6/04/2010
Master Mix for PCR:
cDNA 1 ul
go Taq 12.5 ul
Pf .5ul
Pr .5ul
H20 10.5
95 degree - 10min
95 degree - 15sec - 40x
55 degree - 15sec - 40x
72 degree - 1min - 40x
6/01/2010
Prep PCR
Working Stock: 10uL of Primer + 90uL Water
5/28/2010
Reverse Transcription on Acid shock fish sample.
Sample: 6.2.1-6.2.8, 6.7.1, 6.7.3-7.7.8, 6.10.1-6.10.4, 6.10.6, 6.10.8
Spreadsheet for the sample Acid Shock Spreadsheet
Master Mix:
5 uL 5x Buffer (M-MLV RT Buffer) x24 = 120 ul
1.25 uL 10mM dNTPs x24 = 30
0.5 uL M-MLV RT per ug of RNA x24 = 12
5/11/2010
RNA isolation from pH treated fish

5/4/2010
RNA isolation from pH treated fish

4/15/2010
Isolated Plasmid from Oyster bacteria.

4/6/2010
Can't merge files using galaxy because galaxy is not reading the ''Herring selected file" as data so this is not listed as an option when trying to join the files together.
3/8/2010
Extract RNA and Spec'd samples

3/1/2010
Extract RNA and Spec'd the samples

2/25/2010
Extract RNA (per tube):
- 500ul of Tri-reagent in RNA tube
- Add severed fish head and grind as much as possible
- Transfer 100ul into another RNA tube
- Then add 900ul of Tri-reagent to get to 1ml
- Follow the RNA Protocol
- Take the aqueous out and spin for a couple of seconds to separate.
- Get as much aqueous out
- Then add 30ul of +H2O
- Spec'd the samples

1/25/10 4hrs
1/
1/14/10 3.5hrs
data entering for Mac
1/13/10 2hrs
1/11/2010 4.5hrs
Worked on Paper
Set alarm on fridge
1/6/2010 2 hrs
Worked on Paper
1/5/2010 2hrs
Worked on Paper
1/4/2010
Dish washing
Checking Pipette accuracy
12/8/09 4hrs
Work on GIS
12/7/09 3hrs
Work Paper
12/3/09 5hrs
| 14.8 C |
Bacteria |
Oyster |
| Control |
7.63 |
7.08 |
| CO2 |
7.19 |
7.29 |
11/30/09
| 15.2 |
Bacteria |
Oyster |
| Control |
7.69 |
7.14 |
| CO2 |
6.93 |
7.24 |
11/24 3hrs
Made gel for PCR
150uL of tae buffer
1.5g agar
1.5uL ethidium bromide
Added extra water
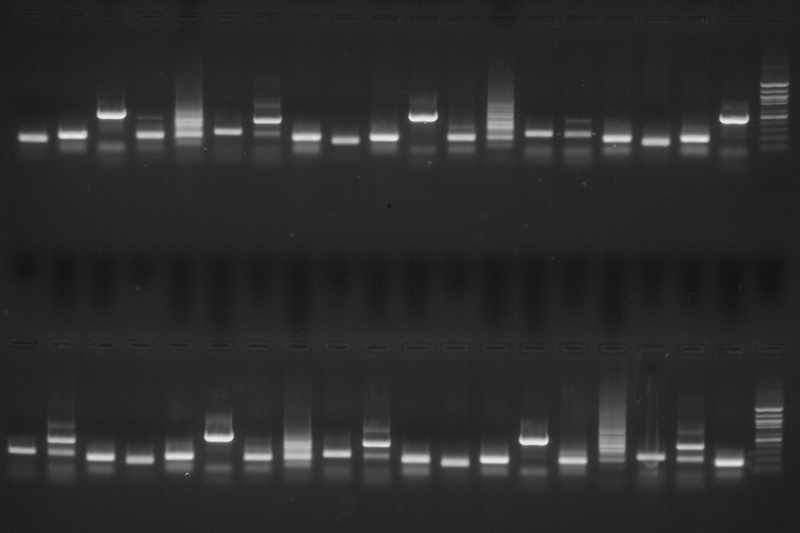
Top (left to right): A8, H7, G7, F7, E7, D7, C7, B7, A7, H6, G6, F6, E6, D6, C6, B6, A6, H5, G5, Ladder
Bottom (left to right): D10, C10, B10, A10, H9, G9, F9, E9, D9, C9, B9, A9, H8, G8, F8, E8, D8, C8, B8, Ladder
Measured pH for Oyster and Bacteria
| 14.5 C |
Bacteria |
Oyster |
| Control |
7.61 |
7.18 |
| CO2 |
7.06 |
7.21 |
11/23 4 hrs
Made gel for PCR
150uL of tae buffer
1.5g agar
1.5uL ethdium bromide
Run PCR
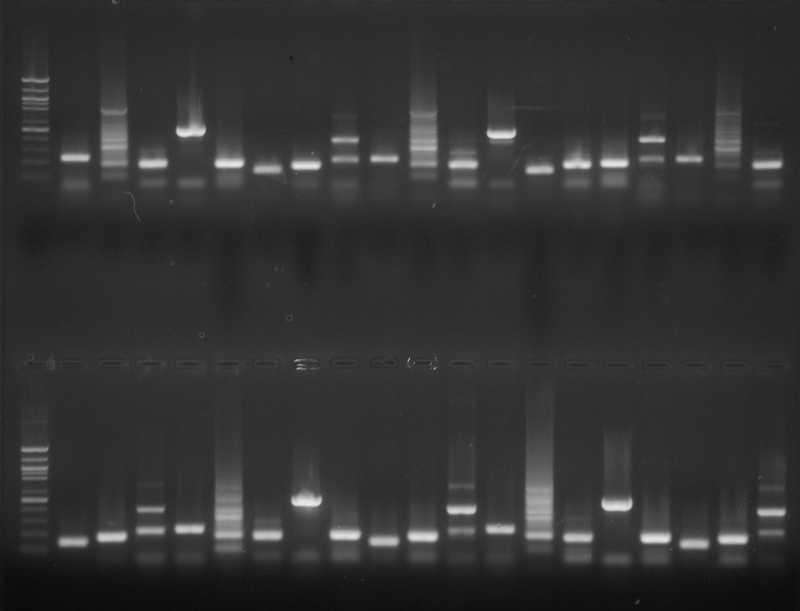
Top( from left to right)
Ladder, A1, B1, C1, D1, E1, F1, G1, H1, A2, B2, C2, D2, E2, F2, G2, H2, A3, B3, C3
Ladder, D3, E3, F3, G3, H3, A4, B4, C4, D4, E4, F4, G4, A5, B5, H4, C5, D5, E5, F5
Measured pH level of Oyster and Bacteria.
| 16.2 C |
Bacteria |
Oyster |
| Control |
7.65 |
7.16 |
| CO2 |
6.86 |
7.23 |
11/19 5.5hrs
Measured pH level for CO2 affects on Oyster and Bacteria. Spin samples for bacteria and freeze them at 20 C.
| Bacteria |
Oyster |
|
| Control |
7.56 |
7.28 |
| CO2 |
6.96 |
7.18 |
Master mix
DNA - 1uL
2x Apex - (12.5 x 14) = 175uL
Pf/Pr - 7uL
H2O - (10.5 x 14) = 147 uL
Incubator
95 degree 10mins
95 degree 10mins (X40)
60 degree 15 sec (X40)
72 degree 25 sec (X40)
72 degree 10 mins
11/17 2.5hrs
Set up 24 well electroylsis
150 mL of TAE buffer
3 g agar (need to within 2% of TAE buffer)
15 uL of ethdium bromide (1 uL for every 10mL)
Set up electrolysis gel (large), USE GLOVES!
1. 150 ml of TAE buffer
2. 3 g agar
3. heat for 3 min
4. add 11.48 g of water
5. wait 15 mins
6. add 15 ul of ethdium bromide (fridge)
5. pour into plate, rest for 35 min
| Top (left to right) |
x |
Con(6) |
CO2(6) |
x |
Con(7) |
CO2(7) |
x |
Con(9) |
CO2(9) |
x |
Con(13) |
CO2(13) |
x |
Con(14) |
CO2(14) |
x |
Con(15) |
CO2(15) |
x |
Ladder |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bottom (left to right) |
x |
Con(19) |
CO2(19) |
x |
Con(38) |
CO2(38) |
x |
Con(20) |
CO2(20) |
x |
Con(18) |
CO2(18) |
x |
Con(17) |
CO2(17) |
x |
Con(12) |
CO2(12) |
x |
Ladder |
Oyster Tank pHs -
Right Front - 7.39
Left Front - 6.85
Right Back - 7.26
Left Back - 7.05
| Bacteria |
Oyster |
|
| Control |
7.39 |
7.26 |
| CO2 |
6.85 |
7.05 |
11/16 3hrs
Cleaned container for the CO2 project
Worked on Papers
11/12 5hrs
Link Colton pics
Work on Mac's data
Download articles into Papers
11/10 3.5hrs
Work on GIS
Input data for Mac
11/9 3hrs
Link Colton pics to this page
Work on GIS
11/2 3hrs
Download anything related to Puget Sound
10/29 6hrs
Install ArcGIS and play around with it
10/27 2hrs
Import articles to Papers
10/26 3.5hrs
Import articles to Papers
10/20/2009 2hrs
Import articles to Papers
8/14/2009 5hrs
Link colton pictures to website
8/13/2009 3hrs
Colton pictures
8/12/2009 3hrs
Colton pictures
8/10/2009 4hrs
Backup wiki
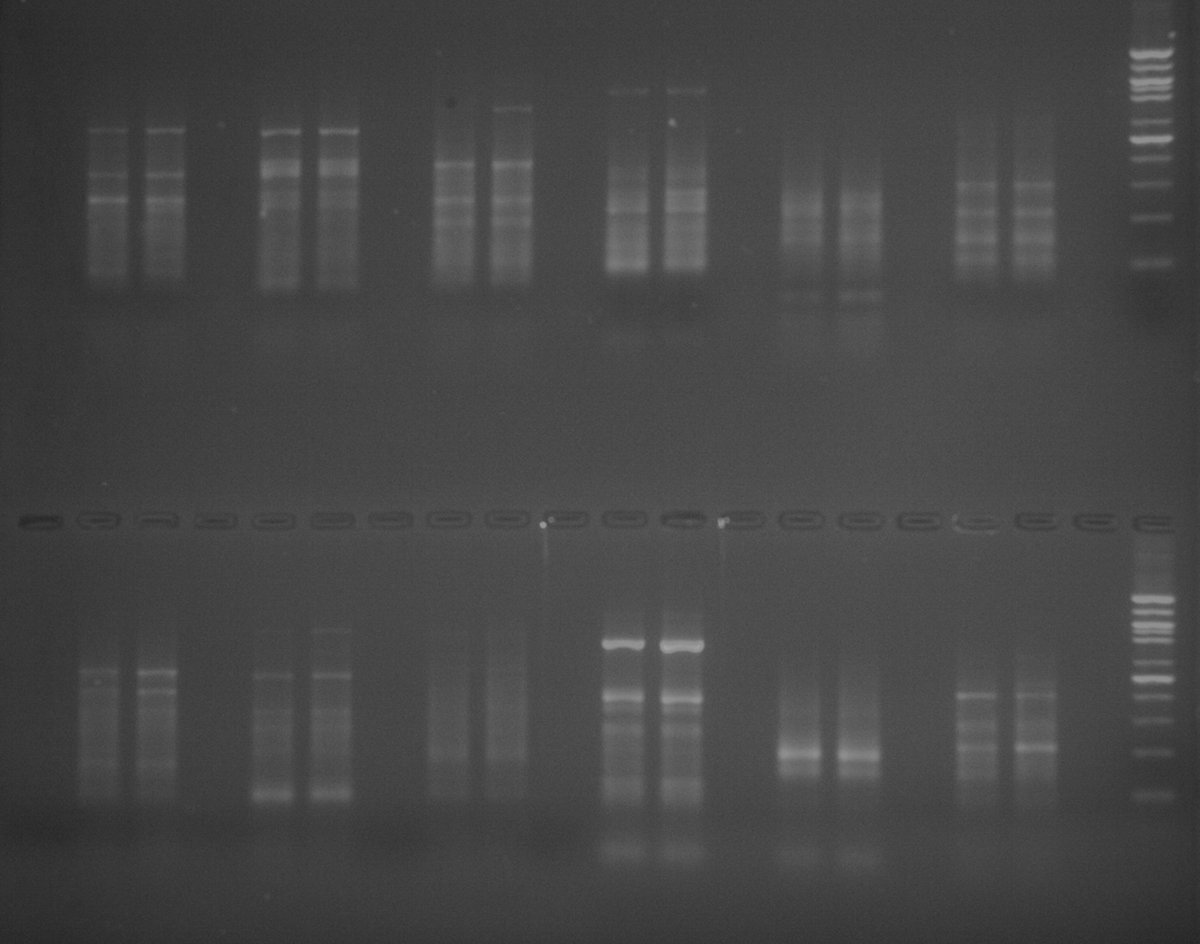
The result of PCR from last week sample.
.
From top left is Water, Water, Sea Male #1, Bay, 7, 6, 5, 4, 3, 2, 1 and ladder. At the bottom is Water, Water, Sea Male #2, Sea,, 14, 13, 12, 11, 10, 9, 8 and ladder. Since this PCR used the primer Sea RV 2, the two sea male samples are positive. From the previous PCR with Bay reverse primer, all but one sample was positive. Meaning that all but one were Bay scallops and the negative sample, #13, was a Sea scallops. So the expected result for this gel should have only sample #13 that is positive while everything else should be negative. The actual result have everything as negative even sample #13. So something went wrong with sample #13, it should've been a positive for the PCR with the Bay reverse primer or positive with the Sea primer. Beside from that the rest of the result was expected.
8/5/2009 3hrs
Incubator
95 degree 10mins
95 degree 10mins (X40)
56 degree 1min (X40)
72 degree 90 sec (X40)
Redo PCR for Hybrid samples with Sea Actin RV 2. Also Sea scallop male 1 and 2 were used for this PCR run.
The gel result of yesterday PCR. Something went wrong because every slot is negative.
8/4/2009 3hrs
Prepare PCR for Hybrid samples with Sea Actin RV 2
Each sample consist (except water samples) of 2uL of gDNA, 12.5uL Go Taq, .5uL Scallop Actin FW, .5uL Sea Actin and 9.5uL water. Water samples have an extra 2uL of water to replace the gDNA amount.
Incubator
95 degree 10mins
95 degree 10mins (X40)
56 degree .30 sec (X40)
72 degree 1min (X40)
8/3/2009
Run PCR for Hybrid samples
Top row (left to right)
Water Water Sea 7 6 5 4 3 2 1 Ladder
Bottom row
Water Water Bay 14 13 12 11 10 9 8 Ladder
From this gel, it seemed that everything is a Bay scallop except for one, sample #13. Since I used the Bay primer, everything that reacted with the primer, the positive, was Bay scallop.
Incubator
95 degree 10 mins
95 degree 10 mins (X40)
53 degree .30 sec (X40)
72 degree 1min (X40)
Prepare PCR for Hybrid samples with Bay Actin RVO
Each sample consist (except water samples) of 2uL of gDNA, 12.5uL Go Taq, .5uL Scallop Actin FW, .5uL Bay Actin RVO and 9.5uL water. Water samples have an extra 2uL of water to replace the gDNA amount.
| Sample |
gDNA |
2x Go Taq |
Scallop Actin FW |
Bay Actin RVO |
Water |
| 1 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 2 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 3 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 4 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 5 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 6 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 7 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 8 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 9 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 10 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 11 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 12 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 13 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| 14 |
2 |
12.5 |
.5 |
.5 |
9.5 |
| Sea Scallop |
2 |
12.5 |
.5 |
.5 |
9.5 |
| Bay Scallop |
2 |
12.5 |
.5 |
.5 |
9.5 |
| Water |
0 |
12.5 |
.5 |
.5 |
11.5 |
| Water |
0 |
12.5 |
.5 |
.5 |
11.5 |
| Water |
0 |
12.5 |
.5 |
.5 |
11.5 |
| Water |
0 |
12.5 |
.5 |
.5 |
11.5 |
Note: To find 2x Go Taq eq: C1*V1=C2*V2
Set up 12 well electroylsis
150 mL of TAE buffer
1.5 g agar (need to within 1% of TAE buffer)
15 uL of ethdium bromide (1 uL for every 10mL)
Set up electrolysis gel (small), USE GLOVES!
1. 100 ml of TAE buffer
2. 1 g agar
3. heat for 3 min, mixing every min
4. add 6 ul of ethdium bromide (fridge)
5. pour into plate, rest for 35 min
7/31/2009
Updated species doc on google
Prepared sample of hybrid scallops and used chelax on sample
7/30/2009
7/17/2009 4hrs
Chelex seaweed and cocopods
7/13/2009 2.5hrs
Update primerdatabase
Link pictures to colton collection
6/17/2009 3.5hrs
Took colton pictures
6/16/2009 3hrs
Link colton pictures to genefish
Fixed the colton collection page
6/12/2009
Journal of Shellfish Research June 2002-April 2009
Marine Biotechnology Feb 2004-June 2009
Journal of the World Aquaculture Society March 2002-March 2005
World Aquaculture March 2008-March 2009
6/5/2009
Took colton pictures
5/22/2009 2hrs
Took Colton Pictures.
5/21/2009 5.5hrs
Continue with citation and worked on ocean acidification
5/19/2009 5.5hrs
Took pictures for colton collection
Did citation for article
5/14/2009 4.5hrs
Did Realtime PCR for Oysters Dermo sample
Worked on ocean acidification sheet
5/12/2009 5hrs
Made new sample for the Dermo experiment.
Filled in ocean acidification sheet
5/7/2009 3hrs
Set up Realtime PCR for Oysters Dermo experiment.
Filled in ocean acidification sheet
5/5/2009 4hrs
Spect the oyster samples
Input data of oyster collection on google
5/1/2009 4hrs
Continued ethanol precipitation for oyster sample.
4/30/2009 4.5hrs
Spect the oyster samples and did ethanol precipitation for the samples.
Mixture for each sample:
DNA 100 mL
3M NaOAc 10 mL
100 l EtOH 220 mL
Incubate 20c
4/28/2009 3.5hrs
Took more photos for the colton collection and upload them.
Filled in ocean acidification sheet on google.
4/23/2009 3hrs
Looked for information about ocean acidification on shellfish or invertebrates.
4/22/2009 4hrs
Set up Realtime PCR for Oysters Dermo experiment.
Here's the set up of the samples
4/21/2009 2.5hrs
Uploaded recent photos to colton page and looked at oyster samples.
4/14/2009 3hrs
Took more pictures for colton pictures
Started the education database on google doc
4/10/2009 -
Ran Ronnie's PCR samples on a gel.
Lane 1 - 100bp ladder
Lane 2 - Sea
Lane 3 - Sea Rv H2O
Lane 4 - Bay
Lane 5 - Bay Rv.2 H2O
Lane 6 - B x S, Bay Rv
Lane 7 - B x S, Sea Rv
Lane 8 - S x B, Bay Rv.
Lane 9 - S x B, Sea Rv.
4/8/2009
Run PCR for Bay/Sea Scallop with Scallop Actin Pf
Lane 1 Ladder
Lane 2 Bay, Bay_Rv2
Lane 3 H2O, Bay_Rv2
Lane 4 Sea, Sea_Rv3
Lane 5 H2O, Sea_Rv3
There are no bands in the water sample so that means nothing is contaminated. Sea and Bay gDNA reacted with their reverse primer so they have bands in their lanes. But Sea and Bay band appear in different spot of the gel, this could be used to identify the two species.
Set up PCR for Bay/Sea Scallop Hybrid with Scallop Actin Pf
| Test Tube |
gDNA |
2x GoTaq |
Pf |
Pr |
H2O |
| Bay, Bay_Rv2 |
4ul |
12.5 |
1 |
1 |
6.5 |
| Bay x Sea, Bay_Rv2 |
4ul |
12.5 |
1 |
1 |
6.5 |
| Bay x Sea, Sea_Rv3 |
4ul |
12.5 |
1 |
1 |
6.5 |
| Sea, Sea_Rv3 |
4ul |
12.5 |
1 |
1 |
6.5 |
| Sea x Bay, Bay_Rv2 |
4ul |
12.5 |
1 |
1 |
6.5 |
| Sea x Bay, Sea_Rv3 |
4ul |
12.5 |
1 |
1 |
6.5 |
| H2O, Bay_Rv2 |
0 |
12.5 |
1 |
1 |
10.5 |
| H2O, Sea_Rv3 |
0 |
12.5 |
1 |
1 |
10.5 |
95 C - 45 sec.
55 C - 45 sec.
72 C - 1.30 min.
72 C - 10 min.
4 C ----
4/7/2009
Set up PCR for Bay/Sea Scallop with Scallop Actin Pf
| Test Tube |
1 - Bay, Bay_Rv2 |
2 - H2O, Bay_Rv2 |
3 - Sea, Sea_Rv3 |
4 - H2O, Sea_Rv3 |
| gDNA |
4 ul |
0 ul |
4 ul |
0 ul |
| 2x GoTaq |
12.5 ul |
12.5 ul |
12.5 ul |
12.5 ul |
| Pf |
1 ul |
1 ul |
1 ul |
1 ul |
| Pr |
1 ul |
1 ul |
1 ul |
1 ul |
| H2O |
6.5 ul |
10.5 ul |
6.5 ul |
10.5 ul |
4/1/2009
Work on OpenOffice database
Run PCR for Bay/Sea Scallop
Lane 1 Ladder
Lane 2 Bay Rv.2
Lane 3 Bay H20
Lane 4 Sea Rv.3
Lane 5 Sea H20
3/13/2009
Play with OpenOffice and entered some database
Set up PCR for Bay/Sea Scallop
Test Tube
| 1 - Bay, Bay_Rv2 |
2 - H2O, Bay_Rv2 |
3 - Sea, Sea_Rv3 |
4 - H2O, Sea_Rv3 |
|
| gDNA |
1 |
0 |
1 |
0 |
| 2x GoTaq |
12.5 |
12.5 ul |
12.5 ul |
12.5 ul |
| Pf |
1 |
1 ul |
1 ul |
1 ul |
| Pr |
4 |
0 ul |
4 ul |
0 ul |
| H2O |
6.5 |
10.5 ul |
6.5 ul |
10.5 ul |
95 C - 10min.
95 C - 45sec.
50 C - 45sec.
72 C - 1.5min.
72 C - 10min.
3/12/2009
Run PCR for Kelvin's Bay/Sea Scallop
Lane 1 Ladder
Lane 2 Bay
Lane 3 Sea
Lane 4 Bay (Female) x Sea (Male) Sea Rv
Lane 5 Bay (M) x Sea (F) Bay Rv
Lane 6 Bay (M) x Sea (F) Sea Rv
Lane 7 Bay (F) x Sea (M) Bay Rv
Lane 8 H2O Sea Rv
Lane 9 H2O Bay Rv
3/11/2009
Scan Papers
Took colton pictures
3/6/2009
Run PCR - Remainder Bay/Sea Scallop samples
Lane 1 Ladder
Lane 2 Bay Actin Primer Reverse with H2O
Lane 3 Bay Actin Primer Reverse with Bay x Sea Scallop gDNA
Lane 4 Bay Actin Primer Reverse with Bay Scallop gDNA
Results:
Bay Reverse Primer reacted with Bay gDNA but not with hybrid. There's nothing going on in Lane 2 with the water sample.
3/5/2009
Took shellfish pictures
Run PCR - Bay/Sea Scallop samples
(From the left)
Lane 1 Ladder
Lane 2 Sea Actin Primer Reverse with Sea Scallop gDNA
Lane 3 Sea Actin Primer Reverse with Bay Scallop gDNA
Lane 4 Sea Actin Primer Reverse with H2O
Lane 5 Sea Actin Primer Reverse with Sea x Bay Scallop gDNA
Lane 6 Sea Actin Primer Reverse with H2O
Lane 7 Sea Actin Primer Reverse with Bay x Sea Scallop gDNA
Lane 8 Sea Actin Primer Reverse with H2O
Lane 9 Bay Actin Primer Reverse with Sea Scallop gDNA
Lane 10 Bay Actin Primer Reverse with H2O
Lane 11 Bay Actin Primer Reverse with H2O
Lane 12 Bay Actin Primer Reverse with Sea x Bay Scallop gDNA
Results:
The Sea Reverse Primer didn't react with the Sea Scallop DNA but it reacted with Bay Scallop DNA which is very unusual. Sea Reverse Primer didn't react with the hybrid samples. There is also something in the water for Lane 10 and 11 but for the other two lanes of water it looks normal.
2/27/2009
PCR - Bay/Sea Scallop samples
PCR work up here.
95 C - 10 min.
95 C - 45 sec.
55 C - 45 sec.
72 C - 1 min.
72 C - 10 min.
4 C ----
2/20/2009
Took shellfish collection pictures
Label photos
2/19/2009
Took snapshot of shellfish collection
Worked on shellfish collection
Reconciled paper
Did snp
2/18/2009
Took snapshot of shellfish collection
2/13/2009
Worked on fish collection
2/12/2009
Worked on snp form, end at 1234 so anything above 1234 is not done yet
Worked on fish collection
2/11/2009
Worked on snp form
Reconciled paper
Worked on fish collection
2/6/2009
Worked on fish collection
2/5/2009
Worked on the fish collection
2/4/2009
Learned how to reconciled paper and work the autoclave. Did some reconciling and scanning.