Editing paper and presentation, phylogenetic trees. Presenting Tuesday, May 31, 4:30pm
May 23, 2011
Update: Paper is almost complete, only missing phylogenetic trees, which I will make today. Presentation is halfway done.
May 17, 2011
First draft of paper is finished, on to working on presentation!
May 11, 2011
Meeting with Steven lead to my next steps:
1. Qualitatively analyze gel from May 4/5 using 1-10 scale and ImageJ software (assumption of expression in how bright bands are)- will be picking bands around 222 and 300-368.
2. Create phylogenetic trees using individual DNMT3 sequences- compare lizard, zebrafish, human, maybe others?
3. Finish paper and presentation.
May 6, 2011
Running PCR again with same protocol, setup, but changing annealing temperature (previously 55C) to 63C.
Running gel as well. Used Jason's diluted cDNA samples and used 2 uL of template instead of 1 uL.
First 10 lanes are primer 222, C1-C4, control, A1-A4, control. No amplification for these, only primer dimer.
Second set of 10 samples (368) same arrangement- looks like there is some amplification for C3 and the acid-shocked samples...as well as the negative control. Will probably try different annealing temperature and run again, but it looks like i get lots of bands when there is amplification...happy weekend.
May 4/5 2011
Running conventional PCR with cDNA samples to run on gel- qPCR was unsuccessful because of random peaks everywhere, including water...
Master Mixes for each primer w/10 reactions each (222, 368) (x10.1 for totals)
1 uL template/water
12.5 uL 2X Apex MasterMix (126.25 uL)
.5 uL Fprimer (5.05 uL)
.5 uL Rprimer (5.05 uL)
10.5 uL PCR water (106.05 uL)
24 uL/reaction plus template or water
Thermocycler parameters:
95C - 10mins
40 cyles of:
95C - 30s
55C - 30s
72C - 2 min
72C -10 min
Running for 4 controls, 4 acid shocked juvenile sockeye salmon and 2 negatives (each primer).
Reactions:
C1, C2, C3, C4, A1, A2, A3, A4 neg, neg
Finally, running 1.2% agarose gel (180 TA buffer, 2.2 g agarose for large gel).

Tomorrow- running everything again because gel shows multiple bands for all samples- at least the waters are clean!
Will be using same procedure, except with 63C annealing temp instead of 55C.
May 3, 2011
Running qPCR for samples C1-C4, A1-A4, 2 negatives with annealing temp reduced to 50deg (primers 222, 368)- setup is the same as the last qPCR (April 22).
Results can be found here
April 27, 2011
Looking at qPCR results and they're not as awesome as I hoped. Having trouble linking to document?Can be found in the sites folder (and dropbox) under qPCRApr22.
For both primers, there is amplification in A3 and A4 (368 amplified C4) and primer dimer on all the negatives. I increased the annealing temperature by 5degC and also used Sso Fast EvaGreen instead of the CytoGreen.\
Results can be found here
April 22, 2011 (happy earth day!)
Today, I'm running qPCR for DNMT3 primers *222 and *368 (stressed and control cDNA) with 12.5 uL GoTaq MasterMix and 1 ug template/sample. Will be running for samples C1-C4, A1-A4, and two negatives for each primer. Trying to optimize by increasing annealing temp.
Master Mixes (per rxn, x10.1 for all)
1 uL template/water
10 uL Sso Fast EvaGreen
.5 uL Forward Primer
.5 uL Reverse Primer
8 uL PCR water
| Primer 222 |
C1 |
C2 |
C3 |
C4 |
A1 |
A2 |
A3 |
A4 |
neg |
neg |
| Primer 368 |
C1 |
C2 |
C3 |
C4 |
A1 |
A2 |
A3 |
A4 |
neg |
neg |
March 1, 2011
Decided not to present this quarter since qPCR results show promise for better optimization. Plus it makes more sense to spend more time perfecting and completing the project rather than speeding and stressing just to present. Will spend less time doing actual lab work next quarter, more time writing and researching.
Next steps:
Continue working on paper- specifically, literature research that describes DNMT3 gene in other species- location? mechanism? comparison to sockeye?
Optimize qPCR: look at negatives' melting curves, see which primer gives the best melting curve
primer 222: looks good, melt temp~85.5degC, clean negatives
primer 228: negatives spike inconsistently with rest of samples..
primer 585: ok, range 76.5-82degC, clean negatives
Salmo368: good negatives, consistent curves
salmodanio: inconsistent, will not use
February 28, 2011
This week: Going over qPCR results and paper with Steven tomorrow. Planning to present March 9 at 3:15pm. This week: finalizing paper, presentation.
qPCR results can be accessed here
February 23, 2011
Summary: This month I have been working on writing the first draft of the paper. Today, running qPCR for all DNMT3 primers: 222, 228, 585, Salmo368, and SalmoDanio.
Run with 12.5 uL SytoGreen, 1 ug template/sample. Thermocycler was set with extension temp 72 for 30 sec instead of 15 b/c of product sizes.
Plate order:
| 222 |
228 |
585 |
Salmo368 |
SalmoDanio |
|
| 1 |
2 |
3 |
4 |
5 |
|
| A |
C1 |
C1 |
C1 |
C1 |
C1 |
| B |
C2 |
C2 |
C2 |
C2 |
C2 |
| C |
A1 |
A1 |
A1 |
A1 |
A1 |
| D |
A2 |
A2 |
A2 |
A2 |
A2 |
| E |
neg |
neg |
neg |
neg |
neg |
| F |
neg |
neg |
neg |
neg |
neg |
January 31-Feb 4, 2011
Summary: Will be working on writing methods this week!
January 27, 2011
Summary: Running gel with PCR salmo368 and products from 3 dnmt3 RNA-based primers on cDNA samples 3 and 4.
January 26, 2011
Summary: Re-running PCR/gel because negatives are positive. Also running products from salmo368 primer. Also working on methods.
January 21, 2011
Summary: Running 1.2% agarose gel with cDNA PCR samples from yesterday.

I got bands for cDNA (which is good) but I also got bands for EVERY negative control. I will be running PCR again on one cDNA sample/primer plus negatives.
January 20, 2011
Summary: Running PCR on cDNA samples with new primers and with Salmo368 to confirm that they work.
Primers to use:
1. Salmo368 (working stock in my -20 box, 100uM stock in primer box #7)
2. DNMT3.228
3. DNMT3.222
4. DNMT3.585
Primer reconstitution: In TE buffer for 100uM concentration, then 10uL in 90uL of buffer as 10uM working stock.
4 MasterMixes for 4 reactions each (2 cDNA- used control1 and control2 from juvenile samples, 2 negative controls each).
*Note- only running one negative control, would only need 2 if running genomic too.
per MasterMix: (4 per MasterMix +10% -> all x 4.1)
1 uL template/water
51.25 uL 2X Apex MasterMix
2.05 uL Fprimer
2.05 uL Rprimer
47.25 uL PCR water
--> 24 uL/rxn plus 1uL cDNA template/PCR water
Next steps: Run PCR products on 1.2% agarose gel tomorrow. If I get decent bands, then I'll run qPCR on control/acid treated juvenile cDNA.
January 18, 2011
Summary: Last week I extracted RNA and reverse transcribed it into cDNA. I ordered primers (based off RNA) to run with control/acid treated juvenile cDNA samples. Primers not here yet to run qPCR for DNMT3! Talked to Steven about Geneious analysis to talk about in my paper. Today I'm compiling that info and starting on writing methods.
3 sets of sequences based off primers for DNMT3/DNMT3b:
1. OMYKISS
- No overlap with salmodanio/salmo368 sequences or genes of reference
- Blastn'd sequence- no results
Outcome: omykiss is not worth looking at or talking about because the one sequence I have is terrible quality and doesn't align with anything
2.SALMODANIO
- One high quality sequence to work with
- Aligned w/ O. nerka, A. carolinensis, and H. sapiens.
Salmodanio and O. nerka
- Aligned with closest gene of reference (EZ815731)- Oncorhynchus nerka (Sockeye salmon) DNMT3 sequence (alignment #410)
- Good alignment from ~100-300 bp = about 150 amino acids align

Salmodanio and A. carolinensis
- Aligned with A. carolinensis DNMT3A mRNA gene (GQ121005) because of good alignment with salmo368 (alignment #440)
- Good alignment from ~2010-2070. Looks a little choppy- sequence was gDNA and and gene is mRNA- definitely introns!

Salmodanio and H. sapiens
- Choppy alignment ~10,850-11,058
- Salmodanio is cDNA, H.sapiens is full gene= choppiness due to introns.

3. SALMO368
- 4 high quality sequences to work with
- Combined into contig consensus sequence and aligned with D.rerio (closest gene of reference), A. carolinensis and Homo sapiens.
Salmo368 and D.rerio
- Alignment ~532-2513.
- Choppy- salmo368 sequences combined gDNA and cDNA, D. rerio gene is gDNA.- choppiness=introns

Salmo368 and A. carolinensis
- High homology ~1510-1653 bp.
- Again a little choppy- sequences are combined cDNA and gDNA, so also introns. Very interesting! Sample was sockeye, primer Atlantic salmon, but lines up the best with a lizard aka American chameleon.

Salmo368 and H. sapiens
- Sequences align w/ human gene from ~7000-7520 bp.
- Very choppy - this is the full gene for humans, including introns and exons, so makes sense that alignment is a little choppy. Good to know it's there though!

Overall:
There is homology between my DNMT3 sequences and the A. carolinensis DNMT3a gene. Salmo368 spans from 1506-1653 bp and Salmodanio spans from 2010-2070 bp. Basically this means that the DNA that I have has the DNMT3 gene in it, but the primers are only splicing in these areas. If I wanted to determine if the whole gene was there, I would pick primers off of A. carolinensis and run them on my sockeye samples, sequence, and check where they align. In the interest of time, I will not be doing that. The same could be done with the human gene, where Salmodanio lies between 10,850 and 11,058. Salmo368 lies between 7,000 and 7,520. Because there are so many introns (choppiness) I am not sure if it is worth doing this with the gene that I am using now. Even though the alignment is choppy, it's worth noting where the gene is actually located.

Next steps:
-Talk to Steven/Mac about intron problem
-Run qPCR on cDNA acid shocked/control juveniles for DNMT3
-Look at DNMT1 sequences (when ready) and align with closest genes, A. carolinensis, and human
-Write paper!
January 14, 2011
Summary: Reverse transcription of RNA samples into cDNA.
Protocol: *Note: I used 1ug of RNA per reaction.
A single reaction volume = 25uL. The volume of RNA, primer(s) and M-MLV RT used are variable and will be specific to your current experiment. The directions below apply to a reaction using 1ug of total RNA. You may need to make changes to accommodate your own conditions.
- Use as much RNA as possible, max volume of RNA = 17.75uL. Generally, identify the RNA sample with the lowest concentration and multiply by 17.75uL. Use this quantity (ug) of RNA for each and every sample.
- Transfer calculated volume(s) of RNA to 0.5mL snap cap tubes or PCR plate. Adjust volumes of individual samples to 17.75uL with H2O.
- Add appropriate amount of primer to sample. Use 0.25ug primer per 1ug of RNA in sample (= 0.5uL of Promega oligo dT in this example). Total volume (RNA + primers) should equal 18.25uL.
- Heat samples at 70C for 5 min in thermocycler.
- Place samples on ice IMMEDIATELY.
- Make Master Mix:
PER RXN
5 uL 5x Buffer (M-MLV RT Buffer)
1.25 uL 10mM dNTPs
0.5 uL M-MLV RT per ug of RNA
7. Mix well.
8. Add 6.75uL of master mix to each reaction.
9. Mix well, but do not vortex.
10.Spot spin.
11.Incubate @ 42C for 1hr in thermalcycler for oligo dT primers OR @ 37C for random primers.
12.Heat inactivate @ 95C for 3 min.
13.Spot spin.
14.Store @ -20C.
January 13, 2011
Summary: RNA extraction of AK control/acid shocked juveniles. Used samples 24/9-24/6 (treated) and 0/8-0/5 (control).

Concentrations and 260/280 look pretty good! concentrations for A2-A4 are lower because the fish were really tiny.
January 11, 2011
Summary: Compiling usable data, summarizing meeting/next steps, finding primers for qPCR of stressed/non-stressed juveniles
Graphs/pictures to use in final paper/presentation:
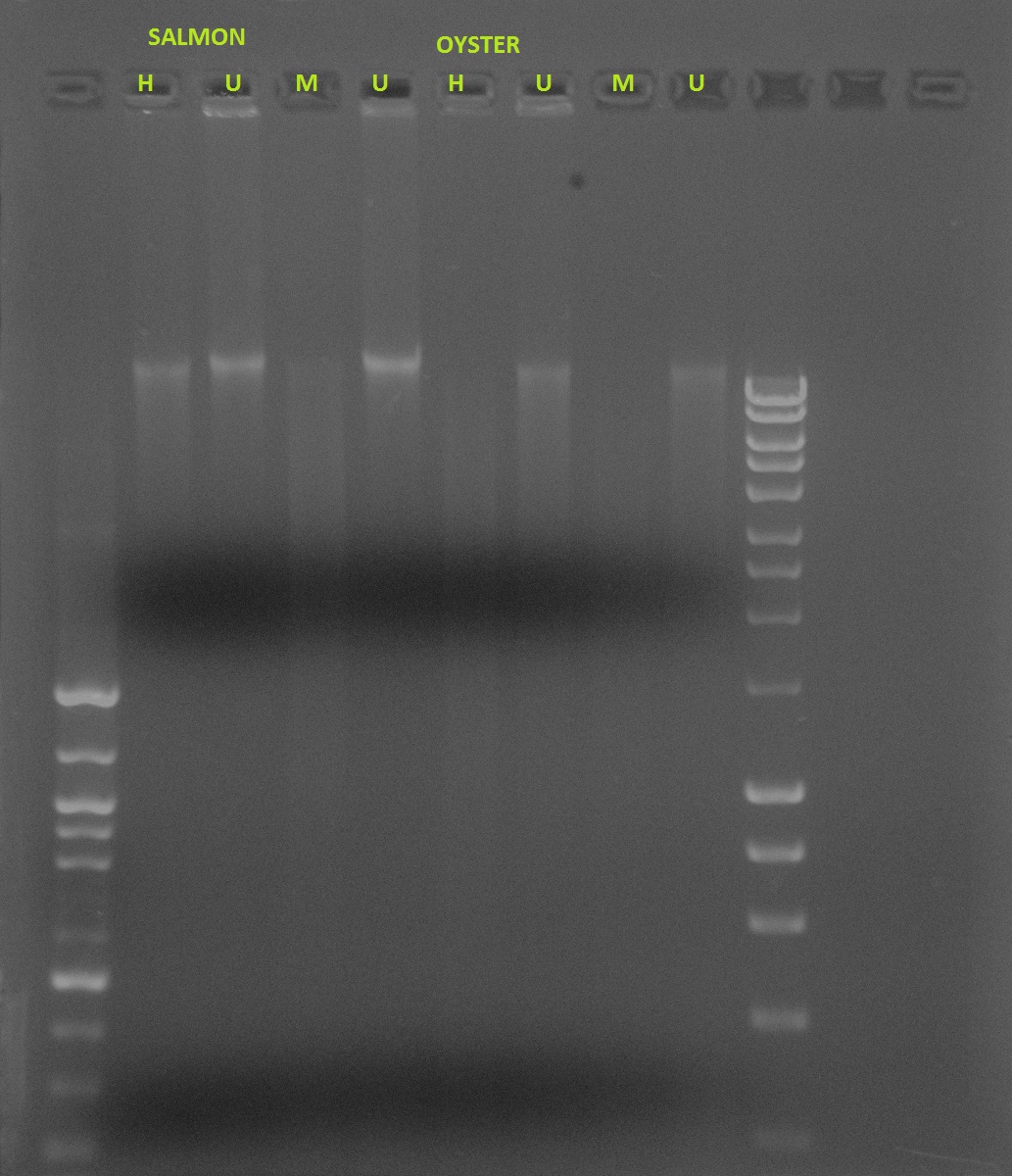
Salmon vs. Oyster methylation- Restriction digest using HPAII/MSPI
If DNA is methylated:
Undigested: Will show a band
HPAII: Will show a band because it doesn’t cut
MSPI: Will not show specific band, will show smear because it cuts at various places
If DNA is not methylated:
Undigested: Will show a band
HPAII: Will not show a specific band, will show smear because it’s cutting at non-methylated areas
MSPI: Will not show a specific band, will show smear because it cuts at various places

Salmon methylation for development stages- missing undigested for 11/14, but very interesting for 1/14.
Development info:
| Date |
Sampling Day # |
Life History Day # |
Characteristics |
| 14 Nov |
1 |
14 |
BT embryo |
| 20 Nov |
7 |
20 |
BT embryo- eyes visible |
| 2 Nov |
11 |
24 |
Eyes + spinal cord |
| 28 Nov |
15 |
28 |
None |
| 4 Dec |
21 |
34 |
None |
| 14 Dec |
31 |
44 |
Hatch- Alevin |
| 24 Dec |
41 |
54 |
Yolk sac smaller |
| 1 Jan |
49 |
61 |
Yolk sac almost gone |
Note: 14 dec- Hatch date!!!!! spike in methylation?
The gel shows smears for both HPAII and MSPI for 11/14 (lanes 2 and 3). There are no distinct bands for the undigested DNA for 11/28, which is a little suspicious (lanes 4 and 5), and HPAII and MSPI also show smears (lanes 6 and 7). The two undigested samples for 12/14 show bands (lanes 8 and 9), with a band and a smear for HPAII and a smear for MSPI (lanes 10 and 11). All samples for 1/1 show smears.
Because the undigested samples are not showing distinct bands as they should, I am concerned that this data is futile. I expected to see less methylation in the earlier samples with an increase in methylation with age. It is clear that the samples will need to be run again for more accurate results.

Primers for DNMT3b- ran genomic DNA on gel, then sequenced
ELISA
ELISA results can be found here
There are several problems...
1. The blank has a concentration of .18, which is higher than most of the actual samples.
Solution: Add a wash to each step to ensure that samples are clean are that the blank reads at 0.
2. The standard curve blows out at the 100 ng mark, indicating that going up to 200 ng is too much- I assumed that because vertebrates have a higher global methylation percentage than inverts, I would use a higher curve than what Mac used for her analysis. Maybe the antibody is limiting with such a high concentration of DNA.
Solution: Keep the standards from 5-100ng and decrease the max of 200ng in samples to 120ng or 100ng max (120 worked better anyway).
3. Most samples fell outside range of standard curve- only sample 1, 5, and 9 have concentration high enough to be within range, all others are below blank.
Solution: Add washes to ensure blank is at 0.
Development info:
| Date |
Sampling Day # |
Life History Day # |
Characteristics |
| 14 Nov |
1 |
14 |
BT embryo |
| 20 Nov |
7 |
20 |
BT embryo- eyes visible |
| 2 Nov |
11 |
24 |
Eyes + spinal cord |
| 28 Nov |
15 |
28 |
None |
| 4 Dec |
21 |
34 |
None |
| 14 Dec |
31 |
44 |
Hatch- Alevin |
| 24 Dec |
41 |
54 |
Yolk sac smaller |
| 1 Jan |
49 |
61 |
Yolk sac almost gone |
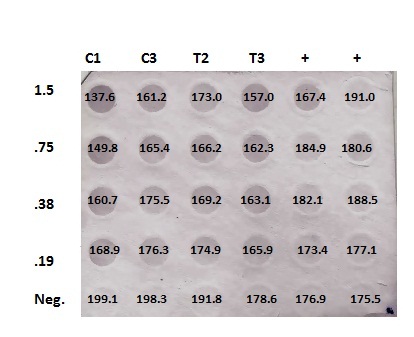
DotBlotMeasurements.xlsx
Numbers are inverted (low #= high methylation). No significant differences in concentrations between Control and Treatment. Furthermore, negative controls aren't really negative, and positives aren't positive... maybe worth running a restriction digest with Control/Treated just to have visual.
Notes from meeting w/ Steven:
Planning on completing Capstone this quarter.
Need to:
1. Make sure lab notebook is complete, organized, and compile data usable for paper.
2. Find primers and extract RNA/make cDNA for qPCR of DNMT3b for stressed/non-stressed juveniles.
3. Describe DNMT3b
4. Sequence DNMT1 samples and do the same.
Timeline:
Feb 4- Methods
Feb 11- Results
Feb 18- Discussion
Feb 25- Intro
March 4- Full draft, changes by March 18th
This week:
Tuesday (today): Fix lab notebook, compile data, find primers for qPCR
Thursday: RNA extraction from fish heads (in -80freezer), reverse transcription for cDNA
Friday: reverse transcription/qPCR if primers are in
Primers (designed for mRNA- not genomic because cDNA based of RNA strand):
||
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
GTCGGCGAACAGGCTGCTGA |
Plus |
20 |
1682 |
1701 |
59.70 |
65.00% |
| Reverse primer |
GCTTCCCTGCGTCTCTGGGC |
Minus |
20 |
2266 |
2247 |
59.77 |
70.00% |
| Internal oligo |
Plus |
||||||
| Product length |
585 |
||||||
||
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
CCCACGCTTTGCAGCCCCTT |
Plus |
20 |
1476 |
1495 |
60.18 |
65.00% |
| Reverse primer |
CAGCCTGTTCGCCGACTGCA |
Minus |
20 |
1697 |
1678 |
59.98 |
65.00% |
| Internal oligo |
Plus |
||||||
| Product length |
222 |
||||||
||
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
TACCCACGCTTTGCAGCCCC |
Plus |
20 |
1474 |
1493 |
59.62 |
65.00% |
| Reverse primer |
TCAGCAGCCTGTTCGCCGAC |
Minus |
20 |
1701 |
1682 |
59.70 |
65.00% |
| Internal oligo |
Plus |
||||||
| Product length |
228 |
||||||
December 7, 2010
Summary: Slow week (finally). Waiting on sequences for DNMT1 so I can align on Geneious- will probably have to wait until next quarter since I'll be gone starting next week... This week, continuing sequence alignments for DNMT3 and researching more on related proteins.

Primer 1 (danio) and Primer 3 (salmo) worked for both gDNA and cDNA. Primer 2 (danio2) did not work for cDNA..I was still able to cut out bands for all 3 primers for sequencing.
Geneious notes:
It looks like my sequences for DNMT3 are solid- good alignment for compiling a single sequence. However, kind of weird that the forward/reverse sequences don't line up with each other. Will research literature and see what I can find!
December 2, 2010
Summary: Running DNMT1 PCR products on 1.2% agarose gel, then cutting bands out for sequencing.
Samples loaded in the following order:
Hyperladder 1, Primer 1: gDNA, cDNA, neg, neg, Primer 2: gDNA, cDNA, neg, neg, Primer 3: gDNA, cDNA, neg, neg
Will edit gel picture at home and then post!
Products at appropriate length cut out and set up in sequencing plates.
November 30, 2010
Summary: So far, I have run my restriction digest test and ordered primers for DNMT1. Next steps are to test the DNMT1 primers (PCR/gel) and sequence products. Then compare DNMT1 sequences to DNMT3 sequences using Geneious. Finally, qPCR on control/acid shocked juveniles to determine expression.
Today:
Run DNMT1 primers on gDNA, cDNA, and two negative controls (following October 28 protocol).
Ran each primer for gDNA (C1 from Alaska control/acid shocked), cDNA (pooled from sockeye brain samples), and two negatives for each.
Primer stocks stored in PrimerBox7.
Primer reconstitution (addition of TE buffer):
#1: DNMT1_Danio
F: 202 uL
R: 381 uL
#2: DNMT1_Danio2
F: 304 uL
R: 308 uL
#3: DNMT1_salmo
F: 319 uL
R: 300 uL
Analyzing sequences on Geneious today..
Next steps: Thursday run PCR products on gel and prep for sequencing. Friday- compare DNMT3 sequences.
Geneious for DNMT3:
1. Blastx all sequences
2. Align to each other and to Danio rerio
3. Figure out what the final sequence is
November 22, 2010
Summary: Running restriction digests from Friday on 1.2% agarose gel @120 volts. 10uL loading dye added to each sample, then 24uL/well plus 20uL 100bp ladder.
*NOTE: Forgot to run undigested DNA (and it's only Monday...).
It looks like C1 is methylated (band at H, smear at M, band at both controls). T1 is a little harder to tell since there is a smear at all of them.
Methylated:
Undigested- band
HPAII- band
MSPI- smear
Unmethylated:
Undigested- band
HPAII- smear
MSPI - smear
November 18, 2010
Summary: Restriction digests for HPAII and MSPI in control/treated juveniles. Samples in 37deg water bath overnight, will run gel on Monday.
| Sample |
Mix |
DNA (1ug) |
10x Buffer (1 or 4) |
Enzyme |
Water (to 50 uL) |
| C1 |
HPAII |
4.23 |
5 |
1 |
39.77 |
| C1 |
HPAII control |
4.23 |
5 |
0 |
40.77 |
| C1 |
MSPI |
4.23 |
5 |
.5 |
40.27 |
| C1 |
MSPI control |
4.23 |
5 |
0 |
40.77 |
| T1 |
HPAII |
15.10 |
5 |
1 |
28.9 |
| T1 |
HPAII control |
15.10 |
5 |
0 |
29.9 |
| T1 |
MSPI |
15.10 |
5 |
.5 |
29.4 |
| T1 |
MSPI control |
15.10 |
5 |
0 |
29.9 |
November 16, 2010
Summary: Finished DNA extraction and quantification, DNMT1 primers ordered (thanks Sam).
Next steps for DNA: restriction digests w/ HPAII and MSPI (run on gel) - Thursday, PCR- next week
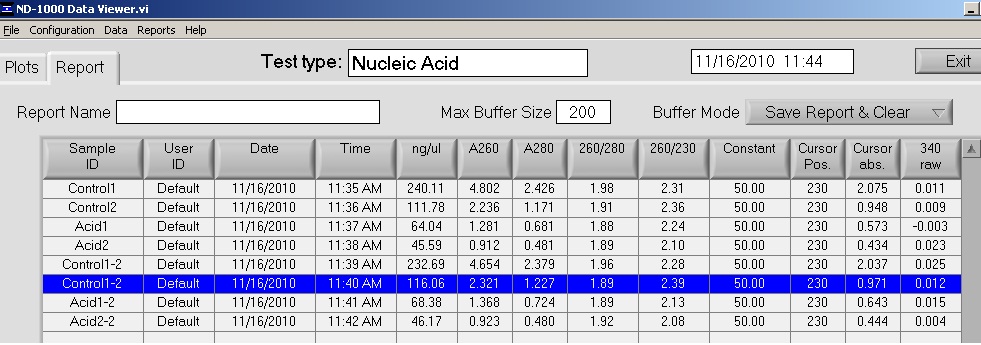
Averages:
Control 1: 236.4 ng/ul
Control 2: 113.92 ng/ul
Acid 1: 66.21 ng/ul
Acid 2: 45.88 ng/ul
November 15, 2010
Summary: Performing DNA extraction on 4 test samples for restriction digests and PCR for methylation fingerprinting, finding primers for DNMT1 on NCBI.
Samples from Caroline (AK):
Control
0_13_8
0_13_9
Acid shocked
24_10_7
24_10_8
Primers: 3 for DNMT1 in Danio rerio, Salmo salar
Details on PrimerDatabase
November 10, 2010
Goals:
For DNMT3b:
- Use Geneious to make trees and figure out if there is overlap with sequences
DNMT1:
- Find primers on NCBI
- Do PCR, run on gel, sequence
- Follow up w/ comparison to DNMT3b
Restriction Digest:
- Extract DNA from acid shocked/control juveniles (tails)
- Run restriction digest with HPAII/MSPI
- Run products on gel
- PCR for methylation fingerprinting
Gene expression:
- Extract RNA from acid shocked/control juveniles (heads)
- Reverse transcribe to make cDNA
- run qPCR to see gene expression for both DNMT3b and DNMT1
TO DO:
1. Review protocols (below)
2. Find and order primers for DNMT1, analyze sequences in Geneious
3. Tissue extraction, DNA extraction part I (1-2h, leave overnight)
4. DNA extraction part II (1-2h)
5. Restriction digests w/ DNA, make agarose gel (1-2h, leave overnight)
6. Run digests on gel, cut products out for PCR (1-2h)*
7. RNA extraction (2-3h)
8. Reverse transcription for cDNA (1-2h)
9. qPCR for cDNA (1-2h prep, ~4h run)
Protocols
DNA Extraction (Qiagen Kit)
1. 25 mg of tissue in 1.5uL tube.
2. 180 uL of ATL buffer added
3. 20 uL of Proteinase K added, vortexed for ~10 seconds
*Note: Enzymes should NOT be vortexed before use and should be placed in cooler while in use.
4. Samples kept at 55 degree hot plate overnight for lysis (~16h)
1. Samples were taken from 55 degree plate and vortexed for 15 seconds.
2. 200 uL AL buffer added, mixed by pipetting and then vortexed
3. Incubated at 70 degrees for 10 minutes
*Note: precipitate/gelatinous precipitate may form but will dissolve during incubation
4. 200 uL 100% (200 proof) ethanol added, then vortexed twice for 15 sec.
5. Samples loaded onto DNeasy Mini spin column + centrifuged for 1 min @ 8000 rpm.
*Note: Make sure to apply all of precipitate to spin column
6. Place spin column onto new 2 mL collection tube
7. Add 500 mL buffer AW1, centrifuge for 1 min @ 8000 rpm
8. Place spin column in new 2 mL collection tube and add 500 uL buffer AW2, centrifuge for 3 min @ 14,000 rpm to dry membrane
9. Spin columns placed in 1.5 mL centrifuge tube, 100 uL of buffer AE was added to membrane of each, centrifuged for 1 min @8000 rpm.
10. Samples stored at -20 freezer.
Restriction Digest
For HPAII:
1 ug DNA (1/concentration x .001)
5 uL 10X Buffer 1
1 uL HPAII enzyme
Water up to 50uL
For MSPI:
1 ug DNA
5 uL 10X Buffer 4
1 uL MSPI enzyme
Water up to 50uL
Place in 37degC waterbath overnight, heat inactivate, store at 20degC.
Run products on 1.2% agarose gel
Primer reconstitution/PCR
Reconstituting primers:
In TE buffer for 100uM concentration, then 10uL in 90uL of buffer as 10uM working stock.
MasterMix per reaction
- 1 uL template (PCR water for negative controls)
- 12.5 uL 2X Apex MasterMix
- .5 uL Forward Primer
- .5 uL Reverse Primer
- 10.5 uL PCR water
RNA Extraction
1. Add 500uL of TriReagent to the 1.5mL snap cap tube containing 25mg tissue and store on ice
2. Homogenize the tissue using a disposable pestle. If the tissue is difficult to homogenize, carefully close the tube tightly and briefly vortex the sample.
3. After the sample is completely homogenized, add an additional 500uL of TriReagent to the tube and close the tube tigh. Vortex vigorously for 15s.
4. Store at -80 if necessary.
1. Turn on heating block to 55°C.
2. Incubate your homogenized tissue at room temperature (RT) for 5 mins.
3. In the fume hood, add 200uL of chloroform to sample and close the tube.
4. Vortex vigorously for 30s. Should turn milky.
5. Incubate tube at RT for 5 mins.
6. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
7. Gently remove tube from microfuge. Be sure not to disturb the tube.
8. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
9. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
10. Add 500uL isopropanol to the new tube containing RNA and close the tube.
11. Mix by inverting the tube numerous times until the solution appears uniform.
12. Incubate at RT for 10 mins.
13. Spin in refrigerated microfuge at max speed for 8 mins. When placing your tube in the microfuge position the tube hinge pointing up, away from the center of the microfuge.
14. A small, white pellet (RNA and salts) should be present.
15. Remove supernatant.
16. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube.
17. Spin in refrigerated microfuge at 7500g for 5mins.
18. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
19. Briefly spin tube (~15s) to pool residual EtOH.
20. Using a small pipette tip (P10 or P20 tips), remove remaining EtOH.
21. Leave tube open and allow pellet to dry at RT for no more than 5mins.
22. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
23. Incubated tube at 55C for 5mins. to help solubilize RNA.
24. Remove tube from heat, flick a few times to mix and place sample on ice.
25. Quantitate RNA yield using Nanodrop spectrophotometer
REVERSE TRANSCRIPTION
Mix stock RNA sample by inverting tube several times.
- In a 0.5 ml PCR tube mix:
- 5 μl of YOUR total RNA (extracted and quantified in lab last week)
- 1 μl of oligo dT
- 4 μl of nuclease free H2O
- Incubate the mixture for 5 min at 70C on the thermocycler then immediately transfer to ice. Briefly centrifuge tube and the add the following:
- 5 μl of M-MLV 5X Reaction Buffer
- 5 ul of dNTPs
- 1 μl of M-MLV RT
- 4 μl of nuclease free H2O
- Incubate the mixture for 60 min at 42C and then heat inactivate at 70C for 3 min on the thermocycler.
- Spin down the sample in a desk top centrifuge.
- Store on ice or at -20C
qPCR
| Component |
Volume |
Final Conc. |
| Master Mix, 2X (Immomix) |
25µL |
1x |
| Syto-13 dye (50uM) |
2µL |
2µM |
| upstream primer, 10μM |
2.5μl |
2.5μM |
| downstream primer, 10μM |
2.5μl |
2.5μM |
| Ultra Pure Water |
16uL |
NA |
2. Add mastermix to wells of a white PCR plate
3. Thaw cDNA samples.
4. Add 2uL cDNA template to each reaction.
5. Add 2uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. If necessary, spin the strips to collect volume in the bottom of the wells.
8. Ensure the lids are clean and place strips on ice.
9. Load the plate, verify the PCR conditions and start the run
November 8, 2010
Summary: Set up selected PCR products from last week's gel to sequence. Order on plate is on sequence_log. All bands cut out of gel, bands in red are going to be sequenced (picked b/c of product size).

November 4, 2010
Summary: Loaded PCR products in gel, had problems, most products got flushed out but running it anyway @ 110 volts for 1h30min. Cut out products for sequencing, stored in 4deg.
Repeated PCR as on October 28 to run samples again tomorrow.
Made 1.2% agarose gel for tomorrow.
Also made 1x modified TAE buffer for carboy. (1000 mL 10x TAE buffer in 9000 mL DI water)
Gel picture:
Hyperladder 1, primer 1: gDNA, cDNA, negative, negative, primer 2: gDNA, cDNA, negative, negative, primer 3: gDNA, cDNA, negative, negative
Cut out all products visible (all except for cDNA in primer 2) Seems to be some contamination into negative controls.
November 3, 2010
Summary: Made 1.2% agarose gel to run PCR results tomorrow.. would have run them today but messed up on gel mold seals and had to start over :(
October 28, 2010
Summary: Reconstituting primers in TE buffer and running PCR on sockeye gDNA and cDNA samples to determine if primers work!
Next steps: Continue to play with Geneious to make trees w/ DNMT3 sequences. If PCR works, then will have to discuss what samples to use for gene expression portion of project. Since running other animals on Geneious, it would be beneficial to run primers on other animals...human, rat, mouse, etc.
Primer reconstitution:
Reconstituting primers in TE buffer for 100uM concentration, then 10uL in 90uL of buffer as 10uM working stock.
Primer working stocks are in my -20deg box, master stocks are stored in PrimerStock box 7, #61-66.
Primer set #1: omykissDNMT3b
Forward (sr_ID 1014): 288 uL TE buffer
Reverse (sr_ID 1013): 234 uL TE buffer
Primer set #2: salmoDNMT3
Forward (sr_ID 1018): 302 uL TE buffer
Reverse (sr_ID 1017): 295 uL TE buffer
Primer set #3: salmo_danioDNMT3b
Forward (sr_ID 1016): 325 uL TE buffer
Reverse (sr_ID 1015): 298 uL TE buffer
PCR Methods:
I made one master mix per primer set, each containing the following:
MasterMix per reaction
- 1 uL template (PCR water for negative controls)
- 12.5 uL 2X Apex MasterMix
- .5 uL Forward Primer
- .5 uL Reverse Primer
- 10.5 uL PCR water
Total: 25uL
MasterMix total (4 reactions + 10%, x4.4)
- 55 uL 2X Apex MasterMix
- 2.2 uL Forward Primer
- 2.2 uL Reverse Primer
- 46.2 uL PCR water
Total: 105.6
Total of 4 templates for each primer set containing gDNA, cDNA, negative control 1, negative control 2.
gDNA was Control
#4 from acid shocked juvenile sockeye, cDNA was pooled from 4 samples of sockeye brain cDNA.
Load into thermocycler with following profile:
95C for 10 min
40 cycles
-95C for 30 sec
-55C for 30 sec
-72C for 90 sec
-72C for 3 min
Store at 4degC
October 27, 2010
Summary: Primers are here! Will run PCR tomorrow since I have more time. Today I will be running different sequences for genes listed below in fish and mammals to determine any overlap (using Geneious)
October 20, 2010
Finished proposal! Turned in to Greg Jensen.
Ready to order following primers: (info also on PrimerDatabase doc)
DW554405.1: Homolog for DNMT3a in Atlantic salmon
salmo_DNMT3F: TCCAACATGGGTCGCGGTGC
salmo_DNMT3R: CCCCCTATGGCAGGGCTGGT
GE838101.1: DNMT3b in Oncorhynchus/Danio reio
salmo_danio_DNMT3bF: TGGGGGAACCTGCCTGGCAT
salmo_danio_DNMT3bR: GCCTGGCCAGCCGACTCATG
EZ813239.1: DNMT3b in Oncoryhnchus mykiss
omykiss_DNMT3bF: TGAGGGCACTGGTCGGCTGT
omykiss_DNMT3bR: GAAGTAGCGGGCGCGGTGAG
October 12, 2010
Second draft of proposal is finished. Searching DNMT3b genes and primers- closest to salmon I found is Danio reio (zebrafish):
http://www.ncbi.nlm.nih.gov/nucest?Db=gene&Cmd=retrieve&dopt=full_report&list_uids=30659#geneGeneral%20protein%20info
Primers can be found under links in "General Gene Information"- need to confirm before ordering
October 11, 2010
This month I will focus on finishing up my proposal/introduction section and obtaining primers for DNMT3b. I will do a conventional PCR on salmon and oysters to test the primers, then conduct gene expression analysis.
August 20,2010
Because I do not have replicates for the DotBlot, I will combine the controls, treatments, and positive controls to do T-tests/ANOVA. Based on only the graphs, it looks like there was something wrong with dilutions since the measurements are not proportional for each and even reversed for positive controls. Furthermore, error bars indicate no significant difference between concentrations. Next steps might include focusing on doing several replicates of one control and one treatment. If there are no differences, then maybe trying the same for juvenile vs. adult.
August 19, 2010
Summary: Quantifying DotBlot using ImageJ software, then statistics on data.

| DNA [] |
Area |
Mean |
Min |
Max |
|
| C1 |
1.5 |
611 |
137.645 |
118 |
188 |
| 0.75 |
611 |
149.807 |
129 |
196 |
|
| 0.38 |
611 |
160.686 |
144 |
197 |
|
| 0.19 |
611 |
168.935 |
149 |
200 |
|
| Neg. |
611 |
199.062 |
181 |
208 |
|
| C3 |
1.5 |
611 |
161.187 |
147 |
201 |
| 0.75 |
611 |
165.365 |
147 |
202 |
|
| 0.38 |
611 |
175.481 |
158 |
188 |
|
| 0.19 |
611 |
176.314 |
165 |
198 |
|
| Neg. |
611 |
198.298 |
186 |
207 |
|
| T2 |
1.5 |
611 |
173.036 |
135 |
197 |
| 0.75 |
611 |
166.223 |
158 |
198 |
|
| 0.38 |
611 |
169.191 |
150 |
192 |
|
| 0.19 |
611 |
174.923 |
159 |
203 |
|
| Neg. |
611 |
191.774 |
181 |
199 |
|
| T3 |
1.5 |
611 |
156.974 |
146 |
184 |
| 0.75 |
611 |
162.288 |
131 |
199 |
|
| 0.38 |
611 |
163.149 |
148 |
189 |
|
| 0.19 |
611 |
165.876 |
151 |
183 |
|
| Neg. |
611 |
178.552 |
168 |
193 |
|
| Pos. Control |
1.5 |
611 |
167.373 |
106 |
201 |
| 0.75 |
611 |
184.884 |
177 |
197 |
|
| 0.38 |
611 |
182.141 |
157 |
198 |
|
| 0.19 |
611 |
173.39 |
155 |
191 |
|
| Neg. |
611 |
176.915 |
168 |
184 |
|
| Pos. Control |
1.5 |
611 |
191.005 |
182 |
203 |
| 0.75 |
611 |
180.614 |
172 |
205 |
|
| 0.38 |
611 |
188.52 |
179 |
198 |
|
| 0.19 |
611 |
177.118 |
169 |
190 |
|
| Neg. |
611 |
175.514 |
164 |
191 |
August 4, 2010
Summary: Finally running Western Blot!
DotBlot/Western Blot picture (adjusted to increase contrast, saturation)
Order by column:
1. C1 - 1.5, .75, .38, .19, Negative control
2. C3 - 1.5, .75, .38, .19, Negative control
3. T2- 1.5, .75, .38, .19, Negative control
4. T3- 1.5, .75, .38, .19, Negative control
5. Oyster- 1.5, .75, .38, .19, Negative control
6. Oyster- 1.5, .75, .38, .19, Negative control
I will most likely use software to quantify!
August 2, 2010
Summary: Will be running DotBlot today with C1, C3, T2, T3, negative control, and the oyster positive control (x2).
The samples will be loaded in the following order for the DotBlot:
C1, C3, T2, T3, Positive, Positive
1.5
.75
.38
.19
Negative
I will be running the Western Blot Wednesday morning (since it takes FOUR hours).
July 30, 2010
Summary: Repeating DotBlot with positive controls (oyster) @ 103 ug/uL
Will be using C1, C3, T2, T3 (see table from July 2 for dilutions), negative controls for each, and a positive control. Made dilutions today, will run DotBlot and Western Blot on Monday.
|| Positive Control || || || || ||
|| Target (ug) || conc. Sample || uL sample || uL H2O || uL 20x SSC ||
|| 1.5 || 103 || 14.563107 || 125.4369 || 60 ||
|| 0.75 || || 7.2815534 || 132.7184 || 60 ||
|| 0.38 || || 3.6893204 || 136.3107 || 60 ||
|| 0.19 || || 1.8446602 || 138.1553 || 60 ||
|| || total vol. || 27.378641 || || ||
July 20, 2010
It looks like the DotBlot/Western Breeze did not work at all :( Looks like the DNA did not bind because, once dried, the membrane looks all the same color (pale purple). Not worth quantifying.
Possible problems:
- DNA dilutions too low- but Mac performed w/ same concentrations and had no problem
- Did not use kit's antibody wash, used 1x TBST instead and did not place on rocker while washing
- Pipetting errors, especially when making primary antibody dilution (1 uL in 10 mL)
- Did not add a positive control
To do:
- Call Invitrogen and ask about TBST as wash buffer
- Repeat DotBlot/Western Breeze adding positive control and placing on rocker while washing
Positive control: C. gigas (oyster) 103 ug/uL
Will run highest dilution of other samples, so 1.5ug of sample.
Note: Because Mac and Sam are both gone a lot this week, I will wait to run DotBlot/Western Breeze until next week and will finish up my proposal and introduction this week.
July 16, 2010
Summary: Western Breeze today!
Notes: I did not put the membrane on a rocker for washes, so that might account for some background noise. Membrane developed for ~10min total and it looks like there's some purple on the non-DNA areas (that could be due to bad washing).
Will scan and quantify on Monday!
July 12, 2010
Summary: Prepping for Western Blot- going over protocol, dilutions, and TBST buffer recipe. I will make dilutions and the buffer tomorrow and perform the Western Blot on Thursday.
July 6, 2010
Summary: Made DNA dilutions and performed DotBlot.
Samples were loaded in the following order:
| C2 |
C4 |
T1 |
T4 |
| 1.5 |
1.5 |
1.5 |
1.5 |
| 0.75 |
0.75 |
0.75 |
0.75 |
| 0.38 |
0.38 |
0.38 |
0.38 |
| 0.19 |
0.19 |
0.19 |
0.19 |
| 6X blank |
6X blank |
6X blank |
6X blank |
July 2, 2010
I have decided that running both groups of samples (juvenile controls/acid treated and WA/RU populations) is too much for a first run, especially with dilutions. I'm only going to run the control/acid treated juveniles because they showed the most differences in the ELISA from April 6/7 and they have the highest quantity of DNA. I might even run two controls and two treatments instead of running all four for both groups.
Dilution Calculations (to be made on Monday, prior to DotBlot Tuesday am)
| CONTROLS |
||||
| C1 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
63.88 |
23.481528 |
116.5185 |
60 |
| 0.75 |
11.740764 |
128.2592 |
60 |
|
| 0.38 |
5.9486537 |
134.0513 |
60 |
|
| 0.19 |
2.9743269 |
137.0257 |
60 |
|
| total vol. |
44.145272 |
|||
| C2 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
132.94 |
11.283286 |
128.7167 |
60 |
| 0.75 |
5.6416428 |
134.3584 |
60 |
|
| 0.38 |
2.8584324 |
137.1416 |
60 |
|
| 0.19 |
1.4292162 |
138.5708 |
60 |
|
| total vol. |
21.212577 |
|||
| C3 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
75.44 |
19.883351 |
120.1166 |
60 |
| 0.75 |
9.9416755 |
130.0583 |
60 |
|
| 0.38 |
5.0371156 |
134.9629 |
60 |
|
| 0.19 |
2.5185578 |
137.4814 |
60 |
|
| total vol. |
37.3807 |
|||
| C4 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
91.35 |
16.420361 |
123.5796 |
60 |
| 0.75 |
8.2101806 |
131.7898 |
60 |
|
| 0.38 |
4.1598248 |
135.8402 |
60 |
|
| 0.19 |
2.0799124 |
137.9201 |
60 |
|
| total vol. |
30.870279 |
|||
| TREATED |
||||
| T1 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
52.39 |
28.631418 |
111.3686 |
60 |
| 0.75 |
14.315709 |
125.6843 |
60 |
|
| 0.38 |
7.2532926 |
132.7467 |
60 |
|
| 0.19 |
3.6266463 |
136.3734 |
60 |
|
| total vol. |
53.827066 |
|||
| T2 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
46.14 |
32.509753 |
107.4902 |
60 |
| 0.75 |
16.254876 |
123.7451 |
60 |
|
| 0.38 |
8.2358041 |
131.7642 |
60 |
|
| 0.19 |
4.117902 |
135.8821 |
60 |
|
| total vol. |
61.118336 |
|||
| T3 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
44.92 |
33.392698 |
106.6073 |
60 |
| 0.75 |
16.696349 |
123.3037 |
60 |
|
| 0.38 |
8.4594835 |
131.5405 |
60 |
|
| 0.19 |
4.2297418 |
135.7703 |
60 |
|
| total vol. |
62.778272 |
|||
| T4 |
||||
| Target (ug) |
conc. Sample |
uL sample |
uL H2O |
uL 20x SSC |
| 1.5 |
46.69 |
32.126794 |
107.8732 |
60 |
| 0.75 |
16.063397 |
123.9366 |
60 |
|
| 0.38 |
8.1387877 |
131.8612 |
60 |
|
| 0.19 |
4.0693939 |
135.9306 |
60 |
|
| total vol. |
60.398372 |
July 1, 2010 (Halfway through 2010!)
Summary: Today I'll be playing around with the DotBlot/Western Breeze apparatus and will be doing the actual DotBlot on Tuesday with Mac.
Helpful protocol of manifold assembly can be found here
Protocol for DotBlot (same as listed below) can be found here
June 29, 2010
I'm back!
Summary of next steps: Will try DotBlot with first DNA samples used (sockeye salmon juvenile, juvenile stressed, adult WA, adult RU). If DotBlot is successful, then I will stay on project. If not, I will consider alternatives. Still waiting to hear from Fall internship.
Today: Review DotBlot/Western breeze protocols and make sure I have DNA samples desired- otherwise, plan for extraction this week.
DotBlot Analysis for 5-MeC in DNA Protocol Using Uncharged Nylon Membrane
Summary: DNA is heat denatured and applied to membrane in a salt buffer. After blotting, membrane is treated with denaturation and neutralization solutions, DNA is immobilized by UV irradiation (for nylon).
Note: Denaturation solution, neutralization solution, and 6x/20x SSC already made by Mac (stored at room temp)
1. Cut a piece of nylon membrane to size of the manifold.
2. Pour 6X SSC to depth of ~.5 cm in a glass dish- place membrane on surface and allow to submerge. Leave for 10 min.
3. Cut piece of Whatman 3MM filter paper to size of the manifold and wet in 6X SSC.
4. Place Whatman 3MM paper in manifold and lay nylon membrane on top of it. Assemble manifold according to manufacturer's instructions (ensure there are no air leaks)
5. Add 20X SSC and water to every DNA sample to give a final concentration of 6X SSC in a volume of 200-400 uL.
Can refer to Mac's dilutions here
6. Denature DNA by placing in water bath or oven for 10 min at 100degC, then place in ice.
7. Switch on suction to manifold defice, apply 500 uL 6X SSC to each well and allow it to filter through, leaving suction on.
*Note: wells that are not being used can be blocked off with masking tape*
8. Spin DNA samples in microcentrifuge for 5 sec, apply to wells - avoid touching membrane with pipet. Allow samples to filter through.
9. Soak piece of Whatman 3MM paper in denaturation solution- dismantle apparatus and place membrane of Whatman paper. Leave for 10 min.
10. Soak piece of Whatman 3MM paper in neutralization solution- transfer membrane to new Whatman paper. Leave for 5 min.
11. Place membrane on dry piece of Whatman paper and allow to dry.
12. Wrap dry membrane in UV-transparent plastic wrap, place DNA-side-down on UV transilluminator, and immobilize DNA by irradiating for appropriate time- 2min at 120 kJ for crosslinking.
13. Store membrane dry between sheets of Whatman paper for months at room temp, in desiccator at room temp or 4degC for long term storage.
Invitrogen Western Breeze Chromogenic Immunodetection Protocol (Nitrocellulose membrane)
Note: Will use protocol for nitrocellulose membrane but will keep nylon membrane from DotBlot- DNA binds better to nylon and protocol works just fine with it!
1. Place membrane in 10mL of appropriate Blocking Solution (follow protocol for 20mL total volume) in covered, plastic dish provided by kit.
2. Incubate for 30 min on rotary shaker set at 1 rev/sec, decant Blocking Solution.
3. Rinse membrane with 20 mL water for 5 minutes, decant. Repeat once.
4. Incubate membrane with 10 mL of Primary Antibody Solution for 1 hour, then decant- 1:5000 and 1:10,000 dilution of primary 5-mC Antibody
5. Wash membrane for 5 min with 20 mL of Antibody Wash, then decant. Repeat 3 times.
6. Incubate membrane in 10mL of Secondary Antibody solution for 30 min, then decant.
7. Wash membrane for 5 min with 20 mL of Antibody Wash, then decant. Repeat 3 times.
8. Rinse membrane with 20 mL water for 2 minutes, then decant. Repeat twice.
9. Incubate membrane in 5 mL of Chromogenic Substrate until purple bands develop- complete in 1-60 minutes.
10. Rinse membrane with 20 mL of water for 2 minutes. Repeat twice.
11. Dry membrane on a clean piece of filter paper to open air, by stream of slightly warm air, or under infrared lamp.
Samples to use:
Sockeye salmon SA1-SA8 from April 1 (1-4 WA, 5-8 RU)- All samples available, concentrations from 40-66.7 mg/uL (dilutions made for ELISA)
Sockeye salmon C1-C4, T1-T4 (Control vs. acid stressed juveniles) from March 15
There's definitely enough DNA for the C1-C4 and T1-T4 samples, but depending on dilutions I may have to extract more DNA from other samples- can get RU samples from Caroline and use WA samples from January 19.
June 4, 2010
Results are murky.. can't see distinct bands for any of the undigested samples. It looks like I may have used too little DNA...Will need to run yet again to improve results.
June 2, 2010
Summary: Running restriction digests from yesterday.
10uL 5x Loading Dye added to each sample.
24 uL of each sample run in the following order @110 volts (15uL ethidium bromide added to positive end):
Hyperladder 1 (5uL)
11/14: HPAII undigested, MSPI undigested, HPAII, MSPI
11/28: HPAII undigested, MSPI undigested, HPAII, MSPI
12/14: HPAII undigested, MSPI undigested, HPAII, MSPI
1/1: HPAII undigested, MSPI undigested, HPAII, MSPI
June 1, 2010
Summary: Re-doing restriction digests for 11/14, 11/28, 12/14, and 1/1. Making 1.2% agarose gel and storing it for use tomorrow.
Running restriction digests with .5 ug of DNA instead of the usual 1 ug (I may run out if I use more!). Samples set in 37degC water bath overnight.Will run samples on gel tomorrow.
Having crazy problems with formatting... table I inserted merged with table from May 11....visually unappealing but still correct.
| Sample |
Mix |
DNA |
10Xbuffer (1for hpa, 4 for msp) |
Enzyme |
Water |
|
| 11/14 |
HPAII |
11.7 |
5 |
1 |
32.3 |
|
| HPAII-Control |
11.7 |
5 |
0 |
33.3 |
||
| MSPI |
11.7 |
5 |
0.5 |
32.8 |
||
| MSPI-Control |
11.7 |
5 |
0 |
33.3 |
||
| 11/28 |
HPAII |
4.5 |
5 |
1 |
39.5 |
|
| HPAII-Control |
4.5 |
5 |
0 |
40.5 |
||
| MSPI |
4.5 |
5 |
0.5 |
40 |
||
| MSPI-Control |
4.5 |
5 |
0 |
40.5 |
||
| 12/14 |
HPAII |
0.7 |
5 |
1 |
43.3 |
|
| HPAII-Control |
0.7 |
5 |
0 |
44.3 |
||
| MSPI |
0.7 |
5 |
0.5 |
43.8 |
||
| MSPI-Control |
0.7 |
5 |
0 |
44.3 |
||
| 1/1 |
HPAII |
0.5 |
5 |
1 |
43.5 |
|
| HPAII-Control |
0.5 |
5 |
0 |
44.5 |
||
| MSPI |
0.5 |
5 |
0.5 |
44 |
||
| MSPI-Control |
0.5 |
5 |
0 |
44.5 |
||
May 25, 2010 Summary: Extracting DNA from 11/14 black salmon sample (ran out last time) and re-specking 11/28, 12/14, and 1/1. Next steps: Re-do restriction digests and run on gel. Can't get file to work... Concentrations in ng/uL: 11/14: 42.65 11/28: 111.46 12/14: 734.54 1/1: 1015.28 Concentrations are almost the same as before. Problems with gel are probably due to pipetting errors. May 18, 2010 Summary: Working on proposal and introduction and possible oyster DNA extraction. May 13, 2010  The gel shows smears for both HPAII and MSPI for 11/14 (lanes 2 and 3). There are no distinct bands for the undigested DNA for 11/28, which is a little suspicious (lanes 4 and 5), and HPAII and MSPI also show smears (lanes 6 and 7). The two undigested samples for 12/14 show bands (lanes 8 and 9), with a band and a smear for HPAII and a smear for MSPI (lanes 10 and 11). All samples for 1/1 show smears. Because the undigested samples are not showing distinct bands as they should, I am concerned that this data is futile. I expected to see less methylation in the earlier samples with an increase in methylation with age. It is clear that the samples will need to be run again for more accurate results. As a reminder: Methylated: Undigested- band HPAII- band MSPI- smear Unmethylated: Undigested- band HPAII- smear MSPI - smear May 12, 2010 Summary: Gel electrophoresis of restriction digest samples. Samples taken out of water bath at 12:30 pm. Samples loaded in the following order: Hyperladder I 11/14: HPAII, MSPI 11/28: Undigested, HPAII, MSPI 12/4: Undigested, HPAII, MSPI 1/1: Undigested, HPAII, MSPI 10uL of loading dye added to each, 5uL Hyperladder I and 30uL of each sample in wells. May 11, 2010 Summary: Re-quantified black salmon samples DNA and restriction digests (HPAII and MSPI). |
Sample |
Mix |
DNA |
10Xbuffer (1for hpa, 4 for msp) |
Enzyme |
Water |
| 11/14 |
HPAII |
43.18 |
5 |
1 |
.82 |
|
| MSPI |
43.18 |
5 |
.5 |
1.32 |
||
| 11/28 |
HPAII |
9.59 |
5 |
1 |
34.41 |
|
| MSPI |
9.59 |
5 |
.5 |
34.91 |
||
| 12/4 |
HPAII |
1.36 |
5 |
1 |
42.64 |
|
| MSPI |
1.36 |
5 |
.5 |
43.14 |
||
| 1/1 |
HPAII |
1.0 |
5 |
1 |
43.0 |
|
| MSPI |
1.0 |
5 |
.5 |
43.5 |
Next steps: Run a gel!

May 8, 2010 -
Ran 0.5ug of Sonia's Black family salmon gDNA samples (isolated 5/6/2010) on a 1.2% agarose TAE gel.
Hyperladder --- 11/14 --- 11/28 --- 12/14 --- 1/1

May 6, 2010
Summary:We have a new DNeasy extraction kit! DNA extraction of Sockeye Salmon (Black family), samples 11/14(embryo BT), 11/28(embryo), 12/14(alevin w/ yolk sac), 1/1(alevin)
Next steps: Restriction digest with MSPI/HPAII to determine methylation visually on gel.

May 5, 2010 -
Summary: Ran the remaining half samples that Sonia prepared for loading on the gel yesterday. Loaded on the same gel that Sonia used yesterday (gel was stored O/N @ 4C).
Hyperladder, Roberts Kit, Horner-Devine Kit

Because on both occasions the gel isn't showing a distinct band for DNA we can assume that our kit is inaccurate!
*Note: need to upload concentrations..
May 4, 2010
Summary: Our DNeasy Blood and Tissue Extraction kit is showing weird bands when samples are run on gel. I'm running the same sample with our kit and with the Horner-Devine lab's kit to compare. Sample used was 1/1 Green alevin (fish cut in half).
Samples quantified and run on gel. Used each kit's AE buffer to blank.


Hyperladder, Roberts, Horner-Devine -
Samples were run too long, by that dope, Sam. Will run remaining half tomorrow to be able to visualize the full size range.

April 29, 2010
Summary: Going to extract DNA from samples of two salmon populations (green and black)- samples are in developmental stages- embryo through alevin. Research on salmon developmental life stages!
Both populations have samples for the following dates:
The only distinct developmental stage I have found is eyes @ day 20 and hatching of alevin @ day 40, so I will assume "Life History Day #" around Day 20, 20 Nov.
| Date |
Sampling Day # |
Life History Day # |
Characteristics |
| 14 Nov |
1 |
14 |
BT embryo |
| 20 Nov |
7 |
20 |
BT embryo- eyes visible |
| 2 Nov |
11 |
24 |
Eyes + spinal cord |
| 28 Nov |
15 |
28 |
None |
| 4 Dec |
21 |
34 |
None |
| 14 Dec |
31 |
44 |
Hatch- Alevin |
| 24 Dec |
41 |
54 |
Yolk sac smaller |
| 1 Jan |
49 |
61 |
Yolk sac almost gone |
April 28, 2010
Summary: Conclusion of DNA extraction and quantification.

The DNA concentrations are terrible and will probably not be used. Will talk to Caroline about other possibilities.
April 27, 2010
Summary: Looking for genes with CpG islands on Inquiry Bioinformatics Portal and DNA extraction from senescent/pre-senescent salmon for Caroline.
DNA extraction:
Samples are stored in TriReagent from RNAlater protocol, which may cause some problems. I'm running 2 trial samples to see if DNA concentration is adequate.
Samples left in 56degC incubator overnight to lyse.
Samples were washed 3 times with 1x PBS, then Qiagen kit protocol was followed (see 1/19 and 1/20 for protocol).
Sample info can be found here
April 22, 2010
Summary: Analysis of gel pictures from 4/20. Also, trying out iNquiry Bioinformatics Portal to search for genes with CpG islands.
Next steps: Compare methylation among life stages, then pick specific genes to compare. Technique still undecided because ELISA is unreliable.
g11 (lane 2) shows signs of degradation and NB02 (lane 4) shows slight signs of degradation. All other samples seem to be normal.
Literature Consulted
- I will be keeping a running list of literature I come across. Thanks Mac Gavery and Kerensa King for providing access to many of these!
Baccarelli, A. and Bollati, V., 2009. Epigenetics and Environmental Chemicals. Current Opinion in Pediatrics 21(2); 243-251.
*Review of environmental toxins and proposed effects on methylation.
Dolinoy, DC., Huang, D., and Jirtle, R.L., 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences of the USA 104; 13056-13061.
*Mouse study- epigenetic changes through maternal line.
Dupont C, Armant DR, and Brenner CA, 2009. Epigenetics: Definition, Mechanisms and Clinical Perspective. Seminars in Reproductive Medicine 27; 351-357.
Elango N, Hunt BG, Goodisman M, and Yi S, 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proceedings of the National Academy of Science 106: 11206-11211.
*Methylation differences in honeybee castes.
Fronk J, Tank GA, and Langmore JP, 1992. DNA methylation pattern changes during development of a sea urchin. PubMed: Biochem J 283: 751-753.
Ho DH and Burggren WW, 2009. Epigenetics and transgenerational transfer: a physiological perspective. Journal of Experimental Biology 213: 3-16.
Laird PW, 2010. Principles and challenges of genome-wide DNA methylation and analysis. Nature Review Epigenetics 11: 191-203.
*Pros and cons of different techniques.
Rodriguez-Osorio N, Wang HF, Rupinski J, Bridges SM, and Memili E, 2010. Comparative functional genomics of mammalian DNA methyltransferases. Reproductive Biomedicine Online 20; 243-255.
Tweedie S, Charlton J, Clark V and Bird A, 1997. Methylation of genomes and genes at the invertebrate-vertebrate boundary. American Society for Microbiology 17: 1469-1475.
*Evolutionary perspective of methylation between vertebrates and invertebrates.
Weber M and Schübeler D, 2007. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Current Opinion in Cell Biology 19: 273-80.
Angers, B., E. Castonguay, and R. Massicotte. 2010. Environmentally induced phenotypes and DNA methylation: how to deal with undpredictable conditions until the next generation and after. Molecular Ecology 19: 1283-1295.
Blouin, M.S., V. Thuillier, B. Cooper, V. Amarasinghe, L. Cluzel, H. Araki, and C. Grunau. 2010. No evidence for large differences in genomic methylation between wild and hatchery steelhead (Oncorhynchus mykiss). Canadian Journal of Fisheries and Aquatic Sciences 67:217-224.
*Steelhead- no difference in methylation between hatchery and wild.
Meaney, M.J. Epigenetics and the biological definition of gene x environment interactions. Child Development 81(1)41-79.
Zhang, T.Y. and M.J. Meaney. 2010. Epigenetic and the environmental regulation of the genome and its function. Annual Review of Psychology 61:439-466.
Aubin-Horth, N and S.C.P. Renn. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Molecular Ecology 18: 3763-3780.
Bossdorf, O., C.L. Richards, and M. Pigliucci. 2008. Epigenetics for Ecologists. Ecology Letters 11: 1060115.
Crews, D. 2008. Epigenetics and its implications for behavioral neuroendocrinology. Frontiers in Neuroendocrinology 29: 344-357.
Fagiolini, M., C.L. Jensen, and F.A. Champagne. 2009. Epigenetic influences on brain development and plasticity. Current Opinion in Neurobiology 19: 207-212.
Jablonka, E. and G. Raz. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. The Quarterly Review of Biology 84(2): 131-176.
Weaver, I.C.G., N. Cervoni, F.A. Champagne, A.C. D’Alessio, S. Sharma, J.R. Seckl, S. Dymov, M. Szyf, and M.J. Meaney. 2004. Epigenetic programming by maternal behavior. Nature Neuroscience 7(8): 847-854.
Rando, O.J. and K.J. Verstrepen. 2007. Timescales of genetic and epigenetic inheritance. Cell 128:655-668.
Richards, E.J. 2008. Population epigenetics. Current Opinion in Genetics and Development 18:221-226.
Araki, H., B.A. Berejikian, M.J. Ford, and M.S. Blouin. 2008. Fitness of hatchery-reared salmonids in the wild. Evolutionary Applications 1:342-355.
Liu, Z.J. and M. Maekawa. 2003. Polymerase chain-reaction-based methods of DNA methylation analysis. Analytical Biochemistry 317:259-265.
Last updated: April 27, 2010.
April 20, 2010
Summary: Troubleshooting for degraded DNA by running gel using adult gigas gDNA.
1 ug total DNA in dilution plus 5 uL of loading dye for total of 30uL to load. Samples loaded in same order as below with 5 uL Hyperladder I in first well. Gel run for 1 hour at ~175 Volts.
Sample dilutions for total 25 uL
- ID From [ug/uL] uL DNA (for 1 ug) uL Water
2 SB02 Mac 0.4102 2.4 22.6
3 NB02 Mac 0.3363 3 22
4 LC02 Mac 0.5684 1.8 23.2
5 DH13 Sam 0.3973 2.5 22.5
6 DH14 Sam 0.4657 2.1 22.9
7 DH15 Sam 0.5128 2 23
8 BB13 Sam 0.5462 1.8 23.2
9 BB14 Sam 0.3256 2 23
10 BB15 Sam 0.4216 2.4 22.6
(can't get Excel formatting to work on table...)
April 15 2010
Summary: Analysis of results for ELISA
*Note: I was sick the last week!
ELISA results can be found here
There are several problems...
1. The blank has a concentration of .18, which is higher than most of the actual samples.
Solution: Add a wash to each step to ensure that samples are clean are that the blank reads at 0.
2. The standard curve blows out at the 100 ng mark, indicating that going up to 200 ng is too much- I assumed that because vertebrates have a higher global methylation percentage than inverts, I would use a higher curve than what Mac used for her analysis. Maybe the antibody is limiting with such a high concentration of DNA.
Solution: Keep the standards from 5-100ng and decrease the max of 200ng in samples to 120ng or 100ng max (120 worked better anyway).
3. Most samples fell outside range of standard curve- only sample 1, 5, and 9 have concentration high enough to be within range, all others are below blank.
Solution: Add washes to ensure blank is at 0.
Next steps:
- Talk to Steven/Mac about how to improve ELISA and possible alternatives
- Start searching literature to write introduction and proposal
- If still on ELISA route, prep next samples + order new kit
April 6/7, 2010
Summary: Continuation of calculations for dilutions- parameters changed as I encountered problems.
Need to target 200ng and 120ng of DNA in a 20 uL dilution (in water). 3uL of dilution will go into 27 uL of DNA binding solution.
200/3=66.7
120/3=40.0
For 200ng: [sample ng/uL](V1 uL)=(66.7 ng/uL)(20uL)
à
1334/[sample]
For 120ng: [sample ng/uL](V1 uL)=(40.0 ng/uL)(20uL)
à
800/[sample]
| Dilutions for samples at 200 ng |
||||
| Sample |
Desired [] in diluent |
Sample [ng/uL] |
uL DNA |
uL Water |
| SA 1 |
66.7 |
245.86 |
5.425852 |
14.57415 |
| SA 2 |
66.7 |
498.43 |
2.676404 |
17.3236 |
| SA 5 |
66.7 |
120.12 |
11.10556 |
8.894439 |
| SA 6 |
66.7 |
151.68 |
8.794831 |
11.20517 |
| Dilutions for samples at 120 ng |
||||
| Sample |
Desired [] in diluent |
Sample [ng/uL] |
uL DNA |
uL Water |
| C1 |
40 |
63.88 |
12.52348 |
7.476518 |
| C2 |
40 |
132.94 |
6.017752 |
13.98225 |
| C3 |
40 |
75.44 |
10.60445 |
9.395546 |
| C4 |
40 |
91.39 |
8.753693 |
11.24631 |
| T1 |
40 |
52.39 |
15.27009 |
4.72991 |
| T2 |
40 |
46.14 |
17.33853 |
2.661465 |
| T3 |
40 |
44.92 |
17.80944 |
2.190561 |
| T4 |
40 |
46.69 |
17.13429 |
2.86571 |
| SA 3 |
40 |
772.69 |
1.035344 |
18.96466 |
| SA 4 |
40 |
296.38 |
2.699237 |
17.30076 |
| SA 7 |
40 |
101.79 |
7.859318 |
12.14068 |
| SA 8 |
40 |
146.72 |
5.452563 |
14.54744 |
| Dilutions for Standards |
||||||
| # |
Well |
Final [] ng |
Final [] ng/uL |
uL Stock |
uL Binding Solution |
Water |
| 1 |
A1 |
0 |
0 |
0 |
27 |
3 |
| 2 |
B1 |
13 |
0.43 |
0.26 |
29.25 |
2.75 |
| 3 |
C1 |
25 |
0.83 |
0.5 |
29.5 |
2.5 |
| 4 |
D1 |
50 |
1.66 |
1 |
29 |
2 |
| 5 |
E1 |
100 |
3.33 |
2 |
28 |
1 |
| 6 |
F1 |
200 |
6.66 |
4 |
26 |
Imprint Methylated DNA Quantification Kit (ELISA) Protocol (to be performed Thursday, April 8 2010):
Preparation:
To make 1X Wash Buffer (stored @ RT):
- Add 11 mL of 10X Wash Buffer (stored @ 4degC) to 99 mL of Water, Molecular Biology Reagent (does not come with kit)
- Store at RT for 6 months
DNA dilutions:
- Dilute DNA in Binding Solution (stored @ 4degC)
- Max dilution: 200 ng of DNA
Procedure:
DNA Binding
1. Add 30 uL of diluted DNA to each well and tilt plate from side to side to ensure bottom is coated.
- DNA Binding solution can be used as blank.
2. Cover with plate seal and incubate at 37degC for 60 min.
3. Add 150 uL Block Solution (-20degC) to each well.
4. Cover with plate seal and incubate at 37degC for 30 min.
5. Remove DNA and Block Solution from each well down the sink. (DNA binds to bottom of wells)
6. Wash 3 times with 150 uL of 1X Wash Buffer by smacking it on sink and then patting dry on paper towels (smack on sink technique - SOST)
Methylated DNA Capture
1. Dilute Capture Antibody (4degC) 1:1000 in 1X Wash Buffer. (1 uL Capture Antibody, 999 uL Wash Buffer) Diluted 1.15 uL antibody in 1150 uL wash buffer
2. Add 50 uL of diluted Capture Antibody to each well.*Accidentally added 150 to first row (standard curve) but removed 100 a few minutes later- Mac says won't affect results.
3. Cover with plate seal and incubate at RT for 60 min.
4. Remove diluted Capture Antibody from each well down the sink.
5. Wash 4 times with 150 uL of 1X Wash Buffer using SOST.
6. Dilute Detection Antibody (4degC) 1:1000 in 1X Wash Buffer (1 uL Detection Antibody, 999 uL Wash Buffer) Diluted 1.15 uL antibody in 1150 uL wash buffer
7. Add 50 uL of Detection Antibody to each well.
8. Cover with plate seal and incubate at RT for 30 min.
9. Remove diluted Detection Antibody from each well down the sink.
10. Wash 5 times with 150 uL of 1X Wash Buffer using SOST.
Detection
1. Add 100 uL of Developing Solution (4degC) to each well.
2. Cover with plate seal and incubate at RT
away from light
for 1-10 min. max and monitor color change - should turn
BLUE
.
3. Add 50 uL Stop Solution (4degC) to each well. Color should change to
YELLOW
.
4. Read absorbance at 450 nm on Seeb Lab plate reader.
5. Calculate relative global methylation levels.
April 2, 2010
Summary: Further research on ELISA assay and verification of availability of all materials and reagents.
Note: Very little DNA binding solution and stop solution left, 10X buffer at Mac's bench @ RT. Will ask Mac about quantities and where in the -20degC the methylated control and block solution are. (Mac away for the day).
*Note: talked to Sigma Aldrich and they said at least 27 uL of Binding Solution needed, but if necessary can be replaced by water or 1X TA buffer.
April 1, 2010
Summary: DNA quantification of Russia and Washington samples (thanks Caroline Storer)
Note: the highlighted SA 2 is actually SA 3.
Need to talk to Mac about making dilutions!

March 30, 2010
Summary: Discussion of Capstone project and research on Imprint protocol.
Doing 3 strips of ELISA assay
4 from Russia population: 2 at max DNA concentration, 2 at below max
4 from Washington population: 2 at max DNA, 2 at below max
4 non-treated juveniles
4 treated juveniles
Next steps: Can use MeDIP to quantify methylation at gene level.
Expected Timeline:
3/30/10: Research Imprint technique and preparation
4/1/10: Quantify DNA of Russia and Washington samples and calculate dilutions for all samples (2 from each group at 200 ng, 2 from each group at 150 ng?)
4/2/10: Research literature and check availability of all necessary materials and reagents for Imprint:
4degC:
10 X Wash Buffer
DNA Binding Solution
Capture Antibody
Detection Antibody
Developing Solution
Stop Solution
8-well Assay Strips
Juvenile Samples
WA + RU Samples
-20degC:
Methylated Control DNA (50ng/uL)
Block Solution
RT:
1X Wash Buffer
Water, Molecular Biology Reagent
Plate Seals
4/6/10: Perform DNA dilutions and make 1X Wash Buffer dilution (if necessary)
4/8/10: Perform Imprint (takes 5-6 hours: Schedule with Caroline)
4/9/10: Analysis of results and discussion of next steps
Imprint Methylated DNA Quantification Kit (ELISA) Protocol (to be performed Thursday, April 8 2010):
Preparation:
To make 1X Wash Buffer (stored @ RT):
- Add 11 mL of 10X Wash Buffer (stored @ 4degC) to 99 mL of Water, Molecular Biology Reagent (does not come with kit)
- Store at RT for 6 months
DNA dilutions:
- Dilute DNA in Binding Solution (stored @ 4degC)
- Max dilution: 200 ng of DNA
Procedure:
DNA Binding
1. Add 30 uL of diluted DNA to each well and tilt plate from side to side to ensure bottom is coated.
- DNA Binding solution can be used as blank.
2. Cover with plate seal and incubate at 37degC for 60 min.
3. Add 150 uL Block Solution (-20degC) to each well.
4. Cover with plate seal and incubate at 37degC for 30 min.
5. Remove DNA and Block Solution from each well down the sink. (DNA binds to bottom of wells)
6. Wash 3 times with 150 uL of 1X Wash Buffer by smacking it on sink and then patting dry on paper towels (smack on sink technique - SOST)
Methylated DNA Capture
1. Dilute Capture Antibody (4degC) 1:1000 in 1X Wash Buffer. (1 uL Capture Antibody, 999 uL Wash Buffer)
2. Add 50 uL of diluted Capture Antibody to each well.
3. Cover with plate seal and incubate at RT for 60 min.
4. Remove diluted Capture Antibody from each well down the sink.
5. Wash 4 times with 150 uL of 1X Wash Buffer using SOST.
6. Dilute Detection Antibody (4degC) 1:1000 in 1X Wash Buffer (1 uL Detection Antibody, 999 uL Wash Buffer)
7. Add 50 uL of Detection Antibody to each well.
8. Cover with plate seal and incubate at RT for 30 min.
9. Remove diluted Detection Antibody from each well down the sink.
10. Wash 5 times with 150 uL of 1X Wash Buffer using SOST.
Detection
1. Add 100 uL of Developing Solution (4degC) to each well.
2. Cover with plate seal and incubate at RT
away from light
for 1-10 min. max and monitor color change - should turn
BLUE
.
3. Add 50 uL Stop Solution (4degC) to each well. Color should change to
YELLOW
.
4. Read absorbance at 450 nm on Seeb Lab plate reader.
5. Calculate relative global methylation levels.
Calculations
1. Average control, sample, and blank absorbances.
2. Subtract average blank from average sample and from average control (separately).
3. Divide background subtracted average sample (sample minus blank) by background subtracted average normal sample (control minus blank).
4. Multiply by 100.
5. Percentage indicates level of methylation compared to control.
[(Average Sample - Average Blank) / (Average Control - Average Blank)] x 100
Example:
Average blank: 0.090
Average absorbance for 60 ng control DNA: 0.785
Average absorbance for 60 ng sample DNA: 0.654
[(0.654 - 0.090) / (0.785 - 0.090)] x 100 = 81.15%
Sample DNA has global methylation level that is 81.15% of the control DNA.
Questions:
1. What is methylation % of control?
2. Does percentage indicate sample concentration or how much more methylated it is than the control?
3. How do I make and interpret a linear regression/standard curve?
4. At what concentrations should I make my DNA dilutions?
Website of interest: GeneImprint
March 15, 2010
Summary: Quantification of juvenile salmon DNA samples.

Samples T1-T2 were repeated because I did not mix them the first time around.
Final Concentrations (T1-T4 Averages):
C1: 63.88
C2: 132.94
C3: 75.44
C4: 91.39
T1: 52.39
T2: 46.14
T3: 44.92
T4: 46.69
Next steps:
DNA isolation of adult salmon from different populations (to be determined), then ELISA assays! I will be out 3/18 - 3/28.
March 12, 2010
Summary: New project! (possible Capstone)
I will be performing ELISA assays for different salmon populations, including juveniles treated with HCl, in order to quantify methylation and compare among populations. Juveniles were treated with 10 mL HCl over 3 hours.
Controls @ pH 7:
6_2_1 (Labeled C1)
6_2_2 (Labeled C2)
6_2_3 (Labeled C3)
6_2_4 (Labeled C4)
Treatments @ pH 4
24_10_1 (Labeled T1)
24_10_2 (Labeled T2)
24_10_3 (Labeled T3)
24_10_4 (Labeled T4)
Today:
1. Cut off salmon caudal fins.
2. DNA extraction using QIAGEN kit (look at 1/19 and 1/20 for protocol)
March 8, 2010

Summary: Analysis of gel and research on DNA methylation.
The first four lanes (sample 0002) show methylation because there is a specific band at HPAII and a smear at MSPI. The oyster sample, however, does not show a specific band at HPAII, indicating that it is either completely unmethylated or that its methylation is not global (therefore forming a mosaic pattern).
Methylation occurs at CpG islands:
HPAII: Can’t cut methylated
MSPI: Cuts both methylated and unmethylated
If DNA is methylated:
Undigested: Will show a band
HPAII: Will show a band because it doesn’t cut
MSPI: Will not show specific band, will show smear because it cuts at various places
If DNA is not methylated:
Undigested: Will show a band
HPAII: Will not show a specific band, will show smear because it’s cutting at non-methylated areas
MSPI: Will not show a specific band, will show smear because it cuts at various places
CONCLUSIONS!
All the gels for salmon are pointing in the right direction: vertebrates have globally methylated DNA. Therefore, my results are in accordance with what is expected for methylated DNA: a band at undigested DNA, a band at HPAII samples, and a smear at MSPI samples. The oyster samples, seen above (lanes 6-9) do not show the expected pattern for global methylation but rather a pattern for non-methylated DNA: band at undigested, smear at HPAII, smear at MSPI. However, looking at Mac's gel pictures for oysters, this is what is expected. Unlike vertebrates, invertebrates do not have a global DNA methylation pattern but rather a mosaic one, meaning that methylation occurs in groups of genes at a time. The reason for the smear at HPAII is because the genome has groups of methylated genes and unmethylated genes.
Next steps:
In order to determine where methylation occurs on the genome, I would have to proceed with a Southern Blot or an ELISA assay, which would cut the DNA into specific regions of interest.
website of interest:
http://mcb.asm.org/cgi/content/abstract/17/3/1469
March 4, 2010
Summary: Gel electrophoresis of Sockeye Sample 0002 and Oyster Sample restriction digests. Gel was loaded using 10 uL of loading dye in each sample and 24 uL per well in the following order:
20 uL 100 bp loading dye, 0002 HPA II, 0002 HPA II control, 0002 MSP I, 0002 MSP I Control, Oyster HPA II, Oyster HPA II control, Oyster MSP I, Oyster MSP I control, 5uL hyperladder 1.
@ ~110 Volts
March 2, 2010
Summary: Agarose gel was made with 60 mL TAE buffer, .73 g agarose, 6 uL EtBr. Digests from yesterday will be run on March 4.
March 1, 2010

The gel above was run for approximately two hours at 110 V in an attempt to improve the quality and 20 uL of ethidium bromide were added to the positive side of the buffer. It looks like there is no change in this one from the previous except for better contrast due to EtBr put in the running buffer.
Summary for today: Restriction digests for another sample plus including oyster DNA as a control.
Oyster sample gDNA BB 13 5/19/08 from 4degC fridge with .5462 ug/uL of DNA.
Samples were set in PCR thermocycler for 16 h at 37degC, 20min at 80degC, and then 1 day at 4degC. Stored in 4degC freezer.
| Sample 0002 |
DNA (uL) |
Buffer (uL) |
Enzyme (uL) |
Water (uL) |
| Hpa II (buffer 1) |
9.77 |
5 |
1 |
34.2 |
| Hpa II control |
9.77 |
5 |
0 |
35.2 |
| MspI (buffer 4) |
9.77 |
5 |
.5 |
34.7 |
| MspI Control |
9.77 |
5 |
0 |
35.2 |
| Oyster |
||||
| Hpa II |
1.83 |
5 |
1 |
42.2 |
| HpaII control |
1.83 |
5 |
0 |
43.2 |
| MspI |
1.83 |
5 |
.5 |
42.7 |
| MspI control |
1.83 |
5 |
0 |
43.2 |
February 24, 2010

The gel shows a distinct band for HpaII (lane 2) and a more smeared band for MspI (lane 4).
If the sample is methylated, then bands would show on undigested (control) DNA, on HpaII treated DNA, but would show smearing for MspI. If the sample is unmethylated, then bands would show on undigested (control) DNA, but smearing would occur for both HpaII and MspI. Because this gel is difficult to analyze and because the two controls do not look the same, the gel will be run again with a few modifications: the wells will be smaller in width and ethidium bromide will be added to the buffer to prevent the "glowing" at the top of the gel.
Gel was made using 60 mL TAE buffer, .73 g agarose, and 6 uL ethidium bromide and stored in 4degC fridge.
February 23, 2010
Summary: Restriction enzyme reaction heat inactivation (thanks Sam) and gel electrophoresis.
Restriction enzyme reactions from yesterday were heat inactivated and put in -20degC freezer.
MspI: 80C for 20mins
HpaII: 65C for 20mins.
1. Gel from yesterday was placed on tray, filled up with 1X TAE buffer (enough to cover top of gel-wells on black+ side).
2. 10 uL of loading dye put into each sample to make 1:5 concentration (50 uL total of sample).
3. Samples were loaded using 20 uL of 100 bp ladder, 5 uL of hyperladder I, and 24 uL of each sample in the following order:
100 bp ladder, HpaII sample, HpaII control, MspI sample, MspI control, hyperladder I.
Gel was run for ~1 hour at 110 V.
February 22, 2010
Summary: Agarose gel preparation and repetition of restriction digests from February 3.
Agarose gel preparation: See January 29.
Note: 50 uL TA buffer used with .70 g of agarose. Microwaved for 1.5 min, 5 uL of ethidium bromide.
*Note: Restriction digests from February 3 could not be used because water was not added to bring total volume to 50 uL. Digest was repeated:
Sample 0008
For HPA II:
9.63 uL DNA
5 uL 10X Buffer 1
1 uL of enzyme or 1 uL of nanopure water (control)
34.37 uL of water
For MSP I:
9.63 uL DNA
5 uL 10X Buffer 4
.5 uL of enzyme or .5 uL of nanopure water (control)
34.37 uL of water
Samples were placed in 37degC water bath overnight.
February 4, 2010
After approximately 19hrs of incubation, restriction enzyme reactions from yesterday were heat inactivated and placed in -20degC.
MspI: 80C for 20mins
HpaII: 65C for 20mins.
February 3, 2010
Summary: Restriction digests of one DNA sample for HPA II, MSP I, and control.
Sample used: 0008 (highest DNA concentration)
For HPA II:
9.63 uL DNA
5 uL 10X Buffer 1
1 uL of enzyme or 1 uL of nanopure water (control)
For MSP I:
9.63 uL DNA
5 uL 10X Buffer 4
.5 uL of enzyme or .5 uL of nanopure water (control)
Samples were placed in 37degC water bath overnight (set at 3:50 pm).
February 1, 2010
Summary: Gel electrophoresis of HPA II and MSP I restriction digests.
Gel electrophoresis:
1. Gel from Jan. 29 was used.
2. Gel was placed on tray, filled up with 1X TAE buffer (enough to cover top of gel-wells on black+ side).
3. 10 uL of loading dye put into each sample to make 1:5 concentration (50 uL total of sample).
Gel was loaded in the following order using 20 uL of 100 bp ladder, 5 uL of hyperladder I, and 24 uL of each sample:
| 0008 |
0008 |
0002 |
0002 |
0007 |
0007 |
0001 |
0001 |
0003 |
0003 |
0004 |
0004 |
0005 |
0006 |
0006 |
||
| 100bp Ladder |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
MSPI |
HPAII |
Hyperladder I |
| 20uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
24uL |
5uL |
Gel ran for 1 hour at 110-111 Volts.

If the sample is methylated, then bands would show on undigested (control) DNA, on HpaII treated DNA, but would show smearing for MspI. If the sample is unmethylated, then bands would show on undigested (control) DNA, but smearing would occur for both HpaII and MspI.The gel shows normal bands at HPA II lanes and lowered, more scattered bands at MSP I lanes. However, because no control was run one sample will be picked to be run individually (0008). Note: the first four samples' DNA was extracted using a QIAGEN kit and the last four were extracted using Chelex, which resulted in a much lower DNA concentration.
January 29, 2010
Summary: Agarose gel preparation and gel electrophoresis of restriction enzyme digestion products.
Agarose gel preparation:
*Note: For genomic DNA, higher agarose concentration than normal, 1.2-1.5%, is desired (normally .8-1%) for better band resolution.
1. 180 uL of modified 1X TAE buffer put in 500 mL Erlenmeyer flask
2. 2.2 g of agarose added to make ~1.2% concentration
3. Microwaved for 3 min total
4. Water added to bring volume back up to account for evaporation
5. Gel allowed to cool down total 20 min, then 18 uL of Ethidium Bromide added.
6. Gel poured onto plate.
7. Let gel harden for 1 hour.
8. Gel was wrapped up for electrophoresis on Monday.
January 26, 2010 -
After 16hrs of incubation, restriction enzyme reactions from yesterday were heat inactivated and put in Sonia's 20C box.
MspI: 80C for 20mins
HpaII: 65C for 20mins.
January 25, 2010
| Restriction Enzyme Mixes |
|||||
| Amounts (uL) |
|||||
| Sample |
Mix |
DNA |
10X Buffer (1 for HPA II, 4 for MSP I) |
Enzyme |
Water |
| 8 |
HpaII |
9.63 |
5 |
1 |
34.37 |
| MspI |
9.63 |
5 |
0.5 |
34.87 |
|
| 2 |
HpaII |
9.77 |
5 |
1 |
34.23 |
| MspI |
9.77 |
5 |
0.5 |
34.73 |
|
| 7 |
HpaII |
13.25 |
5 |
1 |
30.75 |
| MspI |
13.25 |
5 |
0.5 |
31.25 |
|
| 1 |
HpaII |
11.84 |
5 |
1 |
32.16 |
| MspI |
11.84 |
5 |
0.5 |
32.66 |
|
| 3 |
HpaII |
15.46 |
5 |
1 |
28.54 |
| MspI |
15.46 |
5 |
0.5 |
29.04 |
|
| 4 |
HpaII |
23.85 |
5 |
1 |
20.15 |
| MspI |
23.85 |
5 |
0.5 |
20.65 |
|
| 5 |
HpaII |
14.96 |
5 |
1 |
29.04 |
| MspI |
14.96 |
5 |
0.5 |
29.54 |
|
| 6 |
HpaII |
21.98 |
5 |
1 |
22.02 |
| MspI |
21.98 |
5 |
0.5 |
22.52 |
|
Mixtures were made, then set in 37degC water bath overnight.
January 22, 2010
Summary: Quantification of DNA extraction (Qiagen Kit) and extraction/quantification of DNA from sockeye salmon fin samples using Chelex.
DNA Quantification Results from Qiagen Kit extraction (nanodrop):

DNA Extraction Using Chelex:
Samples used: SPICK_08_0003 SPICK_08_0004
SPICK_08_0005
SPICK_08_0006
1. Ethanol was drained and samples were washed once with 500 uL of PBS, then were placed in screw cap tubes.
2. 500 uL of 10% Chelex (found in 4deg freezer) added in each.
3. Samples set at 95degC for 30 min, vortexed 3x throughout.
4. Samples centrifuged for ~1 min, then DNA quantified using nanodrop.
5. Samples stored at 4degC.
DNA Quantification Results using Chelex extraction:
 Next steps: Restriction digests.
Next steps: Restriction digests.January 20, 2010
Summary: Conclusion of DNA extraction from sockeye salmon fin samples.
1. Samples were taken from 55 degree plate and vortexed for 15 seconds.
2. 200 uL AL buffer added, mixed by pipetting and then vortexed
3. Incubated at 70 degrees for 10 minutes
*Note: precipitate/gelatinous precipitate may form but will dissolve during incubation
4. 200 uL 100% (200 proof) ethanol added, then vortexed twice for 15 sec.
5. Samples loaded onto DNeasy Mini spin column + centrifuged for 1 min @ 8000 rpm.
*Note: Make sure to apply all of precipitate to spin column
6. Place spin column onto new 2 mL collection tube
7. Add 500 mL buffer AW1, centrifuge for 1 min @ 8000 rpm
8. Place spin column in new 2 mL collection tube and add 500 uL buffer AW2, centrifuge for 3 min @ 14,000 rpm to dry membrane
9. Spin columns placed in 1.5 mL centrifuge tube, 100 uL of buffer AE was added to membrane of each, centrifuged for 1 min @8000 rpm.
10. Samples stored at -20 freezer.
*Note: All flow-through and membranes were saved.
January 19, 2010
Summary: Overview of lab safety and organization + first steps of DNA extraction (using Qiagen Kit) from sockeye salmon fin samples for future DNA methylation quantification analysis.
Dna Purification from Animal Tissues (Qiagen Kit)
1. Four samples of 25 mg of sockeye salmon fin tissue in EtOH taken from -80 freezer.
2. EtOH drained and samples washed twice with PBS (500 uL each time)
3. PBS drained, samples transferred to 1.5 mL PCR tubes
4. 180 uL of ATL buffer added
5. 20 uL of Proteinase K added, vortexed for ~10 seconds
*Note: Enzymes should NOT be vortexed before use and should be placed in cooler while in use.
6. Samples kept at 55 degree hot plate overnight for lysis (~16h)
Sockeye Sample ID's (all fin samples):
SPICK08_0008
SPICK08_0002
SPICK08_0007
SPICK08_0001
| 1st DNA extraction |
2 ng aliquot (315ul) |
||||||||
| Population |
Individual |
Storage |
Tissue |
Data Extracted |
DNA Concentration (ng/μl) |
260/280 |
DNA (μl) |
H20 (μl) |
|
| SHAPI98E |
SHAPI98E_0041 |
Frozen |
Heart |
20090707 |
439.69 |
1.92 |
1.4 |
313.6 |
|
| SHAPI98E |
SHAPI98E_0042 |
Frozen |
Heart |
20090707 |
167.04 |
1.92 |
3.8 |
311.2 |
|
| SHAPI98E |
SHAPI98E_0044 |
Frozen |
Heart |
20090707 |
243.74 |
1.92 |
2.6 |
312.4 |
|
| SHAPI98E |
SHAPI98E_0045 |
Frozen |
Heart |
20090708 |
424.73 |
1.9 |
1.5 |
313.5 |
|
| SHAPI98E |
SHAPI98E_0046 |
Frozen |
Heart |
20090708 |
366.77 |
1.91 |
1.7 |
313.3 |
|
| SHAPI98E |
SHAPI98E_0048 |
Frozen |
Heart |
20090708 |
522.51 |
1.89 |
1.2 |
313.8 |
|
| SHAPI98E |
SHAPI98E_0049 |
Frozen |
Heart |
20090708 |
347.19 |
1.91 |
1.8 |
313.2 |
|
| SHAPI98E |
SHAPI98E_0050 |
Frozen |
Heart |
20090708 |
321.2 |
1.92 |
2.0 |
313.0 |
|
| STIKC01 |
STIKC01_0001 |
Frozen |
Heart |
20090707 |
195.99 |
1.9 |
3.2 |
311.8 |
|
| STIKC01 |
STIKC01_0002 |
Frozen |
Heart |
20090707 |
330.93 |
1.9 |
1.9 |
313.1 |
|
| STIKC01 |
STIKC01_0003 |
Frozen |
Heart |
20090707 |
148.52 |
1.89 |
4.2 |
310.8 |
|
| STIKC01 |
STIKC01_0004 |
Frozen |
Heart |
20090708 |
479.95 |
1.89 |
1.3 |
313.7 |
|
| STIKC01 |
STIKC01_0005 |
Frozen |
Heart |
20090708 |
187.66 |
1.88 |
3.4 |
311.6 |
|
| STIKC01 |
STIKC01_0006 |
Frozen |
Heart |
20090708 |
377.57 |
1.91 |
1.7 |
313.3 |
|
| STIKC01 |
STIKC01_0007 |
Frozen |
Heart |
20090708 |
250.29 |
1.91 |
2.5 |
312.5 |
|
| STIKC01 |
STIKC01_0008 |
Frozen |
Heart |
20090708 |
178.04 |
1.94 |
3.5 |
311.5 |
|
| SUPNK01 |
SUPNK01_0001 |
Frozen |
Liver |
20090715 |
863.31 |
1.92 |
0.7 |
314.3 |
|
| SUPNK01 |
SUPNK01_0002 |
Frozen |
Liver |
20090715 |
524.43 |
1.87 |
1.2 |
313.8 |
|
| SUPNK01 |
SUPNK01_0003 |
Frozen |
Liver |
20090715 |
547.86 |
1.84 |
1.1 |
313.9 |
|
| SUPNK01 |
SUPNK01_0004 |
Frozen |
Liver |
20090715 |
63.29 |
1.84 |
10.0 |
305.0 |
|
| SUPNK01 |
SUPNK01_0005 |
Frozen |
Liver |
20090715 |
638.97 |
1.93 |
1.0 |
314.0 |
|
| SUPNK01 |
SUPNK01_0006 |
Frozen |
Liver |
20090715 |
472.58 |
1.87 |
1.3 |
313.7 |
|
| SUPNK01 |
SUPNK01_0007 |
Frozen |
Liver |
20090715 |
315.09 |
1.91 |
2.0 |
313.0 |
|
| SUPNK01 |
SUPNK01_0008 |
Frozen |
Liver |
20090715 |
528.72 |
1.84 |
1.2 |
313.8 |
|
| SPICK08 |
SPICK08_0001 |
EtOH |
Fin |
20090716 |
28.01 |
1.88 |
22.5 |
292.5 |
|
| SPICK08 |
SPICK08_0002 |
EtOH |
Fin |
20090716 |
49.15 |
1.93 |
12.8 |
302.2 |
|
| SPICK08 |
SPICK08_0003 |
EtOH |
Fin |
20090716 |
113.13 |
1.86 |
5.6 |
309.4 |
|
| SPICK08 |
SPICK08_0004 |
EtOH |
Fin |
20090716 |
41.38 |
1.88 |
15.2 |
299.8 |
|
| SPICK08 |
SPICK08_0005 |
EtOH |
Fin |
20090716 |
92.25 |
1.9 |
6.8 |
308.2 |
|
| SPICK08 |
SPICK08_0006 |
EtOH |
Fin |
20090716 |
51.46 |
1.87 |
12.2 |
302.8 |
|
| SPICK08 |
SPICK08_0007 |
EtOH |
Fin |
20090716 |
58.34 |
1.9 |
10.8 |
304.2 |
|
| SPICK08 |
SPICK08_0008 |
EtOH |
Fin |
20090716 |
29.31 |
1.91 |
21.5 |
293.5 |
|
| SMEZIB06 |
SMEZIB06_0001 |
EtOH |
Axillary |
20090718 |
57.31 |
1.79 |
11.0 |
304.0 |
|
| SMEZIB06 |
SMEZIB06_0002 |
EtOH |
Axillary |
20090718 |
86.27 |
1.85 |
7.3 |
307.7 |
|
| SMEZIB06 |
SMEZIB06_0003 |
EtOH |
Axillary |
20090718 |
48.67 |
1.89 |
12.9 |
302.1 |
|
| SMEZIB06 |
SMEZIB06_0004 |
EtOH |
Axillary |
20090718 |
36.9 |
1.83 |
17.1 |
297.9 |
|
| SMEZIB06 |
SMEZIB06_0005 |
EtOH |
Axillary |
20090718 |
75.98 |
1.88 |
8.3 |
306.7 |
|
| SMEZIB06 |
SMEZIB06_0006 |
EtOH |
Axillary |
20090718 |
47.42 |
1.83 |
13.3 |
301.7 |
|
| SMEZIB06 |
SMEZIB06_0007 |
EtOH |
Axillary |
20090718 |
62.63 |
1.87 |
10.1 |
304.9 |
|
| SMEZIB06 |
SMEZIB06_0008 |
EtOH |
Axillary |
20090718 |
80.05 |
1.86 |
7.9 |
307.1 |
|