Lab 1: Tissue Extraction I
RNA Extraction
Purpose: To isolate and quantify RNA from a specified tissue sample.
Tissue sample used: ~25 mg 37°C heat stressed barnacle
Day 1 Methods:
1. 500 uL of TriReagent were added to the tissue sample in a 1.5 mL snap cap tube and placed on ice.
2. Sample was homogenized using a disposable pestle (some shell remained).
3. An additional 500 mL of TriReagent were added to the tube, then it was vortexed at top speed for 15 seconds.
4. Sample was stored at -80°C.
Protein Extraction
Purpose: To isolate cellular proteins from a specified tissue sample and determine their concentration using a standard curve based on spectrometer readings.
Tissue sample used: ~25 mg 37°C heat stressed barnacle
Methods for extraction:
1. .5 mL of CellLytic MT solution were added to the tissue sample in a 1.5 mL snap cap tube.
2. Sample was homogenized using a disposable pestle and inverted several times (some shell remained).
3. The tube was spinned in a refrigerated microfuge (~ 4°C) for 10 minutes at top speed.
4. Supernatant was transferred to a clean, labeled 1.5 mL snap cap tube and placed on ice.
Methods for quantification:
1. 15 uL of sample and 15 uL of DI water were aliquoted into a fresh 1.5 mL tube (1 part sample to 2 parts whole)
2. 30 uL of DI water were aliquoted into another 1.5 mL tube to serve as a blank reading.
3. 1.5 mL of Bradford reagent were added to each tube and both were inverted several times and incubated at room temperature (26-28°C) for 10 minutes.
4. 1 mL of the “blank” was transferred to a plastic cuvette. The spectrometer was then zeroed.
5. 1 mL of the sample was transferred to a plastic cuvette. The absorbency was measured at 595 nm.
6. Protein sample was stored at -20°C.
*Note: Absorbency in sample should be measured twice and averaged for accuracy. In this case, it was not.
Results:
Sample absorbency: 1.274 A
Back-calculating from a standard Bradford absorbency curve:
y=1011.9x
y= Protein concentration (ug/mL)
x= Absorbency
*Note: sample absorbency was outside the range for the standard curve.
y=1011.9 (1.274)
Answer must be multiplied by 2 to account for 1:2 dilution.
Protein sample concentration: 2578.32 ug/mL
October 13, 2009
Lab 2: Tissue Extraction II
RNA Isolation
Day 2 Methods:
1. Sample previously stored at 80°C incubated at room temperature for 5 minutes.
2. 200 uL of chloroform were added to the sample tube under the fume hood, the sample was then vortexed vigorously for 30 seconds.
*Observation: sample was milky pink
3. Sample was incubated at room temperature for 5 minutes, then spinned in a refrigerated microfuge (4°C) for 15 minutes at maximum speed.
4. The aqueous phase (top, clear portion) was then transferred to a fresh microfuge tube. The remaining organic/interphase solids were disposed of.
5. 500 uL of isopropanol were added to the new tube containing the RNA and mixed by inverting until the solution was uniform. It was then incubated at room temperature for 10 minutes.
6. Sample was then spinned in the refrigerated microfuge at masimum speed for 8 minutes.
*Observation: Pellet formed.
7. Supernatant was removed and 1 mL of 75% EtOH (Ethanol) was added. The tube was vortexed briefly to dislodoge the pellet.
8. The sample was then spinned in refrigerated microfuge at 7500g for 5 minutes. Supernatant was removed.
9. The tube was briefly spinned to pool the residual EtOH and the remaining supernatant was removed.
10. Tube was left open to allow EtOH to evaporate for no more than 5 minutes.
11. The pellet was resuspended in 100 uL of 0.1% DEPC water and mixed by pipetting.
12. Sample was then incubated at 55°C for 5 minutes and flicked a few times to mix.
13. Sample was stored at 80°C.
Protein Gel Methods:
1. Water was set boiling on a hot plate.
2. The protein sample for Lab 1 was thawed and mixed by inverting.
3. 15 uL of the protein sample and 15 uL of 2X Reducing Sample Buffer in a 1.5 mL screw cap tube. Sample was mixed by flicking and briefly centrifuged (10 seconds) to pool liquid in the bottom of the tube.
4. The sample was boiled for 5 minutes, then centrifuged for 1 minute.
5. 30 uL of the sample were then loaded into a previously set up acrilamyde gel using a gel loading tip. Molecular ladder used was SeeBlue Plus2 Prestained Standard 1X (Invitrogen).
6. Gel was run for 30 minutes at 150 V.
7. After elapsed 30 minutes, gel was put on a container with about 150 mL of Coomassie Stain and incubated on rocker for 5 minutes.
8. Gel was then rinsed with 10% acetic acid and then placed in container with about 250 mL of 10% acetic acid and incubated on rocker for 15 minutes. The buffer was changed out until bands became clearly visible.
9. A digital picture of the gel was then taken.
Results and further investigation:
Protein bands on the gel indicated what size the proteins present were. For further analysis, we will use an antibody to determine the presence of a specific protein. Products from RNA isolation will be run on a PCR machine and then run on a gel.
RNA
RNA was successfully isolated and will be quantified using a nanodrop.
Protein
Based on the presence and concentration of the bands seen, we will be able to determine amounts of proteins in our sample.
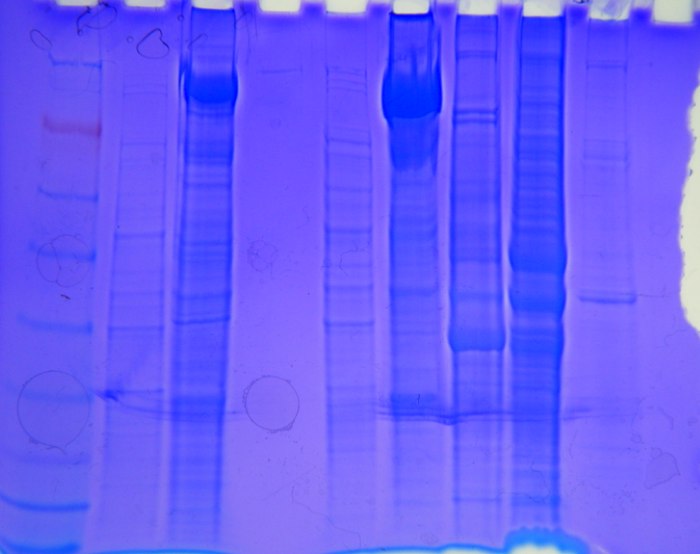
Figure 1: Proteins run on acrylamide gel with standard ladder in well #1 showing possible protein sizes and concentrations. 30 uL of sample were put into each well. The samples in following wells, from 2 to 9 were oyster, heat stressed barnacle, blank, unknown*, heat stressed barnacle, salmon brain, herring brain, and unknown*. *Samples will be further determined.
October 20, 2009
Lab 3: Reverse Transcription and PCR
Purpose: To quantify RNA using a nanodrop, to reverse transcribe RNA to cDNA, and to perform PCR on cDNA samples
Quantification Methods:
*Note: keep RNA sample on ice at all times
1. The RNA sample from Lab 1 and Lab 2 was used
2. 2 uL of DEPC water were pipetted onto Nanodrop pedestal to measure a blank.
3. 2 uL of RNA sample were pipetted onto pedestal and the arm was lowered, then measured.
4. The pedestal was then wiped with a Kim Wipe.
Results:
RNA concentration: 325.1 ng/uL
260/230: 2.10
260/280: 1.85
260 10 mm path: 8.120
280 10 mm path: 4.398
Reverse Transcription Methods:
1. RNA sample was mixed by inverting tube and transfer 5uL to fresh PCR tube.
2. Sample was incubated at 75°C for 5 minutes in thermal cycler.
3. Tube immediately trasnfered to ice and incubated for 5 minutes.
4. The following was added to the sample:
a. 4 uL 5x buffer (AMV RT Buffer)
b. 8 uL dNTP’s (10 mM total)
c. 1 uL AMV RTranscriptase
d. 1 uL Oligo dT Primer
e. 1 uL RNase free water
5. Tube was vortexed, spot spinned, and incubated at RT for 10 minutes
6. Sample was then incubated at 37°C for 1 hour in thermocycler, then heat inactivated at 95°C for 3 minutes.
7. Sample was spot spinned and left on ice.
Primer preparation:
Forward primer: GAGCGGCTGATC GGA GAC GC (32.5 nm)
Reverse primer: TCT TGG GCT TGC CGC TGT CG (36.9 nm)
Length: 166 bp
1. Primers were reconstituted for a working 10uM stock solution by adding 325 uL of water to the forward primer and 369 uL of water to the reverse primer.
2. 10 uL of each primer were added to separate tubes plus 90 uL of water.
PCR Methods:
1. A Master Mix was created using the following recipe (amounts are for one reaction, but a 5X mix was made):
a. 25 uL GoTaq Green Master Mix, 2X
b. 10 uM upstream primer (2.5 uL)
c. 10 uM downstream primer (2.5 uL)
d. 2 uL cDNA (for samples) or PCR water (negative controls)
e. 18 uL PCR water
*Note: the primer amounts were determined using the following equation: (10uM)(Volume)=(.5uM)(50uL)
2 tubes of sample and 2 tubes of negative control were loaded into thermocycler for PCR with the following profile:
95C for 10 min
40 cycles
-95C for 30 sec
-55C for 30 sec
-72C for 90 sec
-72C for 3 min
Stored at 4C.
Agarose Gel Preparation Methods:
1. 2g of agarose were weighed out and mixed with 150 mL 1x TAE buffer in a 1 L flask and microwaved for 2 minutes.
2. Because some buffer evaporated, an additional 30 mL of buffer were added to the solution and microwaved for an extra 30 seconds.
3. Solution was allowed to cool, and 12 uL of ethidium bromide were added to it.
4. Gel mixture was thoroughly mixed and poured onto gel tray. Gel combs were then added.
5. After gel set, it was wrapped in plastic wrap and placed in the fridge.
Results and further investigation:
The products from PCR will be run on a gel to determine if the primers actually worked on our samples. We will therefore be able to determine if the protein coded in our primers is expressed in our organism.
October 27, 2009
Lab 4: Western Transfer – Immunoblots
Purpose:
- To run PCR products from Lab 3 on gel
- To run out protein samples as done previously and transfer them to nitrocellulose membrane
- To probe membrane with antibody (HSP)
Methods for PCR:
1. Gel was paced in box and filled with 1x TAE buffer to fully cover wells.
2. Combs were removed from gel.
3. 7 uL of 100bp ladder were loaded onto first lane.
4. 25 uL of PCR sample were loaded onto following lanes (Top: Rachel and Sonia, Bottom: Sarah and Sophie)
5. Gel was run at 100 V for 1 hour.
6. Gel was visualized on UV transilluminator.
Observations:
Clear bands were seen both for Rachel and Sonia on the sample lanes at approximately 150 base pairs. Some bands were seen in the controls; they could be due to primer dimers. Sarah’s lanes were a bit blurred, but a definite band could be seen as well. Sophie’s bands were very faint.
Methods for protein transfer to membrane:
*Note: the gels used for protein transfer were remade for a more uniform concentration (12 ug/uL).
1. Transfer buffer was cooled to 4°C.
2. The filter paper, membrane, and gel were soaked in Transfer Buffer for 15 minutes.
3. The blotting sandwich was assembled in a semi-dry blotting apparatus as follows:
a. Positive anode
b. Filter paper (about 2 sheets for each gel)
c. Nitrocellulose Membrane
d. Gel
e. Filter paper (2 sheets)
f. Negative cathode
4. The blot was transferred for 30 minutes at 20 V.
5. Gel was removed from sandwich and rinsed with transfer buffer.
Methods for Western Blotting:
1. 20 mL of Blocking Solution were prepared:
a. Ultra filtered water 14 mL
b. Blocker/Diluent (Part A) 4 mL
c. Blocker/Diluent (Part B) 2 mL
2. Membrane was placed in 10 mL of appropriate Blocking Solution in covered, plastic dish and incubated for 30 minutes on rotary shaker set at 1 rev/sec.
3. Blocking Solution was decarted.
4. Membrane was rinsed with 20 mL of water for 5 minutes, then decarted (2X).
5. 10 mL of primary antibody solution (1:3000) was made:
a. Blocking Solution 10 mL
b. HSP 70 antibody 3.3 uL
6. Membrane was incubated with 10 mL of primary antibody solution overnight.
7. The next morning, decant primary antibody.
8. Wash membrane for 5 minutes with 20 mL of prepared Antibody Wash, then decant. Repeat 3 times.
9. Incubate membrane in 10 mL of secondary antibody solution for 30 minutes, then decant.
10. Wash the membrane for 5 minutes with 20 mL of Antibody Wash, then decant. Repeat 3 times.
11. Rinse membrane with 20 mL of water for 2 minutes, then decant. Repeat twice.
12. Incubate the membrane in 5 mL of Chromogenic membrane.
13. Wait 1 to 60 minutes for development, then rinse the membrane with 20 mL of water for 2 minutes. Repeat twice.
14. Dry membrane on clean piece of filter paper to open air or under an infrared lamp.
Results and Conclusions:
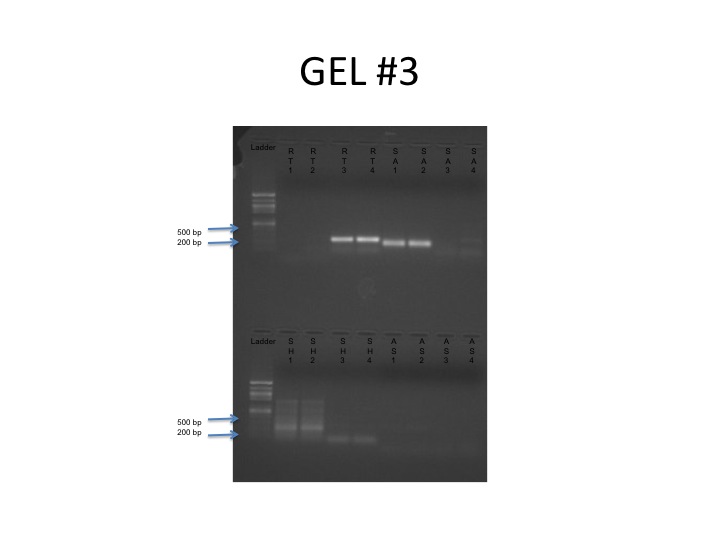 Figure 1: Products from PCR on gel: RT: 1-Negative Control, 2- Negative Control, 3- Sample, 4- Sample. SA: 1-Sample, 2- Sample, 3- Negative Control, 4- Negative Control. SH: 1- Sample, 2-Sample, 3- Negative Control, 4- Negative control. AS: 1-Sample, 2-Sample, 3-Negative Control, 4-Negative Control. Both samples for RT and SA very clearly show bands around 200 bp, which match the expected band size for SA at 166 bp. The samples for SH seem to be blurred and there is presence of a band at the negative controls, which could indicate some contamination. AS did not have any bands, suggesting that her primers did not work on her organism.
Figure 1: Products from PCR on gel: RT: 1-Negative Control, 2- Negative Control, 3- Sample, 4- Sample. SA: 1-Sample, 2- Sample, 3- Negative Control, 4- Negative Control. SH: 1- Sample, 2-Sample, 3- Negative Control, 4- Negative control. AS: 1-Sample, 2-Sample, 3-Negative Control, 4-Negative Control. Both samples for RT and SA very clearly show bands around 200 bp, which match the expected band size for SA at 166 bp. The samples for SH seem to be blurred and there is presence of a band at the negative controls, which could indicate some contamination. AS did not have any bands, suggesting that her primers did not work on her organism.Figure 2: Western blot results: The antibodies successfully binded to proteins for RT and SA (the first four lanes) but did not bind for any others. This could be due to the fact that the antibodies are too specific for the proteins used or that the proteins did not successfully transfer to the membrane.
Figure 3: Gel used for Western blot: Gel shows presence of proteins in samples.
Because the PCR gel shows a strong band at the appropriate size, I can determine that heat stress protein is in fact present in the heat-stressed barnacle. Furthermore, the Western blot results show the expression of HSP-70 due to the binding of antibodies. I can successfully state that heat stress protein is in fact present and expressed in the heat stressed barnacle.
Further investigation:
In order to find solid conclusions for this investigation, many more trials should be done. However, for SA (Sonia Albin) this trial has been successful in that PCR results showed strong bands of the desired protein and that the Western blot results showed successful binding of the antibodies used.
November 3, 2009
Lab 5: Quantitative PCR
Purpose:
To successfully amplify a desired region cDNA using a polymerase, primers, and dNTPs.
Methods:
1. Master Mix was prepared with the following recipe for 8 reactions for a total of 48 uL per reaction (6 total reactions desired, 2 extra for pipetting errors)
a. Master Mix, 2X (Immomix): 25 uL
b. Syto-13 dye (50uM): 3.5 uL
c. Upstream primer (from previous stock): 1.5 uL
d. Downstream primer (from previous stock): 1.5 uL
e. Ultra Pure Water: 16.5 uL
2. 48 uL of Master Mix were added to 6 wells of a PCR plate.
3. 2 uL of cDNA were added to first two wells.
4. 2 uL of ultra pure water were added to second two wells as negative controls.
5. 1:4 dilution was made for RNA with 5 uL of RNA: 15 uL of water.
6. 2 uL of RNA dilution were added to last 2 wells to test for genomic DNA carry-over.
7. The plate was loaded onto a real-time PCR thermocycler.
Results and Conclusions:
Figure 1: qPCR results of cDNA (turquoise and pink), negative controls (yellow and blue), and RNA (green and red). The cDNA was definitely amplified as shown in the peaks above. However, the green sample of RNA was also amplified, indicating that there was some genomic DNA carryover. The cDNA samples seem to be "melting" at a temperature of around 87 degrees Celsius and the RNA sample is "melting" at a temperature of about 84 degrees Celsius.
The reason for a spike in the RNA sample is due to genomic DNA carry-over. The reason that it is expressed in one sample and not in the other can be due to minimal mixing of the sample before loading. However, because the cDNA was amplified, I can determine that my primers were correct for my species and that HSP70 is being expressed. Repeating this process would be beneficial in order to obtain more stable results.
Research Project
Aeolidia Papillosa
Background
A nudibranch’s response to stress can be measured observationally, by focusing on their swimming behavior, as well as molecularly by determining the stress proteins their genes express. Nudibranch response to introduction of a predator and a salinity shock will therefore probably yield a behavioral and a molecular response. Swimming behavior will be assessed by speed, flexing of the nudibranch body and time of termination of swimming. In past studies, the swimming response to introduction of a predator yielded a less extreme response than a 1M KCl salinity shock (Lawrence and Watson, 2002). A response to a predator would be to swim away and a molecular response would most likely result in an expression of a particular stress protein. Introduction of 1M KCl will induce a salinity shock near the nudibranch, causing a molecular stress and therefore a gene response as well as a behavioral response. It is our objective to compare these responses on the behavioral and observational level.
Research Objectives
Our objective is to determine if the nudibranchs’ defense mechanism against a predator matches its defense against an abrupt change in salinity. We will use observations based on their swimming responses to the presence of a sea star and 1 mL of 1M KCl. We will then be able to determine their molecular response based on expression of heat stress protein. By testing whether or not the gene expression is the same, we can compare the nudibranchs’ response not only on the observational level but also on the molecular level.
Methods
Materials for maintenance:
15 nudibranchs (5 for each trial)
1-2 predator sea star (species undetermined)
Holding tank (recirculating)
Hydroids for feeding
Basic Procedure:
1. Control: 5 nudibranchs in stable environment
a. Record natural behavior in stable environment
i. Observation of swimming behavior
ii. Extraction of tissue sample for analysis
2. Salinity response: 5 nudibranchs in stable environment shocked with 1 mL of 1M KCl in proximity
a. Record swimming behavior after salinity shock
b. Extraction of tissue sample after salinity shock for analysis
3. Predator response: 5 nudibranchs in stable environment with sea star in proximity
a. Record swimming behavior before predator introduction
b. Record changing swimming behavior during predator introduction
c. Extraction of tissue sample after predator shock for analysys.
All tissue samples will be analyzed for the presence of stress protein using qPCR for gene expression.
Protein extraction protocol:
1. Add .5 mL of CellLytic MT solution to ~25 mg tissue sample in a 1.5 mL snap cap tube.
2. Homogenize sample using a disposable pestle and invert several times
3. Spin tube in a refrigerated microfuge (~ 4°C) for 10 minutes at top speed.
4. Transfer supernatant to a clean, labeled 1.5 mL snap cap tube and put on ice.
Protein quantification protocol:
1. Alliquot 15 uL of sample and 15 uL of DI water into a fresh 1.5 mL tube (1 part sample to 2 parts whole)
2. Alliquot 30 uL of DI water into another 1.5 mL tube to serve as a blank reading.
3. Add 1.5 mL of Bradford reagent to each tube and invert both several times, then incubate at room temperature (26-28°C) for 10 minutes.
4. Trasnfer 1 mL of the “blank” to a plastic cuvette and zero the spectrometer.
5. Transfer 1 mL of the sample to a plastic cuvette and measure absorbancy at 595 nm.
6. Store at -20°C.
Protein electrophoresis methods:
1. Set water boiling.
2. Thaw protein sample and mix by inverting.
3. Put 15 uL of the protein sample and 15 uL of 2X Reducing Sample Buffer in a 1.5 mL screw cap tube. Mix sample by flicking and briefly centrifuge (10 seconds) to pool liquid in the bottom of the tube.
4. Boil sample for 5 minutes, then centrifuge for 1 minute.
5. Load 30 uL of the sample into a previously set up acrilamyde gel using a gel loading tip. Use SeeBlue Plus2 Prestained Standard 1X (Invitrogen) as a molecular ladder.
6. Run gel for 30 minutes at 150 V.
7. After elapsed 30 minutes, put gel on a container with about 150 mL of Coomassie Stain and incubate on rocker for 5 minutes.
8. Rinse gel with 10% acetic acid and then place in container with about 250 mL of 10% acetic acid and incubate on rocker for 15 minutes. Change out buffer until bands become visible.
Methods for protein transfer to membrane:
1. Cool transfer buffer to 4°C.
2. Soak filter paper, membrane, and gel in Transfer Buffer for 15 minutes.
3. Assemble the blotting sandwich in a semi-dry blotting apparatus as follows:
a. Positive anode
b. Filter paper (about 2 sheets for each gel)
c. Nitrocellulose Membrane
d. Gel
e. Filter paper (2 sheets)
f. Negative cathode
4. Transfer blot for 30 minutes at 20 V.
5. Remove gel from sandwich and rinse with transfer buffer.
Methods for Western Blotting:
1. Prepare 20 mL of Blocking Solution:
a. Ultra filtered water 14 mL
b. Blocker/Diluent (Part A) 4 mL
c. Blocker/Diluent (Part B) 2 mL
2. Place membrane in 10 mL of appropriate Blocking Solution in covered, plastic dish and incubate for 30 minutes on rotary shaker set at 1 rev/sec.
3. Decart blocking solution.
4. Rinse membrane with 20 mL of water for 5 minutes, then decart (2X).
5. Make 10 mL of primary antibody solution (1:3000):
a. Blocking Solution 10 mL
b. HSP 70 antibody 3.3 uL
*This is considering we will use HSP 70
6. Incubate membrane with 10 mL of primary antibody solution overnight at 4C.
7. The next morning, decant primary antibody.
8. Wash membrane for 5 minutes with 20 mL of prepared Antibody Wash, then decant. Repeat 3 times.
9. Incubate membrane in 10 mL of secondary antibody solution for 30 minutes, then decant.
10. Wash the membrane for 5 minutes with 20 mL of Antibody Wash, then decant. Repeat 3 times.
11. Rinse membrane with 20 mL of water for 2 minutes, then decant. Repeat twice.
12. Incubate the membrane in 5 mL of Chromogenic membrane.
13. Wait 1 to 60 minutes for development, then rinse the membrane with 20 mL of water for 2 minutes. Repeat twice.
14. Dry membrane on clean piece of filter paper to open air or under an infrared lamp.
Expected Results
We expect that the nudibranch’s response to any sort of stress will yield in a similar outcome, both with behavior and with gene expression. Therefore, we expect that a salinity stress will yield similar results as a predatory stress.
Expected time line11/10:
Basic procedure for experiment and tissue extraction.
Supplies needed:
15 nudibranchs and 1 sea star (predator)
Enough sea water for 3 tubs (1 control/housing, 2 treatments)
1 M KCl (or solid KCl and DI water to make dilution)
Scale
Syringe or pipette
Salinity meter
Thermometer
Scalpels or blades
1.5 mL snap tubes
Paper towels
11/17:
Protein extraction
Protein quantification
Protein gel electrophoresis
11/24:
Western Blot and data analysis
NOTE: Due to a lack of sufficient antibodies to test for several proteins, we will be instead testing gene expression in RNA for heat shock protein and two others yet to be determined.
Expected Timeline:
11/10:
Supplies needed:
15 nudibranchs and 1 sea star (predator)
Enough sea water for 3 tubs (1 control/housing, 2 treatments)
1 M KCl (or solid KCl and DI water to make dilution)
Scale
Syringe or pipette
Salinity meter
Thermometer
Scalpels or blades
1.5 mL snap tubes
Paper towels
RNA extraction:
1. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.
2. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.*
*to save time, this step has been performed for you.
3. Carefully homogenize the tissue using a disposable pestle.
4. Add an additional 500uL of TriReagent to the tube and close the tube.
5. Vortex vigorously for 15s.
6. If we run out of time we can store at -80C
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube.
Note: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
17. Incubate at RT for 10 mins.
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer (we will be using the Nanodrop).
11/17:
RNA QUANTIFICATION
NOTE: Always keep your RNA samples on ice!
1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedistal and lower the arm.
2. Click "Blank", to zero the instrument
NOTE: steps 1 and 2 only need to be done once for the whole class.
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your A260 absorbance, RNA concentration (ng/µL), A260/280 ratio and A260/320 ratio.
NOTE: The Nanodrop uses the Beer-Lambert Law to calculate RNA concentration for you. See Lab 1 notes on RNA extraction for more information on the calculation and how to evaluate RNA purity using A260/280 and A260/A320 ratios.
5. Raise the arm and wipe off you sample with a Kim Wiple
6. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
7. storage at -80C.
REVERSE TRANSCRIPTION:
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix (MM)
PER RXN
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave cDNA on ice or store at –20C
PCR -
Polymerase Chain Reaction involves amplifying a DNA (genomic or complementary) target using a polymerase, primers (short oligonucleotide), and dNTPs (A, C, T, Gs). In general the reaction is placed in a machine (thermocycler) where a series of temperature changes are performed [Denature (~94C), Anneal (primer specific ~50-60C), and Extention (~72C)].
For this lab you will be using Promega's GoTaq Product. Please Read!
Prepare your samples in duplicate AND make sure to include at least 2 negative controls for each primer (no template).
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
| GoTaq®Green Master Mix, 2X |
25 |
1x |
| upstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| downstream primer, 10μM |
0.5–5.0μl |
0.1–1.0μM |
| DNA template |
1–5μl |
<250ng |
Load reactions into thermocycler.
- prepare Agarose gels here. (Plan on making them earlier- maybe outside of lab time, so we can run PCR here. If not, run next week (11/24)
11/24
Run PCR products on gel
1. place gel in gel box and fill with 1x TAE buffer (to fully cover wells)
2. remove combs from wells
3. load 7uL 100bp ladder in far left lane
4. load 25uL of your PCR sample into the gel (retain the remaining vol at -20ºC)
5. run gel at ~ 100V for ~ 1hr
6. visualize the gel on the UV transilluminator
Further analysis.
11/10: Day 1 of Research Project
Purpose: To conduct full experiment and to extract RNA from nudibranch head samples.
For KCl solution:
74.553 g/mol if molecular mass of KCl
1.864g of KCl in 25 mL of DI water
Start with KCl shock. Measuring 1 liter of sea water in open glass tank.
Acclimate for a bit, and add 1M KCl.
Seawater from same source, and will be using same seawater for each treatment.
Original housing salinity is 38ppm
Table 1. Labeled tubes. Also record where the samples are stored (temperature and location in lab)
||
| Types of treatment |
Control |
KCl |
Predator |
|
| Labels: |
1,2,3,4 |
A, B, C |
D, E, F |
|
| RNA extraction |
RNA 1, 2, 3, 4 Stored at -80C |
RNA A, B, C Stored at -80C |
RNA D, E, F Stored at -80C |
|
| RNA isolation |
||||
| RNA quantification |
||||
| Table for Observations: Table 2: KCl observations, A-C Observations for before sample was taken and approximate time (sec) after KCl shock was induced. All samples are in 1L seawater to begin Nudibranchs have been sitting in tanks for an least ten minutes to acclimate 1 M KCl that was mixed has salinity at 55ppm Taking about 50 mg of tissue sample from head. Tissue samples are suspended immediately in TriReagent, 1mL. Matched videos and images. |
||||
| A |
B |
C |
||
| Start Salinity: 32ppm 2:28pm Video 1 Initial salinity: 35ppm Observations: see Video 1, specimen A rolled up away from place of origin and is still freaking out... Picture 1 (after videos) 153 0237 Sample taken at 16:37 |
Start Salinity: 32ppm 2:30pm Video 2 Initial salinity: 34ppm Same observation, quickly rolled up away from origin of KCl Picture 2 sample taken at 17:33 |
Start Salinity: 32ppm 2:32 Video 3 initial salinity:34ppm same observation, see video 3 Picture 3(after videos. Sample taken at 17:27 |
||
 |
| Salinity%20shock%20test%20setup-1.jpg |
Issues:
-possibly stress right before dissection...?
-carry over of sea star residue or nudibranch residue
-sea star my not be hungry for our nudibranchs...
hopefully nudibranch will be scared
Table 3: Predator response observations for D-F. Time taken after sample taken (Sec). Also record which video number correlates which each sample.
| D |
E |
F |
||||
| Start salinity:32ppm time:2 min 38 Video #4 observations at how many seconds in: Nudibranch and seastar make initial contact and nudibranch seems to go away, and then seastar makes an advance... nudibranch may be stinging the seastar. |
Start salinity:32ppm time:7:38 Video #5 & 6 Similar observations from D. |
Start salinity:32ppm time:2:54 Video #7& 8 (mostly 8) Similar observations from D and E. |
||||
RNA extraction procedure:
1. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.
2. Using a clean razor blade, cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.*
*to save time, this step has been performed for you.
3. Carefully homogenize the tissue using a disposable pestle.
4. Add an additional 500uL of TriReagent to the tube and close the tube.
5. Vortex vigorously for 15s.
6. If we run out of time we can store at -80C
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube.
NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
17. Incubate at RT for 10 mins.
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer (we will be using the Nanodrop).
Observations: Samples remained very "sticky" or "gooey" and were not completely homogenized, especially sample E.
Next steps: DNAse samples and reverse transcribe to get cDNA for qPCR.


 Setup up for treatments:
Setup up for treatments: |
| external image 20091112-jns5rps724c51uj172b84gjcuc.jpg |
Nudibranch:
 |
| external image 20091112-x6q46dafd8s8icq7ddnmi9758a.jpg |
Salinity shock physical response
 |
| external image 20091112-rddxjigtr1s2db7h4un1eg7ggm.jpg |
Sea star and nudibranch
 |
| external image 20091112-89uq2s7jx3y56c7ne5dexjjjra.jpg |
11/17/09, Day 2 of project nudibranch defense.
Purpose: to use DNAse procedure to get rid of any genomic DNA in our RNA sample.
DNAse methods:
- enzyme that will break up any doubled stranded DNA, always keep on ice
0.5mL tubes
1) 2.5uL DNA ase buffer 1uL turbo DNAase and 20.5uL of RNA to make total of 24uL at 37C for 30 minutes
2)+1uL TUrbo DNAase and put at 37C for 30 minutes
3) 2.5uL inactivation reagent, RT for 2 minutes with mixing
4)spin at 10,000rcf for 1.5 minutes
5) transfer supernatent to a new tube and spec, normalize with 0.1% DEPC H2O
RNA was then quantified using a nanodrop.
NOTE: Always keep your RNA samples on ice!
1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedistal and lower the arm.
2. Click "Blank", to zero the instrument
NOTE: steps 1 and 2 only need to be done once for the whole class.
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your A260 absorbance, RNA concentration (ng/µL), A260/280 ratio and A260/320 ratio.
NOTE: The Nanodrop uses the Beer-Lambert Law to calculate RNA concentration for you. See Lab 1 notes on RNA extraction for more information on the calculation and how to evaluate RNA purity using A260/280 and A260/A320 ratios.
5. Raise the arm and wipe off you sample with a Kim Wiple
6. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
7. storage at -80C.
Future steps: to dilute samples and reverse transcribe them.
11/19/09
Purpose: to make all RNA dilutions to 227.06 ug/uL and to reverse transcribe all samples.
RNA QUANTIFICATION- perform dilutions so lowest spec-ed concentration is uniform with all of them....
(normalize with 0.1% DEPC H2O) - dilution: C1V1=C2V2... C2 will be lowest RNA concentration and figure out all the rest accordingly
Spec result is C1, V1 is unknown, C2 is lowest RNA concentration and V2 option (lower amount)
C1V1 = C2V2
(1127 ng/uL) (V1) = (227.06 ng/uL)(20 uL)
V1= 4 uL
New tube: total of 20 uL: 4 uL of RNA, 16 uL water
Table 4: Table summarizes concentrations and dilution factors. Each dilution total volume will add up to 20μl. Each volume below is in μl.
| Sample ID |
Concentration |
RNA for Dilution |
Water for dilution, total is 20 μl |
|||||
| 1 |
838.84 |
5.41 |
14.59 |
|||||
| 2 |
227.06 |
20 |
0 |
|||||
| 3 |
415.51 |
10.9 |
9.07 |
|||||
| A |
744.44 |
6.10 |
13.9 |
|||||
| B |
364.59 |
12.4 |
7.54 |
|||||
| C |
287.47 |
15.80 |
4.2 |
|||||
| D |
1127.51 |
4.03 |
15.97 |
|||||
| E |
387.27 |
11.73 |
8.27 |
|||||
| F |
599.79 |
7.57 |
||||||
| 12.43 |
Reverse transcriptase protocol:
REVERSE TRANSCRIPTION:
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix (MM)
PER RXN
4 ul 5x Buffer (MMLV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul MMLV Reverse Transcriptase
1 ul Oligo dT Primer
1 ul RNase free water
Total = 15 ul
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave cDNA on ice or store at –20
- Samples were stored at -80.
11/24/09 Tuesday
qPCR performed at 4:00pm with cDNA.
Methods:
1. Master Mix was prepared with the following recipe for 10 reactions for a total of 48 uL per reaction, 2uL of sample of control...
10 reactions desired.. make master mix for 12 reactions.
a. 300uL Master Mix, 2X (Immomix): 25 uL
b. 42uL Syto-13 dye (50uM): 3.5 uL
c. 18uLUpstream primer (from previous stock): 1.5 uL
d. 18uL Downstream primer (from previous stock): 1.5 uL
e. 198uLUltra Pure Water: 16.5 uL
2. 48 uL of Master Mix were added to strip tubes
3. 2uL of cDNA for each primer was added to the wells
4. 2uL of pure water were added for the blank negative controls
5. The plate was loaded onto a real-time PCR thermocycler.
Results and next steps:
Results from qPCR are inconclusive. Contamination is present probably in water sample or during some molecular procedure.
Curves are irregular and there is no pattern; COI results are coming up sooner than they should, so dilution of cDNA for COI qPCR is necessary.
Tuesday 12/1/09
A second qPCR was performed
Same procedure from above:
COI primer samples were diluted 1:10 because that may be the source of the irregular curves
Results:
Results from the second qPCR are again, irregular. Contamination is present and amplified curves are not in any identifiable pattern.
Possible reasons:
- contamination
- wrong primers
- wrong species
- wrong tissue samples taken
Next steps:
- Run a gel, this may tell us if anything is even there, possible that we have incorrect tissue.
Friday - 12/4/09
- Run a gel and interpret
Procedure:
1. weigh 2g of agarose and mix with 150mL 1x TAE in a 1L flask
2. microwave solution for ~ 3 minutes (less than 3 minutes- approx. 1:45 min)
3. cool solution (you should be able to touch the flask for a few seconds), then add 12uL ethidium bromide.
4. mix thoroughly by swirling, then pour into gel tray.
5. add gel combs
6. after gel is set, wrap in plastic wrap and place gel in the fridge, sit for at least 1 hour
7. Load samples:
On the far left well, added 20uL of 100bp ladder
Added 5uL loading die to each qPCR product that was 50uL each
From left to right:
Ladder, 1, 2, 3, A, B, C, D, E, F samples
Results:

gel_fail.jpg
Our samples were run in the top section of the gel.
There is no conclusive banding, there is only "intense" primer dimer
Next steps:
Time is a limiting factor.
Ideally, we would like to either duplicate the experiment and homogenize entire nudibranch to ensure that the correct RNA was extracted. GenBank did not have a sufficient number of available primers, possibly use a different species, or approach with a different method for measuring stress response.
Or, with our original RNA, we could perform dilutions again and make more cDNA and run a few more qPCRs and see if there was just a problem with contamination.
