I ran another round of qPCR on my DNA and posted all my results on the oyster group spreadsheet. Good! -
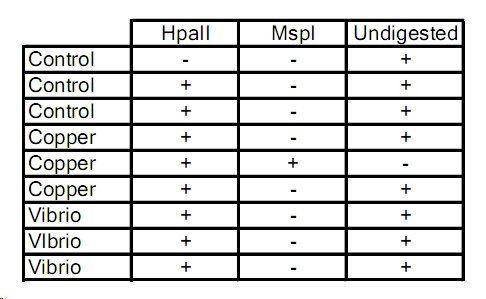
+ denotes DNA that amplified past threshold
So which one is unmethylated? copper? -
12/6/2010
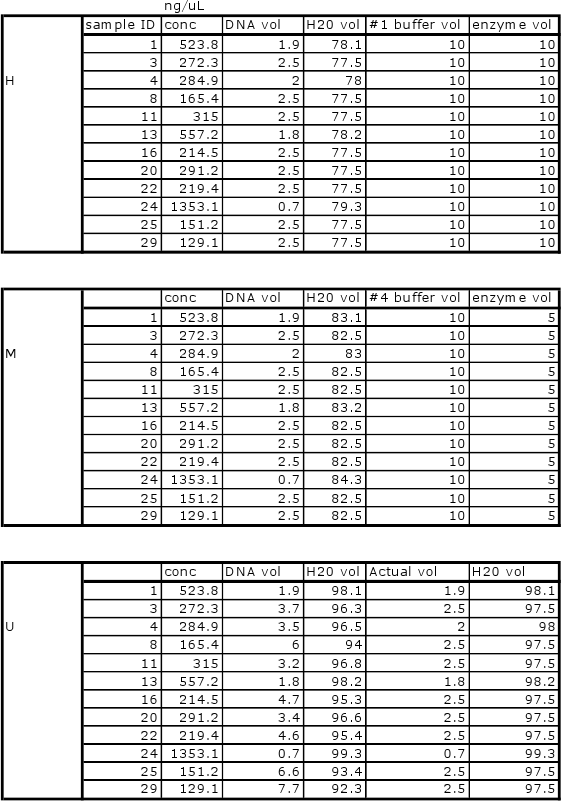
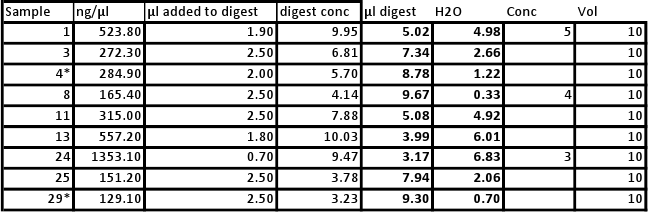
Will be conducting new qPCR due to error in first round of samples. All will be normalized as follows:
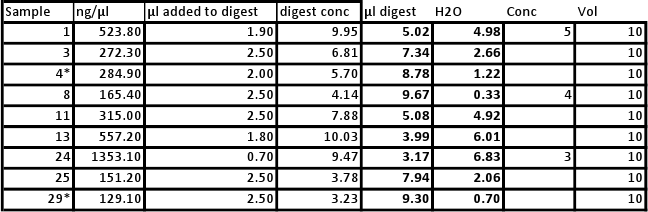
12/5/2010
Results of qPCR need to be reviewed with Mac.
Bolded samples were selected for overnight digestions with methylation-specific restriction enzymes. Samples 16, 20, 22 could not be run due to shortage of enzyme.
Heat bath set to 37C and samples left overnight (15hrs). Bath temperature increased overnight to 45C. Enzymes stopped (H=65C 20min, M=80C 20min)
Samples spun max for 30 sec and placed in 60C water bath for 1 minute prior to Nanodrop.
11/28/2010
Unfortunately due to weather related events I was unable to get any tissue processing done this week. So instead I've spent my time doing literature searches for various stress response papers in invertebrates. Hopefully this week I will be able to complete all the extractions, run all the qPCRs, and begin statistical analysis. Yikes!
11/19/2010
Today we killed a bunch of oysters! They were shucked and samples were taken of hemolymph, gill, and mantle. The copper treated oysters had turned GREEN. Only one oyster was obviously dead before we got to it, and that one was probably dead when it was collected. Now all the samples are sitting pretty in the freezer waiting for processing. I'm hoping to get extractions done by Tuesday, and at least have all my qPCRs set up to go by Wednesday. We'll see how that goes. I need to get the primers and enzymes I need from Mac, dot blot might have to wait.
11/16/2010
Today we added the CuSO4 to the treatments and prepped for sampling on Friday. I put together a labeling scheme for the cryotubes to make sampling easier:
BB – Big beef
BF – Belfair
<line> – control
Cu – copper treated
V – vibrio treated
CuV – copper and vibrio treated
A/B – used to denote oysters in same bucket
M – mantle
G – gill
H – hemolymph
A total of 192 tubes were labeled.
It was good to get this prep work out of the way so hopefully sampling will go easier. I’m probably going to miss a large portion of it due to previous commitments, but I’m hoping to get a good chunk done with the rest of the group. It’s been a crazy week!
11/9/2010
Notes from Oyster Project Meeting:
Collections will be made from two sites: Big Beef Creek (pristine), and Lynch Cove (urban). These two groups will be treated with vibrio and/or copper for response. Oysters will be acclimated over the weekend in buckets held in 12˚C water bath. Treatments will be applied as follows:
Oysters (n=64) 32 from each site.
Copper treated, 2 replicates of 4/site
Vibrio treated, 2 replicates of 4/site
Copper+Vibrio, 2 replicates of 4/site
Control, 2 replicates of 4/site
Copper will be added to treatements 11/16/2010, Vibrio added 11/18/2010, Oysters sacrificed and samples taken (mantle, gill, hemolymph) 11/19/2010.
Samples will be subjected to protein, RNA, DNA extractions and quantified via pPCR, dotblot.
I have chosen to look at methylation patterns of oysters in the various treatments. Not sure if it’s best to do whole genome, specific known sequences or both. I will have to talk to Mac and see what she has to say. The timeline on this project is going to be fairly tight, but I think there are enough of us to accomplish everything we are setting out to do. I need to start reading methylation papers, etc to find likely portions of DNA to look at.
11/2/10
Summary
Lab exercise #5 was performed on 2 November 2010. qPCR was prepped and performed on previously extracted cDNA samples. Dot blot of DNA methylation from previous week was developed.
Materials and Methods
- Blocking soln (prepared last week)
- Dot blot membrane (prepared last week)
- 5-MeC antibody
- TBS-T
- ddH2O
- Secondary antibody soln
- Chromogenic substrate
- Plastic dish
- Rotary shaker
- Serological pipettes
- Optical PCR plates
- 1.5ml microfuge tubes
- Nuclease-free H2O
- Micropipettes (1-1000µl)
- Micropipette tips
- Opticonthermal cycler
- 2X Immomix master mix
- SYTO-13 dye
- Student-designed primers
- microfuge tube racks
- cDNA (prepped in previous labs)
- Microfuge
- Gloves, kim wipes, ice buckets, etc.
Dot blot development
1. Membrane placed in plastic dish on rocker and covered in 10ml blocking soln.
2. Incubated at rev/sec for 30 min.
3. Blocking soln decanted.
4. Membrane rinsed w/ 20ml H2O for 5 min then decanted. Repeat.
5. Primary antibody soln prepared:
a. Blocking soln 10ml
b. 5-MeC antibody 2µl
6. Incubated membrane w/ 10ml primary antibody soln for 1hr @ 1 rev/sec.
7. Primary antibody decanted.
8. Membrane washed w/ 20ml TBS-T for 5 min and decanted X4.
9. Membrane incubated in 10ml secondary antibody soln for 30 min and decanted.
10. Membrane washed w/ 20ml TBS-T for 2min and decanted X3.
11. Membrane rinsed w/ 20ml H2O for 2min and decanted X3.
12. Membrane incubated in 5ml chromogenic substrate until color developed.
13. Membrane rinsed w/ 20ml H2O for 2min X3.
14. Blot dry w/ filter paper.
qPCR
1. Master mix prepared as follows:
For 1 rxn:
Component Volume Final Conc.
Immomix Master Mix 25µl 1X
SYTO-13 Dye (50µM) 2µl 2µM
Primer F (10µM) 2.5µl 2.5µM
Ultra-pure water 16µl -
Master mix made for 7 rxns.
2. 48µl master mix added to each of 6 PCR wells.
3. cDNA and controls as follows:
Tube Contents (2µl)
1 cDNA
2 cDNA
3 RNA
4 RNA
5 Water
6 Water
4. Wells capped and placed on ice.
5. qPCR plate run by SAFS lab tech.
RESULTS
Dot Blot: (RW seastar)
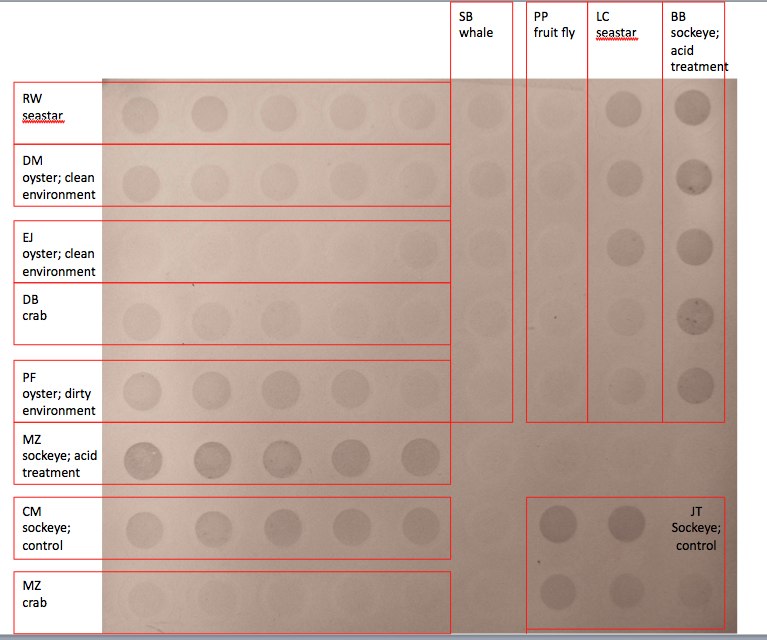
It is clear that the less dilute DNA produced a stronger blot on the membrane with the more dilute samples decreasing in strength with each dilution. My seastar (which had been exposed to pesticides) showed similar amounts of methylation to the ‘dirty oyster’, though the oyster showed stronger methylation across dilutions overall. My seastar sample also showed overall less methylation than that of the other seastar sample.
qPCR:
My qPCR produced positive results for cDNA with a Ct of 26.96 for sample 1, and 26.63 for sample 2. Melting curve analysis indicated that amplification was specific to target cDNA. RNA and water controls both produced negative results.
CONCLUSION
It’s interesting to note that my seastar showed relatively less methylation than the other seastar sample. There are several possibilities for this observed difference; 1. The seastar samples are from two different individuals exposed to different levels of stress. 2. The samples are from the same seastar but at different exposure times. 3. The samples were from two different tissues within the same seastar which methylated differentially in response to the stressor. 4. Operator error. I would like to think that it’s not #4.
The qPCR results demonstrated that the primer I chose worked! This is very encouraging. The difference in Ct between the two cDNA samples is most likely a result of pipetting error (I did notice slight differences in the quantities of fluid that made it out of the pipette tip.) My negative controls were also a resounding ‘success’. This indicates that there was no DNA contamination in my sample (or any DNA contamination was not targeted by my primers.) To determine if GSTO expression as quantified by this procedure indicated a high or low stress environment, an oyster control would need to be subjected to the same set of primers and compared to the above results.
REFLECTION
I found this lab to be rather satisfying as it was the culmination of a number of weeks’ worth of work. I do wish we had time to quantify the methylation dot blot, but hopefully we’ll get a chance to do that later in the quarter with our class experiment. The qPCR was especially exciting as I discovered that the primers I designed worked. Designing primers is such a useful skill and I was glad to get a chance to try it.
A few additional questions come to mind regarding the results of this lab. In the case of the dot blot I began to wonder what sort of differential methylation you would expect to see in different tissues from the same organism. How do you know what tissues might methylate under a particular stress? Do you just test a bunch of different tissues in stressed and control organisms for comparison? Or do we tend to target particular tissues specifically? For the qPCR I wondered what the Ct actually means in real number terms. Can you say how many copies of a particular gene were present from the Ct? Is it all relative? Does is really matter exactly how much of something was there?
10/26/10
Summary
Lab exercise #4 performed on 26 October 2010. Gel electrophoresis was performed to check amplification of cDNA from Crassostrea gigas extracted over the previous labs. Methylation of seastar DNA exposed to Roundup® was also measured using dot blot and chromogenic immunodetection methods.
Materials and Methods
- 10-1000µl micropipettes
- 10-1000µl micropipette tips
- Gel box
- 1X TAE Buffer
- Hyper-I Ladder (Bioline™)
- cDNA
- UV transilluminator
- Seastar DNA
- 1.5ml microfuge tubes
- ddH2O
- 20X SSC
- 72 well dot blot manifold
- Nylon membrane
- Filter paper
- Beakers
- Hotplate
- Microfuge tube ‘floatie’
- Centrifuge
- Denaturation buffer
- Neutralization buffer
- Plastic wrap
- Blocker/Diluent
- Serological pipettes
- Falcon tubes
- Plastic dish
- Rocker
- 5-MeC antibody
- Chromogenic substrate
Gel electrophoresis
1. Placed gel in box and cover with 1X TAE buffer.
2. Removed combs.
3. Loaded 5µl Hyper-I ladder to lane 1.
4. Loaded 25µl cDNA on bottom portion of Gel 1 as follows:
Lane Sample
6 1
7 2
8 3
9 4
5. Gel run as follows: 60min @100v, 15min @ 150v, 20min @ 85v
6. Visualized on UV transilluminator
Methylated cytosine dot blot procedure
DNA dilutions
1. Five dilutions of seastar DNA prepared in 1.5ml microfuge tubes as follows:
Dilution Target conc. µl H2O µl 20X SSC µl 50ng/µl DNA
1 0.8ng/µl 124 60 16
2 0.4ng/µl 132 60 8
3 0.2ng/µl 136 60 4
4 0.1ng/µl 138 60 2
5 0.05ng/µl 139 60 1
Dot blot
1. Nylon membrane cut to fit 72 well manifold.
2. Nylon membrane soaked in 6X SSC for 10min.
3. Filter paper cut to size of nylon membrane and wet in 6X SSC.
4. Manifold assembled w/ membrane on top of filter paper.
5. DNA denatured in boiling water bath for 10min and placed on ice.
6. 500µl 6X SSC added to each manifold well and run through with vacuum.
7. DNA spun down for 5sec.
8. 200µl DNA added to wells as follows:
Well Sample conc.
A1 0.8ng/µl
A2 0.4ng/µl
A3 0.2ng/µl
A4 0.1ng/µl
A5 0.05ng/µl
9. Samples filtered through manifold.
10. Filter paper cut to size and soaked in denaturation buffer.
11. Filter paper cut to size and soaked in neutralization buffer.
12. Once filtered, membrane transferred to paper soaked in denaturation buffer and left for 10min.
13. Membrane then transferred to neutralization soaked filter and left for 5min.
14. Membrane transferred to dry filter paper and dried in hood.
15. Dried blot wrapped in plastic wrap and set DNA side down on UV transilluminator for 2min at 120kJ.
Chromogenic immunodetection
1. Prepared 20ml blocking soln.
- 14ml ddH2O
- 4ml Blocker/Diluent (A)
- 2ml Blocker/Diluent (B)
2. Membrane placed in 10ml blocking soln in covered dish.
3. Incubated for 30min on shaker @ 1rev/sec.
4. Blocking soln decanted
5. Membrane rinsed w/ 20ml water for 5min. Repeat.
6. 10ml Primary antibody soln prepared (1:5000 dil.)
- 10ml blocking soln
- 2µl 5-MeC antibody
7. Membrane incubated w/ 10ml PAS 1hr.
8. Decanted and washed for 5min w/ 20ml TBS-T. Repeat 3X.
9. Membrane rinsed w/ 20ml water for 2min. Repeat 2X.
10. Membrane incubated w/ 5ml Chromogenic substrate until developed.
11. Rinsed w/ 20ml water for 2min. Repeat 2X.
12. Membrane dried on clean filter paper.
Results
Gel visualization produced strong bands between 400-600bp. Some weak banding was also observed around 200bp. Controls also showed banding under 200bp, though this may be due to primer dimers. I was unable to upload the gel image into this notebook. Dot blot was not completed by the end of lab, so results from the procedure are yet to be determined.
Conclusion
It appears that PCR was successful indicating expression of cytochrome oxidase P450 in the C. gigas source tissue. qPCR and use of a comparative control are needed to determine if expression in the sample tested are relatively high or low. High expression is likely due to oxidative stress, low expression may indicate absence of a stressor or impaired response to stress. When the dot blot results are in we will hopefully be able to draw conclusions as to the methylation-state of the Roundup® seastar.
Reflection
I was glad to practice my gel-pipetting technique in this lab. I think it was one of the first times that my shaky hands didn’t get PCR product all over the place. The dot blot procedure was interesting, but seemed a bit involved. I’m not quite sure what we’re expecting to see either. As I had a seastar soaked in Roundup® would I expect it to be more, or less methylated? Is it simply the darkness of the dots relative to each other that will provide information? Or is it possible to quantify the level of methylation by the dots (like a pH strip)? It will be interesting to see what our results bring next week. I think these techniques will also be very helpful when it comes to designing our projects for the rest of the quarter.
10/19/10
Summary
Lab exercise #3 conducted on 19 October 2010. RNA extracted in previous labs from the gill tissue of C. gigas was subjected to reverse transcription in preparation for end point PCR. PCR was run to amplify the resulting cDNA and an agarose gel was prepped for visualization of the product.
Materials and Methods
- Micropipettes (1-1000µl)
- Sterile filter pipette tips (1-1000µl)
- Tip waste jar
- PCR tubes (0.5ml; thin walled)
- RNA samples
- M-MLV reverse transcriptase
- M-MLV 5X reaction buffer
- Oligo dT
- dNTPs
- Nuclease free water
- Thermal cycler
- Microfuge tube racks
- PCR tube racks
- Ice buckets
- Kim wipes
- 1.5ml microfuge tubes (RNAse free)
- cDNA
- dNTPs
- 2X GoTaq Green Master Mix
- Primers
- 1L flask
- Agarose
- 1X TAE
- Ethidium bromide
- Microwave
- Gel rigs
- Gloves, safety glasses, etc.
Reverse transcription protocol
1. RNA stock mixed by inverting.
2. Add 5µl RNA to 0.5ml PCR tube.
3. Add 1µl oligo dT
4. Add 4µl nuclease free water
5. Incubate for 5min at 70ºC in thermocycler.
6. Immediately place on ice.
7. Briefly centrifuge tube.
8. Add 5µl M-MLV 5X reaction buffer.
9. Add 1µl M-MLV RT.
10. Add 4µl nuclease free water.
11. Incubate for 60min at 42ºC in thermocycler then inactivate at 70ºC for 3mins.
12. Spin down tube in centrifuge.
13. Place on ice.
PCR protocol
1. Make master mix in 1.5ml microfuge tube.
- 250µl GoTaq®Green Master Mix, 2X
- 15µl cytochrome oxidase P450 forward primer
- 15µl cytochrome oxidase P450 reverse primer
- 108µl nuclease free water
2. Pipette 48µl master mix into each of 4 0.5ml PCR tubes.
3. Add 2µl of each template to tubes as follows:
- Tube 1 – cDNA
- Tube 2 – cDNA
- Tube 3 – Nuclease free water
- Tube 4 – Nuclease free water
4. Spin tubes to pool liquid and run in thermocycler as follows:
- Denaturation – 95ºC – 5min – 1 cycle
- Denaturation – 95ºC – 30sec
- Annealing – 55ºC – 30sec
- Extension – 72ºC – 90sec – 40 cycles
- Final extension – 72ºC – 3min – 1 cycle
- Hold – 4ºC
Agarose gel pouring procedure
1. Weigh 2g agarose and mix with 150ml 1X TAE in 1L flask.
2. Microwave solution until clear.
3. Briefly cool.
4. Add 12µl EtBr and mix.
5. Pour gel and add combs.
6. Once set, remove gel from box, wrap in plastic wrap and aluminum foil and hold in refrigerator.
Results
As this is an ongoing lab, no results have yet been produced for this portion.
Conclusions
No conclusions can yet be drawn from the work accomplished in lab 3.
Reflection
I’m glad I got a chance to perform a reverse transcription on the RNA we extracted. I have never run this procedure before and I’m curious to see if I’ll get product when we run the gels next week. I just wonder how much our RNA might have degraded in the couple weeks it took to finally convert it into cDNA? I know that it was held at -80ºC for most of that time, but will the repeated thawing it underwent during our past couple labs prove to be too much? We talk about how fragile RNA is, but how fragile is it really? If we can mess with it as much as we have and still get useable results, I wouldn’t consider it to be that sensitive. I also wonder how long one can hold a gel after making it? I have always used gels I have made immediately. What is the ‘shelf life’ of a gel?
10/12/10
Summary
Performed lab exercise #2 on 12 October 2010. Protein extracted during lab exercise #1 was run on an SDS-PAGE gel which separates proteins by molecular weight, and was visualized. Extractions were completed on RNA prepped during exercise #1 and quantified with a spectrophotometer.
Materials and Methods
- Micropipettes 1-1000µl
- Sterile pipette tips 1-1000µl
- Sterile gel loading tips
- 1.5ml centrifuge tubes (RNase free)
- 1.5ml screw cap tubes
- Microcentrifuge tube rack
- Tube ‘floatie’ (technical term)
- Heating block
- Glass container for heating water w/ ‘floatie’
- Protein gel box
- SDS-PAGE gels
- Staining trays
- Platform rocker
- Plastic wrap 2X SDS reducing sample buffer
- Protein ladder marker
- Coomassie stain
- Gel running buffer acetic acid
- Light box
- Digital camera
- Vortex
- Microcentrifuge
- Tri-Reagent
- Liquid waste container
- Ice bucket
- Hot water bath
- Nanodrop spectrophotometer
- RNase free water
- Chloroform
- Isopropanol
- 0.1% DEPC treated water
- 75% ethanol
- Gloves, etc.
SDS-PAGE gel protocol
1. Water boiled on hotplate.
2. Protein labeled ‘RW’ thawed and inverted several times.
3. 15µl protein extract added to 1.5ml screw top tube along with 15µl 2X reducing sample buffer. Protein stock returned to -20C
4. Sample flicked and centrifuged for 10 sec.
5. Tube placed in boiling bath for 5 min.
6. Centrifuge tube for 1 min at maximum speed.
7. Load entire sample into gel lane.
8. Put lid on gel box and run gel for 45 min at 150V.
9. Add enough Coomassie stain to tray to cover gel.
10. Disconnect power from gel and gently remove gel from cassette.
11. Trim wells from top of gel and mark orientation with notch.
12. Place gel in stain tray.
13. Run rocker for 5 mins.
14. Pour stain back into original container
15. Rinse gel briefly with 10% acetic acid. Dispose liquid down drain.
16. Add enough 10% acetic acid to tray to cover gel. Incubate for 15 mins. Replace fluid and repeat until bands become clearly visible.
17. Take photo of the gel.
RNA extraction (con’t)
1. Turn on heating block to 55˚C.
2. Incubate tissue sample at RT for 5 mins.
3. In fume hood, add 200µl chloroform to sample.
4. Vortex for 30 secs.
5. Incubate at RT for 5 mins.
6. Spin in refrigerated microfuge at maximum speed for 15 mins.
7. Transfer clear aqueous phase of tube to fresh tube.
8. Properly dispose of liquid in spun tube as well as tube itself.
9. Add 500µl isopropanol to tube with aqueous phase
10. Vortex briefly and incubate at RT for 10 mins.
11. Spin in refrigerated microfuge at maximum speed for 8 mins.
12. Remove supernatant being careful not to remove the pellet.
13. Add 1ml 75% EtOH to pellet. Vortex to dislodge pellet.
14. Spin in refrigerated microfuge at 7500g for 5 mins.
15. Remove all supernatant. Allow EtOH to evaporate from opened tube (no more than 5 mins.)
16. Resuspend pellet in 0.1% DEPC water and mix until pellet dissolves.
17. Incubate at 55˚C for 5 mins.
18. Remove tube and flick to mix. Place on ice.
19. Quantify with Nanodrop spectrophotometer.
RNA quantification
1. Pipette 2µl 0.1% DEPC water onto Nanodrop pedestal and set blank.
2. Pipette 2µl RNA sample onto Nanodrop pedestal and measure.
3. Values recorded, pedestal wiped clean and labeled sample ‘RW’ returned to -80˚C.
Results
Unable to upload image of gel. Lane showed 3 strong bands with lighter banding surrounding.How did your sample compare to others? -
RNA quantification results:
Nanodrop absorbance at 260nm = 747.8
260/280 ratio = 2.03
260/230 ratio = 1.47
Proteins of interest & Primer design
Final protein chosen for analysis:
GSTO - hydrocarbon and pesticide response
Accession number - AJ557141
Primer pair 3
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
ACAGGTTCCGCCTGTCCGGA |
Plus |
20 |
49 |
68 |
59.83 |
65.00% |
| Reverse primer |
ACCCGCCCCAGTGGGTTCTT |
Minus |
20 |
230 |
211 |
60.04 |
65.00% |
| Product length |
182 |
||||||
Conclusions
The SDS-PAGE gel showed at least three strong bands of protein in the sample. Some protein extract was lost during the gel pipetting process which may have decreased the intensity of some of the weaker bands. If this were part of a larger project I would likely rerun the samples.
The RNA extraction produced a good absorbance with a nice curve on the Nanodrop (compared to what I’m used to seeing). The ratios, however indicated that contamination of the samples has taken place, either phenol, ethanol, salt or all of the above.
Reflection
The SDS-PAGE gel was very interesting to run as I have never used a pre-fab gel cassette before. I also realized I need to improve my gel pipetting technique, something that I always have trouble with. The results of the RNA extraction were a bit disappointing as my Nanodrop ratios indicate that my sample is likely contaminated with various junk. Though I’m not sure how detrimental the ratios I got would be to actual RNA amplification. Maybe just bad for QPCR? I’m also excited about the protein I have decided to look at with my newly designed primers. GSTO, or ‘omega class glutathione S-transferase’ is a protein that has been found to be expressed (primarily in the digestive gland) of C. gigas when exposed to hydrocarbon stress. I believe that this will make for an interesting line of inquiry for my project.
10/5/10
Summary
Performed lab exercise #1 on 5 October 2010. Two gill tissue samples from a single Pacific Oyster (Crassostrea gigas) individual were prepped for protein and RNA extractions/processing. A Tri-reagent was used for the RNA extraction procedure. Protein concentrations were determined using a Bradford protein assay. Potential primers were sourced from NCBI website to amplify protein coding regions of C. gigas genome.
Materials and Methods
- Micropipettes 1-1000µl
- Sterile pipette tips 1-1000µl
- 1.5ml centrifuge tubes (RNase free)
- Sterile tube pestles
- Vortex
- Microcentrifuge
- Tri-Reagent
- Bradford assay reagent
- CelLytic MT
- Ice bucket
- Spectrophotometer
- Cuvettes
- Gloves, etc.
RNA isolation
1. 1.5ml tube labeled: RW RNA 10/5/10
2. 500µl Tri-reagent added to 1.5ml tube containing tissue.
3. Tissue homogenized with pestle
4. After homogenization, 500µl more Tri-reagent added. Vortexed for 15 sec.
5. Placed in -80˚C for later processing.
Protein extraction
1. 500µl CelLytic MT added to 1.5ml tube containing 18mg of gill tissue.
2. Tissue homogenized with pestle and mixed by inverting tube.
3. Tube spun at ~13,000rpm in refrigerated microcentrifuge for 10 mins.
4. Supernatant removed and placed in 1.5ml tube labeled: RW Protein 10/5/10
5. Tube and contents placed in -80˚C except for portion used in Bradford assay.
Protein quantification
1. 15µl protein supernatant from previous extraction diluted 1:1 with 15µl DI Water in tube.
2. 30µl DI Water added to second tube to serve as blank for spectrophotometer.
3. 1.5ml Bradford reagent added to protein tube and blank tube.
4. Tubes inverted and incubated at room temp for 10 mins.
5. Blank mixed with pipette and 1000µl added to cuvette
6. Blank measured and standardized in spectrophotometer at 595nm.
7. 1000µl protein sample mixed with pipette and measured twice at 595nm, average taken.
8. Protein concentration back-calculated from standard curve.
Results
Spectrophotometer readings: 0.076, 0.078 Average: 0.077
Back calculation of protein concentration:
y = 1013.9 X 0.077 X 2 = 156.1ug/ml
Proteins of interest & Primer design
All information below drawn from NCBI online on 10/8/10. http://www.ncbi.nlm.nih.gov/
Proteins of interest identified via literature search on Web of Science at http://www.isiwebofknowledge.com/
Primer sequences generated by NCBI online primer designer software.
GSTO - hydrocarbon and pesticide response
Accession number - AJ557141 good! It is an mRNA sequence and an interesting gene. -
Primer pair 1 I would choose one of the primer pairs with a product length >150 bp -
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
AGAACCCACTGGGGCGGGTT |
Plus |
20 |
212 |
231 |
60.04 |
65.00% |
| Reverse primer |
TCCCGGGCCTGACGGTAAGG |
Minus |
20 |
350 |
331 |
59.97 |
70.00% |
| Product length |
139 |
||||||
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
CCTTACCGTCAGGCCCGGGA |
Plus |
20 |
331 |
350 |
59.97 |
70.00% |
| Reverse primer |
AGGCGCCTTGCCTTGCTGTT |
Minus |
20 |
499 |
480 |
59.83 |
60.00% |
| Product length |
169 |
||||||
Primer pair 3
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
ACAGGTTCCGCCTGTCCGGA |
Plus |
20 |
49 |
68 |
59.83 |
65.00% |
| Reverse primer |
ACCCGCCCCAGTGGGTTCTT |
Minus |
20 |
230 |
211 |
60.04 |
65.00% |
| Product length |
182 |
||||||
EST9XVIIIb – vibrio response
Accession number - EX956449 Although this sequence has an interesting origin (vibrio challenge) the gene is un-anottaed. For this half the quarter you might want to stick with an mRNA sequence from annotated to gene so that there is a better chance of designing a working gene expression qPCR assay. When designing primers, specify the product length: 150-300bp.-Primer pair 1
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
ACCACGGGCTTCAGATCCACGA |
Plus |
22 |
16 |
37 |
59.92 |
59.09% |
| Reverse primer |
GGGGGCGTGCTCATTGTGGT |
Minus |
20 |
101 |
82 |
59.62 |
65.00% |
| Product length |
86 |
||||||
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
ACCACGGGCTTCAGATCCACG |
Plus |
21 |
16 |
36 |
58.85 |
61.90% |
| Reverse primer |
TGGGGGCGTGCTCATTGTGG |
Minus |
20 |
102 |
83 |
59.62 |
65.00% |
| Product length |
87 |
||||||
Primer pair 3
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
CCACGGGCTTCAGATCCACGA |
Plus |
21 |
17 |
37 |
58.58 |
61.90% |
| Reverse primer |
GTGGTTGGGGGCGTGCTCAT |
Minus |
20 |
107 |
88 |
59.62 |
65.00% |
| Product length |
91 |
||||||
Heat shock protein 90 – thermal stress
Accession number - EF687776 Good! This nucleotide sequence is also a good candidate. -
Primer pair 1
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
GCACTCCGTGACTCGAGCACC |
Plus |
21 |
1810 |
1830 |
59.79 |
66.67% |
| Reverse primer |
GCTGGCGTGTGTTCCTGGCT |
Minus |
20 |
2016 |
1997 |
59.90 |
65.00% |
| Product length |
207 |
||||||
Primer pair 2
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
AGCCAGGAACACACGCCAGC |
Plus |
20 |
1997 |
2016 |
59.90 |
65.00% |
| Reverse primer |
CGTCTCCCTCCAGAGGTGGCA |
Minus |
21 |
2119 |
2099 |
59.64 |
66.67% |
| Product length |
123 |
||||||
Primer pair 3 The product length is to long here! -
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
| Forward primer |
AGCTCTGCAGGCTGGAGCTGA |
Plus |
21 |
345 |
365 |
59.91 |
61.90% |
| Reverse primer |
AAGCGGAGCCCTCCTTGGGA |
Minus |
20 |
1005 |
986 |
59.53 |
65.00% |
| Product length |
661 |
||||||
Conclusions
As this was the first step in a lab which will span several weeks, no definite conclusions can be drawn. It appears that there is sufficient protein in the protein extraction sample to conduct further analyses. There is also sufficient information available from NCBI to create primers to test for many different protein coding genes.
Reflection
This lab seemed to consist of fairly straightforward lab procedures. Protocols were clearly presented and the extractions ran smoothly. I was surprised, however, by the relatively similar sterility protocols between RNA extraction and previous DNA extractions I have performed. I expected RNA sterile technique to be much more rigid and time dependent, but it didn’t deviate much (if at all) from what I have seen done with DNA. How the tissue is preserved is one primary difference. DNA can be extracted from really degraded tissue, but RNA can not be. Also RNA is much less stable at room temperature requiring storage at -80C even if it is only over night. -