Lab 7
Summary:
The purpose of this lab is to extract tissue from the oyster (Crassostrea Gigus) and prepare it for reverse transcription.
Methods and Materials
To prepare for this lab, 40 oyster specimen were separated into 5 groups of 8 specimen:
- Control group-left alone
- Heat group-heated at 30 degree C for 2 hrs *
- Heat & Mechanical-heated at 30 degree C for 2 hrs, placed on roto-shaker at level 5 for 30 minutes.*
- Mechanical & Heat- placed on roto-shaker at level 5 for 30 minutes, then heated at 30 degree C for 2 hrs.*
- Mechanical- placed on roto-shaker at level 5 for 30 minutes.*
To begin lab, the oysters were shucked using blood, sweat, and tears. Once removed, the tissue was obtained from the gills of each specimen. The tissue was put into a solution containing 500 uL of Tri-reagent and homogenized using pestels and lightly vortexing. Then 200 uL chloroform was added to each tube and was mixed again by inverting the tubes and vortexing. Each solution was clearly labeled by group, and placed in the refrigerated centrifuge at max speed for 15 minutes. Once removed from the centrifuge, the mixtures were separated with a clear, aqueous solution sitting on top of a murky, thick solution. The aqueous solution was removed via pipette, placed in a new tube, with 500 uL Isopropyl Alcohol added, and spun at maximum speed in the refrigerated centrifuge for 8 minutes. The solutions concentrated to form a small pellet at the base of the tubes with an aqueous solution on top. The solution was removed from the tube by pipetting, tube with pellet was air dried, and 100 uL of Nanopure water was added to each pellet. Stored on ice.
Results:
We completed all the steps. There are 40 samples ready for reverse transcription.
Conclusion:
It is difficult to determine the success or failure of our work today because we haven’t put any of the samples through any testing. We will find out how successful we were at tissue and RNA extraction when we run the reverse transcriptase and PCR.
The purpose of this lab was to preopare our tissue samples for analysis. Simply having protein saples is not enough to perform any genetic tests on: the tissue must be broken down and prepared. I actually was not confused all day at lab which was really nice, no confusion here this week.
Lab 6
Purpose:
The purpose of this lab was to use the primers we ordered for our experiment and run PCR on them.
Materials and Methods:
First a cDNA template was made for Crassostrea Gisus and C. Virginica by diluting the pure cDNA samples by a factor of 10 4 times, creating 5 templates each (1; 1:10; 1:100; 1: 1,000; 1:10,000). Then
100 uM primer stock was made for each primer (forward and reverse) by diluting the primer 1: 10 nM nuclease free H2O. Then a PCR master mix was made using a ratio of 12.5 ul of SSoFast Mix: 0.5 uL of forward primer stock: 0.5 uL of reverse primer stock: 10.5 uL of nuclease free water. The wells in the PCR loading tray were filled with a mix of 24 uL PCR master mix and 1 uL cDNA for each concentration. The the PCR was run.
Results & conclusions:
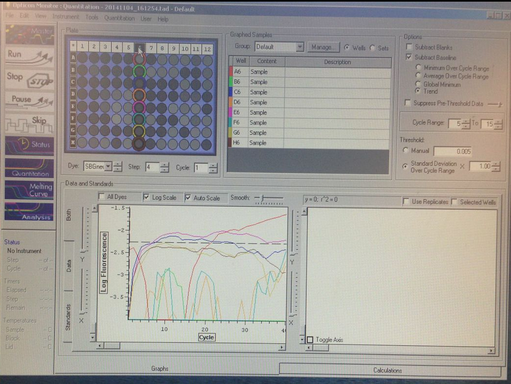
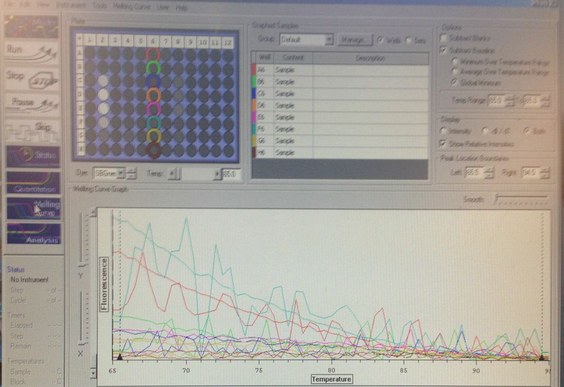
There is DNA dissociation occuring with a melting point of 66 degrees C. The Primers were successfully amplified.
Reflection:
The purpose of this lab was to run a PCR on the primers we ordered. Based on our results, the next step is to begin the experiment by setting up the organisms, applying stress, and taking tissue samples. These methods we are using are useful for experiments just like the one we are attempting: searching for gene expression in an organism's tissue. I still struggle interpreting the results graph, finding information about PCR results and how to interpret them is easy, being able to read it is mind numbing.
10/28
Lab 5
Purpose:
The purpose of lab 5 was to determine what our project will be. Once we determined our project, we learned how to make primers in order to perform an experiment using PCR.
Project guideline:
We will study how crasseostrea gigas responds to heat stress and mechanical stress. We will run PCR amplifying certain genes that we expect to see in response to these stresses compared to the control, non-stressed species.
Maerials and methods and reults:
Using the NCBI database, I searched for a nucleotide sequence to design a primer for heat shock protein 70. The search resulted in multiple potential primers pairs.
(Picture of the 10 Primer products produced and their lenghts)
I chose to use primer pair 2 because it resulted in a desirable product length of 217 BP. We were told to look for a product between 100-500 BP, and other than primer 6, all the other primers resulted in BP lengths much higher than 500.
Primer pair 2
| Sequence (5'->3') |
Template strand |
Length |
Start |
Stop |
Tm |
GC% |
Self complementarity |
Self 3' complementarity |
|
|---|---|---|---|---|---|---|---|---|---|
| Forward primer |
CCTGGCAGAGGTTGACGAAT |
Plus |
20 |
1895 |
1914 |
60.04 |
55.00 |
5.00 |
2.00 |
| Reverse primer |
TGGCAACTCTTGCTCACTTTC |
Minus |
21 |
2111 |
2091 |
59.05 |
47.62 |
5.00 |
0.00 |
| Internal oligo |
Plus |
||||||||
| Product length |
217 |
||||||||
>AB549340.1 Crassostrea gigas hsp70B mRNA for heat shock protein 70B, complete cds product length = 217 Forward primer 1 CCTGGCAGAGGTTGACGAAT 20 Template 1895 .................... 1914 Reverse primer 1 TGGCAACTCTTGCTCACTTTC 21 Template 2111 ..................... 2091
I ran the BLAST, and there were 0 other sequences producing significant results.
Discussion:
I chose to do HSP 70 because I expct to see a large change in expression in the organisms subjected to heat stress. I believe the primer I chose should be sufficient, the BLAST results state that the likelihood the primer will bind at an unintended site is low. What I didn’t fully understand was how the primers were generated from the database. I remember enough of the broad strokes that I was able to get by and produce a viable primer (I hope), however when It came to choose which primer to pick out of the dozen generated, I chose based on product length. I feel like that is not exactly scientific. I guess Im not sure what the differences are between the primers, yet they all attach to the same binding site? Also, I cant upload a JPEG to save my life.
10/21
Lab 4
Purpose:
The purpose of this lab was to run a cPCR with the products from lab 3’s qPCR, run SDS-PAGE gel electrophoresis, and perform Western Blotting on the results looking specifically for HSP70.
Methods and Materials:
Agarose gel electrophoresis: The agarose gel made in lab 3 was placed into a gel box. 2 uL of 10x loading dye was added and mixed to the PCR product from lab 3. Well 1 was filled with 7 uL of a 100 bp ladder, and well 2 was filled with 20 uL of the PCR-dye solution. The gel was run at 100V for one hour. After being run, the gel was removed and visualized under UV light.
SDS-PAGE: A solution of 15 uL protein stock and 15 uL of 2x reducing sampling buffer was placed into a screw cap tube and mixed by centrifuge for 10 sec. the sample was brought to boil for 5 minutes, and quickly centrifuged to pool the liquid. Using a gel loading tip, the entire sample was loaded into well 3 and the gel was run for 35 min at 150V. the gel was then removed and trimmed in preparation for Western Blotting Protocol.
WesternBreeze Chromogenic Western Blot Immunodetection: The gel from SDS-PAGE and a nitrocellulose membrane were soaked in a Tri-Glycine transfer buffer for 15 min. in the semi-dry blotting apparatus, a blotting sandwhich was made. It was composed of; in order: the anode, filter paper, the nitrocellulose membrane, the gel, filter paper, and the cathode. This was run for 30 min at 20V. Then the gel was carefully removed, rinsed with transfer buffer, and placed in a box with approximately 10 mL of blocking solution. The box containing the gel and buffer was placed on a rotary shaker at 1 rev/sec for 30 min. Then the buffer was decanted ($$$) and the gel stored. After being run in the semi-dry blotting apparatus, the membrane is removed and washed twice with approximately 20 mL nanopure H2O for 5 min each time. The membrane is then placed in a box with 10 mL of Primary Antibody Solution for one hour, then the solution was decanted. The membrane was rinsed 3 times in 20 mL of Antibody wash for 5 min, decanting the solution each time. The membrane is then incubated in 10 mL of Secondary Antibody solution for 30 min. it is again washed three times for 5 min each with Antibody wash. The membrane is rinsed twice with nanopure H2O for 2 min each, and finally is incubated in 5 mL of Chromagenic Substrate. Once a purple band appears, the membrane was dried on filter paper.
Results:
The cPCR shows amplification around 200-300 BP.
The Gel-staining of the SDS-PAGE gel shows proteins present.
The Western Blot did not yield results.
Conclusion:
The conclusions were not surprising. The qPCR results show amplification at 200-300 BP, which is reasonable for the elongation factor we chose. The banding in the gel staining of the SDS-PAGE shows proteins were present, however when I researched how to interpret the binding patterns of SDS-PAGE gel staining, it appears that there was certain information necessary for a full interpretation that I don’t have. Also, it appears that the process is fairly sensitive, and many things can goslighty wrong that will have large effects on the results. I am unsure as to why the Western Blotting Protocol didn’t yield results. We were very much crunched for time in this lab, and didn’t exactly perform each step with precise measurements of time. This may have affected our results. The purpose of this lab was to run a cPCR with the products from lab 3’s qPCR, run SDS-PAGE gel electrophoresis, and perform Western Blotting on the results looking specifically for HSP70. These methods could be used to detect whether or not an organism is stressed/ in which ways is it responding to the stress via looking for certain biological markers such as HSP70. What is unclear to me and I wish there was more information on is the interpretation of the results. I have found that is extremely difficult to sift through all the mumbo jumbo online in order to find out how to interpret the pictures that we get as results. I find myself most confused when writing my conclusions sections.
10/14
Lab 3
Purpose: To create cDNA from our RNA and run it through qPCR to interpret the results. There is also a side mission of protein extraction from tissue for a Bradford Assay.
Materials & Methods:
Reverse Transcription: The RNA samples from week one were taken off ice, thawed at room temperature, and mixed by inversion. 5uL of the RNA were then transferred to a 5 mL PCR tube. Added to the RNA in the tube was a master mix (MM1) thta contained a ratio of 0.5 uL oligo dT per 12.25 uL nuclease free H2O. 12.75 uL of the master mix was added to the 0.5 uL of RNA in the PCR tube. The tube was then placed in the thermocycler and incubated at 70 degrees C for 5 minutes, and was then stored in ice. During the incubation period, master mix two (MM2) was created by mixing 5 uL of M-MLV 5X Reaction buffer, 1.25 uL of dNTPs, 0.5 uL of M-MLV RT and 0.5 uL of nuclease free H2O. After the RNA with MM1 was finished incubating and put on ice, 7.25 uL of MM2 was added to the PCR tube. The tube was then incubated at 42 degrees C for 60 minutes, with a 3 minute heat inactivation at 70 degrees C at the end. The sample was spun on a desk top centrifuge and stored at -20 degrees C.
qPCR: A master mix was prepared by combining 12.5 uL SsoFast EvaGreen supermix, 0.5 uL of upstream primer, 0.5 uL of downstream primer, and 10.5 uL of nuclease free H2O. 24 uL of master mix is combined with 1 uL of the cDNA obtained from reverse transcription of the RNA in a PCR well. A control is set up in another PCR well using 24 uL of the master mix, and 1 uL of nuclease free H2O. Then the wells were capped, and loaded into the PCR machine to be run. The PCR conditions set in the machine were:
1. 95 degrees C for 10 minutes
2. 95 degrees C for 15s
3. 55 degrees C for 15 s
4. 72 degrees C for 15 s
5. Cycle steps 2-4 39 times
6. 95 degrees C for 10s
7. Melt curve from 65 degrees C to 95 degrees C, with 0.5 degree C imcrements for 5s
Making Agarose Gel: While the PCR machine was running the PCR, 2g of agarose was mixed with 150 mL of 1x TAE in a 1 L flask. The solution was microwaved for 3 minutes, pausing every 30 seconds to open the door and prevent boiling over. Once the solution is cool to the touch, 12 uL of ethidium bromide was added. The solution was mixed by swirling in the flask, then added to the gel tray. Then the gel combs were added and all bubbles within the gel were popped using a pipette tip. The gel was then wrapped in plastic wrap, labeled and placed in the refrigerator for storage.
Protein Extraction: Whole tissue was obtained in a snap cap tube and weighed by taring a scale with the weight of an empty tube, and using the difference as the assumed tissue weight. 500 uL of Cellytic MT solution to the tube, and homogenized with a pestle. Then the tube was mixed via inversion, and spun in the refrigerated microfuge at max speed for 10 minutes. The supernatant produced was transferred to a new tube and stored on ice for the week.
Results:
Unfrtunately, I left my notebook in class on Friday, therefore I do not know which sample was mine. Fortunately, there are 4 other folks who are performing the same expreiment. Obvioulsly there will be slight differences based upon how the experiment was carried out, but the results of all samples combined is a reasonable guideline for what my sample would be. As a whole, the average efficiency of DNA production was 2.125, including the controls. The average C(t) value was 22.91125 incluing the controls. The products melted between 80-85 degrees C. Assuming that samples E4, E5, and F4 were the control samples due to their extreme values, the average efficiency of the 5 experimental samples was 2.396, and the average C(t) value was 14.368.
Conclusions:
The efficiency rating suggests that the DNA was amplified at above 100%. There are many possible reasons for this, after reviewing the subject, I believe the most likely reason for this is a contamination of the sample due to excess ethanol that was left behind with the cDNA. The average C(t) values omitting the assumed controls shows that the threshold is crossed at the 14th cycle. This, along with the 80-85 degree melting point are within expected parameters for PCR products. The Samples are ready to be run through electrophoresis.
Reflection:
The goal of this lab was to perform a qPCR with our cDNA that was made from our RNA. Most procedures involving measurements were related to the pipetting of solutions. These methods are a classic example of DNA extraction from whole tissue, and how to amplify what is found. I am unclear on what the results mean honestly. Finding information on interpreting PCR results, in understandable English, is tough. For example I know what the C(t) values mean by definition, but I am not clear on why we are interested in when threshold is reached.
October 7th
Lab 1:
The purpose of this lab is to obtain and break down a tissue sample that is to be extracted of its RNA.
Methods and Materials
56 mg of oyster tissue was scraped off the frozen organism and placed in a microfuge tube. 500 uL of Tri-Reagant was added to the tube. The solution was mixed by "vigerous" vortexing as well as being physically stirred by a plastic rod. Once mixed, although the solution did not become uniform, 500 uL more Tri-Reagant was added. Because I missed lab 1, I did not need to store my sample because I completed the tissue extraction right before beginning the RNA extraction.
Results:
The solution contains stringy, fiberous pieces of tissue.
Conclusion:
The sample is ready for its RNA to be extracted.
Reflection:
The purpose of this lab was to prepare our tissue samples for RNA extraction, but it was also helpful in allowing us the students to get familiar with each other, the lab, and the equipment. The only measurable procedure was using the correct pipette setting to measure out 500 uL of Tri-Reagent to be added to the tssue. These methods of tissue extraction could be used in a wide variety of sciences that are interested in biology. I feel very clear about this lab, no confusion. I cannot think of any areas of this lab that I felt there was a lack of information-very straightforward.
10/7
Lab 2:
The purpose of this lab is to extract an usable RNA sample from the tissue sample prepared in lab 1. Once the RNA is extracted, it will go through a Nanodrop spectrophotometer to be assessed for quality and usability in reverse transcript and qPCR.
Materials and Methods
Because I performed lab 1 & 2 consecutively on the same day, my tissue sample was never stored overnight and therefore did not need to be thawed to room temperature because it was already there. 200 uL of chloroform was added to the tissue to create a solution that was vortexed at high for 30s until the solution mixed and reacted until it became a milky emulsion. After incubating at room temperature for 5 minutes, the sample was spun in a refrigerated microfuge for 15 minutes at maximum speed. Once removed, the solution had a top clear layer, containing the RNA, that separated from the milky emulsion. The RNA containing top layer was then transferred to a new microfuge tube. 500 uL of isopropanol was added to the solution containing the RNA and mixed by inverting the tube until the solution is uniform. After incubating at room temperature for 10 minutes, the solution was spun in the refrigerated microfuge for 8 minutes at max speed. Once the tube was removed, a white pellet composed of RNA and salts appeared at the bottom. all the liquid supernatant was removed from the tube, leaving behind just the pellet. 1 mL of 75% EtOH was then added to the tube, and the tube was vortexed on high in attempt to free the pellet from the walls of the tube. My pellet remained stuck, so prof Roberts flicked the bottom of my tube once and the pellet was dislodged. then the solution was spun in the refrigerated microfuge for 5 minutes at 7500g. the liquid supernatant was then removed by using a pipette to get an initial majority out, vortexed briefly to pool the remaining liquid, and that liquid supernatant was removed with the same pipette. To ensure maximum supernatant removal, the tube was left open for 5 minutes. Once isolated and dry, the pellet was suspended in 100 uL 0.1% DEPC-H20 and mixed by pipetting up and down. my sample completely mixed, therefore I did not have to incubate it at 55 degrees for 5 min.Once the pellet was completely dissolved, it was placed in an ice bucket to be transferred to the Nanodrop spectrophotometer. The nanodrop s-meter was zeroed by placing 2 uL of 0.1% DEPC-H2O on the pedestal. then 2 uL of the RNA contining sample was placed on the pedestal, and "measure" button was pressed. The nanodrop used the Beer-Lambert Law to calculate the RNA concentration in the sample. The A260/280 ratio and A260/230 ratios were calculated and recorded. After the sample was analyzed, a KimWipe was used to wipe off the pedestal. After going through the spectrophotometer, the sample was stored at -80 degrees C.
Results:
A260/280=1.99
A260/230=1.63
RNA Concentration: 1,415.9 ng/mL
Conclusions:
The A260/280 ratio being 1.99 confirms that the solution being tested contains RNA.
The A260/230 ratio being 1.63 suggests that there are high levels of organic contaminants present in the solution with the RNA.
The 1,415 ng/mL concentration was much higher than the rest of my peers in lab. this could be due to the fact that I performed the tissue extraction and preparation on the same day and everybody else had their samples in storage for one week. In general, the conclusions confirm that RNA was successfully extracted from the tissue sample.
Reflection:
The purpose of this lab was to extract RNA from the tissue samples prepared in lab 1. We were also introduced to new machinery this lab, using the vortex, the refrigerated centrifuge, and the NanoDrop spectrophotometer. The procedure in this lab that produced a measurement was from the spectrophotometer. It measures the wavelengths absorbed by a solution. Using the wavelengths absorbed, the A260/280 measures whether the solution contains DNA or RNA. The A260/230 measures the amount of organic contaminents in the solution. And the concentration measures…the concentration of RNA in the solution. The methods used are designed for RNA extraction. The only part of the procedure today that I am unclear on is the process that is occurring within the solution with each new chemical added. For example, I knew from the directions that adding chloroform to my solution and mixing it should have resulted in the solution becoming a milky emulsion. What I don’t know is what is actually causing this change in color and consistency.