Electrophoresis was conducted for 55 minutes at 100V, 9 minutes at 150V, and 85V for about 15 more minutes. We tested each gel under the UV transilluminator and continued at the above settings. At 4pm, the gels were photographed as seen here.
For the cytosine methylation and dot blot, 5 concentrations from an oyster DNA (oyster dirty #2) were diluted as follows:
| Dilution |
Target Conc. |
uL of H20 |
uL of 20x SSC |
uL of 50ng/uL DNA (oyster dirty #2) |
| 1 |
0.8 |
124 |
60 |
16 |
| 2 |
0.4 |
132 |
60 |
8 |
| 3 |
0.2 |
136 |
60 |
4 |
| 4 |
0.1 |
138 |
60 |
2 |
| 5 |
0.05 |
139 |
60 |
1 |
Nylon membrane and filter for manifold was cut to fit 72 wells, soaked in 6x SSC for 10 minutes. The manifold was prepared with the filter then the membrane, making sure no bubbles were present. DNA samples were boiled for 10 minutes to denature the DNA and isolate them into single strands. Immediately after samples were transferred to ice. Vacuum was switched on and 500uL of 6X SSC were added to the four wells where my samples will go. This was filtered through after tape covered the used wells. DNA was spun down and all of the DNA was added (50uL) to the wells (row E, 1-5), and these were filtered through. Filter paper was soaked in denaturing buffer and when vacuum was finished and the membrane was transferred to this filter. Another 5 minutes with neutralization, replaced the membrane and transferred nylon membrane onto neutralization filter and placed in hood for 5 minutes. The rest will be done next week.
Results:
The amplification of my DNA using the GnRH primer was a bright band between 800-1000 bp (100 ng/Band) and my two controls which were supposed to have zero amplification had bands existing between 600-800 bp. Next week, we will continue with the epigenetics portion of this lab.
Conclusion:
We expected to see gene expression in sockeye salmon using the GnRH primers to help us better understand this gene’s involvement in reproduction. My bands were not crisp like some of the other students and my controls were slightly contaminated. Whether this was from using the pipette multiple times or possibly the concentrations that I used were contaminated. Having bands span between 800-1000 pb slightly above the controls might also imply that they might not be significant. However, to increase my understanding, I would want to fully analyze these results again, to get cleaner values.
Reflection:
I would like more explanation or modeling of how to go about analyzing our results. I’m not quite confident what this means and what implications it has on reproduction of salmon. How do I quantify this band? Or can you because it is so fuzzy? Why are some students’ bands bright? More pronounced?
Lab 3
October 19, 2010
Summary:
Three tasks were conducted in lab; first we reverse transcribed Sockeye salmon RNA into complementary DNA. Next, ran a Polymerase chain reaction (PCR) with our cDNA, polymerase (GoTaq Green Master Mix, 2x), GnRH primer (Gonadotropin releasing hormone), and dNTPs so that gene expression can be measured. Lastly, Agarose gel was prepared for next week’s lab agarose gel electrophoresis.
Materials and Methods:
Reverse Transcription was started by mixing stock Sockeye salmon RNA sample, extracted 5ul of RNA and placed in a .5ml PCR tube. Added 1ul of oligo dT, 4ul of nuclease free water and incubated in the thermo cycler at 70C for five minutes. Placed on ice, briefly centrifuged and added 5ul of M-MLV 5x Reaction buffer, 5ul of dNTPs, 1ul of M-MLV RT, 4 ul of nuclease free water and incubated at 42C for 60 minutes, heat inactivated at 70C for 3 minutes in the thermo cycler (using protocol 44170). Once done, spun on centrifuge and stored on ice. For the PCR, a reaction master was combined in a 1.5mL micro centrifuge tube containing: 250 ul of GoTaq Green Master Mix, 2x; 15 ul of forward primer GnRH, 10 uM; 15 ul of reverse primer GnRH, 10 uM; and 108 ul of nuclease free water. 48ul of this mixture was added to four .5ml PCR tubes labeled 1, 2, 3, and 4. Next, 2ul of cDNA was added to the PCR tubes 1 and 2 and 2ul of nuclease free water was added to tubes 3 and 4. Hence, each tube contained 50 ul of solution. The liquid was pooled by spinning the tubes and loaded into thermo cycler. The cycling profile consisted of denaturing at 95C for 5 minutes (1 cycle); denature at 95C for 30 seconds, annealing temperature of 55C for 20 seconds (40 cycles), extension at 72C for 90 seconds, final extension at 72C for 3 minutes and then dropped down to 4C. Afterwards tubes were stored at 20C until next week. While waiting for cDNA sample, 2 agarose gels were made. For each, 2 g of agarose were weighed out and mixed with 150 mL of 1xTAE. Swirling and microwaving for about 3 minutes until cloudy appearance disappeared. Once cooled, 12uL of ethidium bromide (EtBr) was micropipetted into the flask and mixed again. This mixture was poured into a gel tray and two combs were added to make wells. After about an hour or so, the gels were wrapped in plastic and stored in the refrigerator until next week where we will do an agarose gel electrophoresis.
Results:
The first cDNA sample I created accidently spilled, so I made a second sample that was used in the PCR. The measurement of gene expression will be conducted next week. Two agarose gels were prepared for next week’s electrophoresis lab.
Conclusion:
I predict that I will see some gene expression, the GnRH used is a key regulator of reproduction in vertebrates. My tissue was extracted from a juvenile sockeye salmon, hence there is chance that this gene is expressed in growth and development preparing for adulthood. It would be interesting to use this primer on eggs, fry, smolt, adults, and spawning salmon, males and females to see if the expression changes. I’m not really confident what to expect to be honest.
Reflections:
Loved learning about reverse transcription, but will we use genomic DNA to run a PCR this quarter? Is that common or do most people use the reverse transcription option? Is there a price difference or ease in procedure? I know there was a protocol to set up the thermo cycler, but if there is any chance to come in prior to lab while you are programming, I would love to see that process. As a teacher, I would need to know those techniques and am enjoying using the different techniques and equipment.
10/12/10
Lab 2: RNA Extraction and Protein Analysis, Part 2Summary:
In order to analyze protein from tissue (sockeye salmon) which is a heterogenous mixture with varying charges and complex in structure; a reducing sample buffer was used to denature the non-covalent bonds, the buffer maintains the appropriate pH level; B-mercaptoethanol removed disulfide bonds to linearize the proteins and SDS ensured they have the same negative charge. Once homogenous, this sample will be exposed to an electrical field and the proteins were separated on the basis of molecular weight on the polyacrylamide gels. This technique will not determine what types of proteins specifically, however comparisons were made to a standard and the different species and treatments the other students were analyzing. Next we finished our RNA extraction from lab 1 by using another organic solvent, chloroform, to separate the phenol, DNA, and RNA. Once isolated and washed, its absorbance was quantified with a spectrophotometer at 260nm and calculated using the following equation: RNA concentration=40ug/mL xA260 x dilution factor.
Materials and Methods:
Protein extract was thawed, 15uL was added to equal parts 2x Reducing Sample Buffer, then boiled for 5 minutes to ensure denaturing and linearization. Next it was centrifuged for 1 minute, then loaded with a gel loading tip into well of polyacrylamide gel plate. Electrophoresis was set to 150V for 45 minutes. During that time, my RNA sample from last week was exposed to room temperature for 5 minutes, then 200uL of chloroform was added to the sample under the fume hood. Shortly after that the sample was microfuged in the refrigerator for 15 min. at the highest speed. Next, the aqueous upper layer was transfered via micropipette to another microfuge tube. Then 500uL isopropanol was added, 10 minutes resting at room temperature, then microfuged int the refrigerator for 8 min. at high speeds. The supernatant was removed. 1m> of 75% EtOH was added to the "pellet" or RNA and salts. Again spun in the refrigerator microfuge at 7500g for 5 minutes. All the liquid was removed again, and all liquid was evaporated for no longer than 5 minutes. Then the pellet was resuspended with 100uL of 0.1%DEPC-H20 until the pellet was dissolved. This solution was incubated at 55C for 5 minutes to solubilize the RNA. Then it was placed on ice and this RNA stock will be used in future labs. We zeroed the Nanodrop machine with 2uL of 0.1%DEPC-H2O and then prepared our samples for the machine. We pipetted 2uL of our RNA sample onto the pedestal of the machine and closed the arm on the sensor. The computer measured the 260nm absorbance, RNA concentration (ng/uL), A260/280 ratio and the A260/230 ratio for RNA clarity. Once finished, our samples were stored at -80C. Then, we removed our gel from the gel box and placed it into shaker. Our TA and others who finished first, added 150mL of Coomassie Stain to container on the shaker with the gel and incubated on this device for 5 minutes. Stain was poured back into the original container and the gel was rinsed with 10% acetic acid. Then 250mL of 10% acetic acid was added to the container with the gel. Incubated in shaker for another 15 minutes. The buffer was changed out and repeated several times until the protein bands became visible. We looked at the gel on a lighted surface and the bands were not fully developed so the process was going to continue. A photo will be taken so that analysis can continue.
Results:
At the end of lab, our gel still needed more time absorbing the stain in order to see the displacement of proteins according to molecular weight. The far left of the gel, where the standard was added many proteins settled down the gel. One very faint band was visible about 2/3 of the way down all across our different tissue samples. My sample, #11 looks like it had a band 3 down from phosphorylase. It looks like this band was evident in all of our samples. Some had larger quantities at this molecular weight, 22kDa. I'm not sure if I had other bands that just were very faint or non existent. I want to know more about Carbonic Anhydrase and what role it has in salmon. In regards to the RNA extraction, I had a hard time detecting a pellet at all. Protocol for the most part was followed, however when I removed the top aqueous layer, I accidently pulled up the interphase layer. I did not transfer that into the sample tube, rather I placed it into the waste and centrifuged for a few seconds in hopes to separate by layers again. Then in rinsing the invisible pellet with EtOH it was honestly blind faith. I was thankful I had placed the hinge facing out and only pipetted from the opposite side. My absorbance measurement at 260nm on the Nanodrop was an RNA concentration of 164ng/uL. My A260/280 was 1.78 (supposed to be between 1.8 and 2.0 and wasn't) and my A260/230 was .39 (was supposed to be between 1.5 and 2.0 and mine was way off). My absorbance ratios were off indicating that I had carryover of either phenol, ethanol, or salt in my sample. Again without knowing if I had a pellet or not, trying to extract the liquid from the tube was not very precise. However, I hope that this sample will have a high enough RNA concentration to run a PCR or other assays.
Reflection:
This was a little unnerving going into this lab, because in lab 1 I wasn't very confident in my homogenizing the tissue. Then when I heard everyone talking about their pellet and I couldn't see one. I was trying to be extra careful in all of the extractions and washes, but didn't have the confidence that I would have liked. It was very cool to see the technology (Nanodrop machine and computer program) and learn a new technique. In regards to the gel work, I am not very sure what the second gel photo was? or how the two photos were different. I want to know more about the molecular weight band that appeared in almost all of our tissue samples and what role that protein might play. Other students had larger concentrations (thicker bands) and more bands settle out on the gel. Due to different exposures and treatments I want to better understand how those stresses might have expressed those proteins. In regards to the other controls like mine, why were they not all the same? Not quite sure what control A, B, and C means and if they were from the same animal? or what?
10/5/10
Lab 1: RNA Extraction and Protein Analysis, Part 1
Summary:
Tissue was selected from Sockeye salmon control B for RNA and protein extraction. RNA was isolated from this teleost tissue using TriReagent, homogenized, placed back into freezer at -80 degrees Celcius, and will be analyzed next week. Protein was also extracted from this tissue, isolated with CelLytic MT, and the concentration of proteins in the sample will be determined using a colorimetric assay, known as the Bradford Assay. The absorbance read from the spectrometer was compared to a standard curve that was prepared for us, due to time constraints. Lastly, we were introduced to NCBI genetic database and taught how to find stress genes associated with our species (O. nerka) of interest.
Materials and Methods:
Obtained 9g of Sockeye salmon control B tissue, put my initials and date on the side and stored it on ice at my station. Using a micropipette, 500uL of TriReagent was added to the tissue, then stored on ice. The tissue was homogenized with disposable pestle and placed on the vortex 2x. An additional amount of 500uL TriReagent was added, placed in vortex for 15 seconds, then stored at -80C until next week.
Obtained another vial of 9g Sockeye salmon control B tissue, labeled it protein with my initials and date. 500uL of CelLytic MT solution was added to the tube and homogenized with pestle, inverted the sample several times, and placed in refrigerated microfuge for 10 min. at highest speed. Meanwhile, labeled another tube Sockeye Salmon B Protein #2, 10/5/10 and micropipetted the supernatant liquid and stored on ice. To quantify the protein, labeled a 2mL tube,
diluted a portion of Protein #2 sample, by pipetting 15uL into the 2mL tube, with 15uL of DI water. Mixed with pipette
Made a control or blank sample; filled another 2mL tube with 30uL of DI, labeled Blank, 10/5/10, PLF. Added 1.5mL of Bradford Reagent to each tube; inverted several times and then incubated on the counter for 10 minutes. Mixed the “Blank” solution with pipette, transferred 1000uL to cuvette and ran it through the spectrometer and measured the absorbance at 595nm. Then the same quantity and procedure from the protein #2 sample two times and averaged the two absorbance values. The protein sample was then stored at -20C. To quantify the protein, the standard curve, y=1013.9x + 0, R2=0.97738 The R^2 value describes how well the line (y=mx +b) fits the points on the graph. The closer R^2 is to 1.0 the better the line fits your data -
Primer design was introduced using a PowerPoint presentation and steps needed to accomplish finding genes of interest, especially stress genes. The links are on the wikipage and we searched for genes and wrote down three accession numbers.
Results:
RNA isolation labeled: Sockeye salmon 9g, RNA, 10/5/10, PLF was stored at -800C for next week. After the protein was quantified via a Bradford Assay. After diluting the protein sample and adding the Bradford reagent, the solution looked a midnight blue color, while the control with DI water and Bradford reagent looked charcoal/brownish color. The absorbance at 595 nm on the spectrophotometer on the first sample was .141 A and .142 A for the second. Using the standard curve provided:
y=1013.9 (.1415) + 0
y=143.46685 (I multiplied this by 2, because I diluted my protein sample in half);
y=286.9337 ug/ml So, the total yeild of protein was approximately 287ug/ml of protein in the sockeye salmon control C sample. I would expect this to be quite low, I had a hard time homogenizing the sample. I would expect the absorbance to be higher possibly if I had continued longer with the pestle.
Primer Design
From Veldhoen et al., 2010, I am interested in three possible genes As we discussed, any of these sequences and genes would be perfect for primer design. -
VTG, NCBI GenBank accession number for Sockeye is FJ226375
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;">ORIGIN
[[#sequence_226316380]] 1 ttcttcagct caacatcaag aagacacaaa acgtctatga gttgcaggag gctggagctc
61 agggagtgtg caagacccac tatgtgatca gggaagatgc caaggcagag cgcatccatt
121 tgaccaagag caaggatctc aataactgcc agcagagaat catgaaggac tttggtctgg
181 cttacacaga gaagtgtgta gagtgccggc agagagggga ggccctgatg ggagctgcca
241 cttacaaata cctcatgaag ccctc</span>
VEPy, NCBI GenBank accession number for Sockeye is FJ226376
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;"> YFSMRLMTADWQYERAGNM"
ORIGIN
[[#sequence_226316382]] 1 tttgaaacac cactggattg gacctatcct ctggacccaa agccagagcc caagattatt
61 gggagctcag agacgagaac ccctgtggct gcccattcag tgagggctga gtgcagggag
121 aacatggtcc acgtggaagc gaagcatgac ctgctgggga tcggccagtt gatccagcta
181 gaagacctca ctttgggaga ctgccctatg tctggattcg acaatgtcaa ccaggtgctc
241 atctttgagt atccgctgca gtcatgtggc agccagctaa ggatgactac cacctccctc
301 atctacatct tcactctatt ttacaaaccc aaacctctgg caaacacccc cctcatcagg
361 acaaatgaag cgatgatcaa tattgagtgc cactatccaa ggaaacacaa tgtgagcagc
421 ctggccctga tcccaacctg gacccctttc tccgctgcga agtatgcaga ggaactcctg
481 tacttctcca tgaggctcat gactgctgac tggcagtatg agagggccgg taacatgta</span>
ERalpha, CBI GenBank accession number for Sockeye is FJ226377
<span style="font-size: 13px; margin-bottom: 0px; margin-top: 0px; overflow-x: visible; overflow-y: visible; white-space: pre-wrap; width: 50em; word-wrap: break-word; zoom: 1;">ORIGIN
[[#sequence_226316384]] 1 tgaagtgggg atggtgaaag gaggcttgcg taaggaccgc ggtgggcggc ttctcaggaa
61 ggataagcgg tattggggcc ctgctggtga cagagagaaa ccctacggtg acctggagca
121 cagggcagcg ccccctcagg acgggggtag gaacagcagc agcagcagtc tcaatggagg
181 tggagtatgg cgtgggccca gaatcaccat gcctcctgaa cagttgctgt tcctgctgca
241 gggggcagag cctccggccc tgtgttctcg tcagaaggtg gcccgcccct acacagaggt
301 caccatgatg accctgctca ccagcatggc tgacaaggag ctggtgcaca tgatcgcttg
361 ggctaagaaa gtaccaggtt tccaggagct gtctctccat gaccaggtgc agctgctgga
421 gagttcctgg ctggaggtgc tgatgatcgg actcatatgg cggtccatcc ccagccctgg
481 gaaactcatc ttcgcccagg acctcatact ggacaggagt gaaggggact gtgt</span>
Reflection: The purpose of this lab allowed us to learn the primary steps to deconstruct chemical bonds and isolate specific molecules for future studies. These methods I assume will allow us to look at gene expressions and other techniques that we can compare how different stressors, look at mechanistic responses, and even evolutionary relationships. I have to admit that I felt a little rushed, but was able to inquire with lab partners and TA to clarify certain steps. The last component I was having a hard time understanding the concept of why were finding primers before we new what stressor genes. I look forward to learning more about vitellogenin, vitelline envelope protein gamma, and estrogen receptor alpha and how it might be expressed in ph, Cu+, or other exposures that other folks have in lab. It would be neat to work with one of these groups to compare expression.
Veldhoen, N., M.G. Ikonomou, C. Dubetz, N. MacPherson, T. Sampson, B.C. Kelly, and C.C. Helbing. 2010. Gene expression profiling and environmental contaminant assessment of migrating Pacific salmon in the Fraser River watershed of British Columbia. Aquatic Toxicology 97: 212-225.