Lab 1: Tissue Extraction 1
Summary:
Doing protein extraction and start RNA extraction. First, did protein extraction, had the sea scallop tissue and smashed it up to extract the protein. Then second thing we did was the RNA extraction, tissue was smashed up again this time to extract the RNA and we added TriReagent to it.
Protein extraction
Use CellLytic to isolate cellular protein, the salts and detergent will break up the cell membranes. And then determine the concentration of proteins in sample with spectrophotometer.
Procedures:
Extraction
1. Get a 1.5 snap cap tube with organism's tissue weighing 25mg.
2. Add 0.5ml of CellLytic MT solution to the snap cap tube.
3. Use a disposable pestle to homogenize the solution.
4. Close cap and mix by turning the tube upside down multiple times.
5. Use a refrigerated microfuge for 10 mins at max speed to separate the contents in the tube.
6. Label new tube with word "Protein", source organism/tissue, initials, and the lab day date.
7. After microfuge transfer the clear liquid in the tube to the new balled tube and store in ice.
Quantification
8. Using 2ml tube dilute the "Protein" sample, at 1:2, 15ul of protein and 15ul of DI water, mix well.
9. In a second tube fill with 30ul of DI water.
10. Add 1.5ml of Bradford reagent to both tubes.
11. Mix tubes multiple times by turning them upside down and incubate at room temperature for 10 mins.
12. Mix "blank" tube and pipette 1ml to disposable cuvette.
13. Use the blank sample to zero the spectrophotometer. Measure absorbance at 595nm.
14. Mix "sample" tube and pipette 1ml into disposable cuvette.
15. Measure absobance of sample and record value. Measure absorbance twice, the second time after mixing.
16. Average the two absorbance.
17. Back calculate the protein concentration.
RNA extraction
We are to isolate RNA using TriReagent. TriReagent uses guanidine isothiocyanate to denature the protein, then phenol that keep proteins insoluble, then pH which keeps DNA out of solution.
RNA isoloation
1. Get a 1.5 ml snap cap tube with organism's tissue weighing 50-100mg.
2. Add 500 ul of TriReagent to tube.
3. With disposable pestle smash the solution to homogenize it.
4. Add 500ul of TriReagent to tube and close tube.
5. Vortex for 15 secs.
6. Store at -80 degrees C.
Results:
Samples are tagged with the date 10/6 and my initials SV.
Absorbance
Blank- 0 nm
Sample- 0.799 nm (did not measure absorbance twice)
Take the absorbance and multiply by the standard curve y=1011.9 ug/ml (0.799 nm)= 808.508. Then take that and mulitiply by two because the sample was diluted, 1:2, 808.508 ug/ml *2= 1617.02ug/ml.
Conclusion:
The absorbance tells me the protein concentration that I have. Considering I was to extract protein I would expect that I'd have protein in my sample. Based on the results we're going run our proteins next week.
10/13/09
Lab 2: Tissue extraction 2
Summary:
We're going to continue with RNA extraction from lab one. And run an Tris HEPES-SDS Page gel to separate proteins by molecular weight.
RNA Isolation (continue from lab 1)
Procedure:
7. Thaw frozen RNA, then incubate the sample at RT for 5 minutes.
8. Add 200uL of chloroform to the sample in the fume hood carefully and quickly and close the tube tightly. Have tube ready for pipetting.
9. Invert tube to mix and vortex for 30s, til it becomes a milky white mixture.
10. Incubate sample at RT for 5 minutes.
11. Microfuge in for 15 minutes in the refrigerated microfuge at max speed.
12. Remove tube from microfuge, do not disturb the sample.
13. Carefully transfer most of the clear aqueous phase to a new microfuge tube. Do not transfer the white interphase, the cell debris and organic phase.
14. Close the tube with the organic phase and interphase. Dispose of the liquid in the liquid waste jar in the fumehood. And tube in the solid waste jar.
15. Add 500uL isopropanol to the new tube sample of RNA and close tube.
16. Mix the solution in tube by inverting till solution appears to be uniform. Solution should not appear viscous/lumpy.
17. Incubate at RT for 10 minutes.
18. Spin in refrigerated microfuge for 8 minutes.
19. Should have a small white pellet (RNA and salts), though the pellet could be too small to see.
20. Remove the supernatant, the clear liquid. Go slowly to not remove the pellet.
21. Add 1ml of 75% EtOH to pellet. Close tube and vortex to loosen the pellet form side of the tube.
22. Spin in microfuge at 7500g for 5 minutes.
23. Carefully remove supernatant. Again go slow because pellet might be loose.
24. Spin quickly ~15s to pool EtOH.
25. Using the P20 or P200 pippet remove the rest of the EtOH.
26. Air dry the tube at RT for 5 minutes ( in the fumehood dries the pellet faster).
27. Resuspend pellet with 100uL of 0.1% DEPC-H2O. Pippet up and down till pellet is dissolved.
28. Incubate tube at 55 degrees C for 5 minutes.
29. Remove tube from heat, flick to mix samples and then place on ice.
30 Quantitate RNA yield next lab.
SDS- Polyacrylamide Gel Electorophoresis (SDS-Page)
Is the process of separating proteins by molecular weight using an electric field that pulls proteins through polyacrylamide matrix to the cathode. Before this is done proteins must be treated because they have different charges, and they have teritary or quartemary sturctures. So the proteins must be treated to a specific fashion to linearized the proteins and to have the same charge.
Protein Gel Protocol
Procedures:
1. Boil water on hot plate.
2. Thaw protein from last weeks lab. Mix by inverting tube several times.
3. In new 1.5 mL SCREW CAP tube add 15uL of my protein and 15uL of 2X Reducing Sample Buffer.
4. Mix sample by flicking and centrifuge ~10 s to pool liquid to bottom.
5. Boil sample for 5 minutes.
6. Observe assembly of gel box and gels. Rinse gel wells thoroughly.
7. When sample is finished boiling, immediately centrifuge for 1 minute to pool liquid.
8. Slowly load the entire sample into well using gel loading tip.
9. Put lid on gel box and plug electrodes to receptacles on power supply.
10. Turn on power supply and set voltage to 150V. Run for 45 minutes.
11. Turn off power supply and disconnect gel box from power supply.
12. Remove lid of gel box. Disengage tension wedge. Remove gel from gel box.
13. Carefully crack open the cassette to expose gel. If able trim wells at top of gel.
14. Notch designated corner to remind of the position of gel.
15. Place gel into container of Coomassie Stain.
16. Incubate in shaker/rocker for 5 minutes.
17. Pour stain back into original container carefully. Careful not to dump the gel.
18. Rinse gel briefly with 10% acetic acid and pour wash down the drain.
19. Add ~250 mL 10% acetic acid to container with gel. Incubate on shaker/rocker for 15 minutes. Change out buffer and repeat til bands become visible. Most likely will need to incubate over night with plastic wrap and left on shaker/rocker.
Recorded.
Name:Sophie Sample: sea scallop
Concentration: 24.2ug in Lane 5
Calculations: 1617.02 ug/mL * .015 mL/ 0.1 uL= 24.2 ug
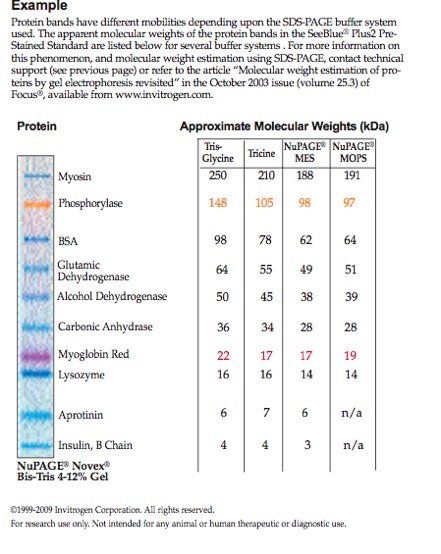 |
| Fig 1. Protein Ladder |
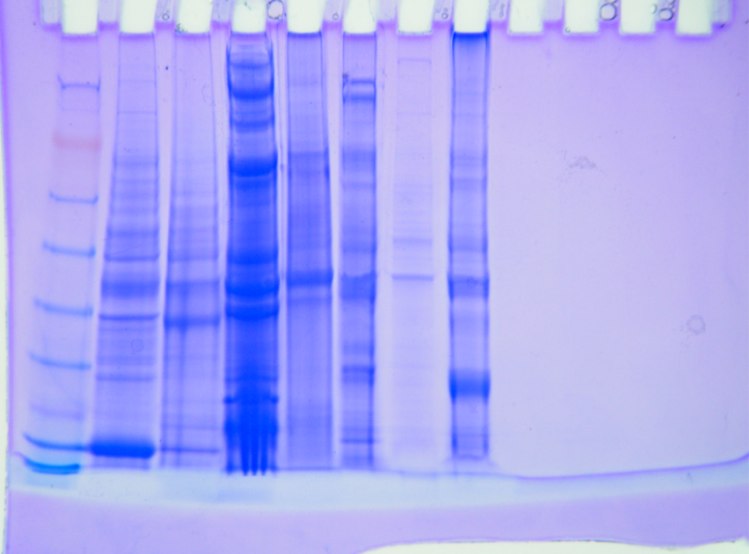 |
| Fig 2. Gel of Proteins |
Results:
My band is in well 5, the sea scallop. I would say that I have five distinct bands. The first band lines up very nicely with the myosin band, so that band of protein must be about 250 kDa. Then there is a second band right after the first band that does not line up with any of the known protein so it is most likely more then 250 kDa. Another line matches up with Alcohol Dehydrogenase about 50 kDA and the last that lines up with lysozyome about 16 kDa. There is also a very broad band in my well that's between Glutamic Dehydrogenase and Carobonic Anhydrase.
Conlcusion:
I honestly did not know what to expect from my gel. I'm just glad that I got distinct lines at all. Though I would like to know what that broad line is between Glutamic Dehydrogenase and Carbonic Anhydrase. There range of molecular weight in my band is from 16-250 kDa. that's a pretty broad range. I also noticed that well four has a lot of protein in it and that most of the bands are really dark. My darkest band is the broad one between Glutamic Dehydrogenase and Carbonic Anhydrase.
Notes:
RNA Isolation -- At step 9 did not mix the sample till milky emulsion. And centrifuged did not separate the interphase from aqueous phase the first time around. So had to mix again then centrifuge a second time for 15 minutes. Then at step 18 and 22 used the smaller centrifuge in the refrigerator.
Protein Gel -- At step 7 used the big centrifuge.
Designing Primers
When designing primers I had wanted to look at C-type lectin. But since my organism sea scallop doesn't have c-type lectin mapped out in the genome, I had to design my primer for it off other organisms. So I put that gene on blast and look for other organism that had c-type lectin genes and find parts of the sequences that are similar in those organisms.
Primer forward- TGGGACCATGTGGGACGGGT
Primer backward- TGTCGTTCCAGTGCGTGCGA
Amplification- 300 bp
10/20/09
Lab 3: Reverse Transcription and PCR
Summary: In lab today we quantified our RNA using the Nanadrop to get our RNA concentration. We also did reverse transcribe RNA to complementary DNA and made agarose gels.
RNA Quantification
Procedure:
1. Pippette 2uL of 0.1% DEPC-H2O on the Nanodrop, the little silver button and lower the arm carefully.
2. Zero the instrument by clicking "Blank". Then with Kim Wipie clean the Nanodrop.
3. The pippette 2uL of own RNA sample to the Nanodrop and lower arm of pedestal carefully.
4. Click "Measure". Record A260 absorbance, RNA concentration (ng/uL), A260/280 ratio and A260/230 ratio.
5. Clean Nanadrop of RNA sample with Kim Wipie.
6. Clearly label stock RNA sample with "RNA", organism, and initials and place on ice.
Reverse Transcription Protocol
Procedure:
1. Mix stock RNA sample, inverting several times.
2. In fresh PCR tube add 5uL of RNA. Spin to pool liquid.
3. Incubate for 5 min at 75 C in thermal cycler.
4. Transfer to ice immediately and incubate for 5 minutes.
5. Make master mix.
Master Mix
Per RXn
1. 4 uL 5x Buffer
2. 8 uL dNTPs
3. 1 uL AMV RTranscriptase
4. 1 uL Oligo dT Primer
5. 1 uL RNase free water
total=15 uL
1. Add MM to diluted RNA (total volume now 20 uL)
2. Vortex and spot spin.
3. Incubate at RT for 10 minutes.
4. In thermocycler incubate at 37C for 1 hour.
5. For 3 minutes heat activate at 98C.
6. Spot spin.
7. Put cDNA on ice.
PCR
Polymerase Chain Reaction is when a target DNA is amplified using primers, polymerase, dNTPs. The amplification happens in a machine thermocycler, the processes goes through denature, anneal, and extension.
Prepare at least 2 samples and have 2 negative controls in microfudge tubes.
Making 50 uL of reaction volume: use C1*V1=C2*V2
1. GoTaq- 125uL
2. Upstream primer- 12.5uL
3. Downstream primer-12.5uL
4. Water- 90uL
Add 48uL of reaction to the 2 sample and 2 negative tubes and put on ice.
To the 2 samples add 5 uL of DNA template
To the 2 blanks add 5 uL of water
Load reaction into thermocycler
Agarose Gels
Procedure:
1. Weigh 2 mg of agarose and mix with 150 mL TAE in a flask
2. Microwave for 3 minutes or until it boils.
3. Cool solution then add 12 uL of ethidium bromide carefully.
4. Mix by swirling then pour into gel tray.
5. Add combs.
6. Wrap gel in plastic and store in frig.
Recorded
RNA Quantification:
A260 absorbance- 6.723
RNA concentration- 268.9
A260/280 ratio- 1.91
A260/230 ratio- 1.80
Calculations: 268.9ng/mL*(10^6ug/10^9ug) = .2689ug/uL
25 ug*(uL/.2689 ug)= 92.97 uL
Notes: I was pippetting wrong for most of the lab. I was going to the second stop on the digital pippett instead of the first, so i ended up with more solution at times and with the wrong concentration of mix. I did not catch this until we were making our 50 uL of reaction to add to the RNA. It was a really bad pippetting day.-
-
Conclusions: I had think I had a good amount of RNA concentration, though I had expected a higher RNA concentration. I think my results for PCR will be thrown off because of my pippetting, I might have more amplification of proteins then I should have because my concentrations for the mixes were wrong. I had more dNTP's, AMV RTranscriptase, and Oligo dT Primer, whichs is more materials for making proteins.
10/27/09
Lab 4: Western Transfer- Immunoblots
Summary:
We ran our PCR products on gel in groups of four, loaded the 4 wells with our two samples and two blanks. Then we transferred our proteins onto membrane, and talked about our projects in classes.
Run PCR products on gel
1. Put gel in gel box filled with 1x TAE buffer (completely cover wells)
2. Remove combs from wells
3. Pippette in 7uL 100bp to furthest left lane (ladder)
4. Load 25uL of PCR sample into gel (refrigerate remaining PCR samples at 20 C)
5. Run gel at about 100 for about an hour
6. Look at gel on UV transilluminator
Principles of Western Blotting
After proteins have been separated by electrophoresis it is then transferred to nitrocellulose membrane. The nitrocellulose membranse binds all proteins no matter the charge, though having a charge is good to strengthen the bond.
Western Blot Procedure
-Blocking
To reduce non-specific interactions between the membrane and antibody membrane is blocked with non- specific proteins like non-fat milk. This covers the areas of membrane where no protein has bound from transfer.
-Primary Antibody
First antibody is applied to be incubated with membrane, it should bind to protein of interest with the appropriate concentrations. In order for this to happen correctly the membrane needed to be coated.
-Second Antibody
Rinse the membrane to remove unbounded primary antibody add second antibody and incubate with membrane. The secondary membrane binds to species specific portions of primary antibody.
-Developing
Wash away secondary antibody, and incubate membrane so positions of membrane bound secondary antibodies will change color or emit light bands. Will be able to see bands of proteins of interest, it will be a dark purple.
Transfer Proteins to Membrane
1. Cool buffer solution to 4 C
2. Soak filter paper, membrane and gel in Transfer Buffer for 15 minutes
3. Put together blotting sandwich on semi-dry blotting apparatus
-Anode (+++)
-Filter paper
-Nictrocellulose Membrane
-Gel
-Filter Paper
-Cathode (---)
4. Transfer blot for 30 minutes at 20V
5. Take gel out of sandwich and rinse with transfer buffer
6. Wipe with cotton swab to remove adhering gel from membrane
Work fast without touching the membrane, keep membranes wet, add solutions to trays slowly at membrane edge avoid bubbles
1.Prepare 20 mL Blocking Solution
Ultra Filter water 14 mL
Blocker/Diluent (Part A) 4 mL
Blocker/Diluent (Part B) 2 mL
Total volume= 20 mL
2. Prepare membrane in 10 mL of appropraite Blocking Solution in covered plastic dish. Incubate for 30 minutes on rotary shaker at 1 rev/sec.
3. Decant Blocking Solution
4. Rinse membrane with 20 mL of water for 5 minutes, repeat one more time
5.Prepare 10 mL of Primary Antibody Solution (1:3000 dilution)
Blocking Solution 10 mL
HSP 70 antibody 3.3 uL
Total volume= 10 mL
6. Incubate membrane with overnight with 10 mL Primary Antibody Solution
Next Day Decant Primary Ab, saved at 4 C
Results:
PCR

I had very little amplification compared to other people in my group. I had very light bands compared to my group members, though my band SV S1 seems to have a band that lines up with the 500 bp bands. I also have very faint lines after the 200 mark in both of my samples.

Western Blot Gel
My western blot gel shows that my protein, in well 6, were amplified and are there to be transferred to the western plot paper to be tested for heat shock protein.
Western Plot
The Western blot shows that my well 6 does not have a purple line. Though three of the people on the same western plot show purple lines. The wells are 2, 3, 4 this means that they have heat shock protein in their gels.
Notes:
During lab three I had pippette wrong through most of the lab and i expected there to be lots of amplification in my PCR gels.
Conclusion:
My PCR gel did not show that I had a lot of amplification of the gene that I was looking for. Which is not what I expected because of my pipetting technique. I had added too much dNTP's, AMV RTranscriptase, and Oligo dT Primer so I thought that instead I would get over amplification of the protein I wanted to amplify. So this probably means that I didn't design a primer that worked for amplifying the gene I wanted for my organism. So next time when designing primers I should have more then one choice of primer and probably experiment with 2 others.
The western blot from the protein transfer did not turn purple when tested for heat shock protein. So that probably means that my sea scallop was not heat stressed when it was killed to become a sample or it could be that my proteins did not transfer well onto the paper. Though 3 other people on the same Western blot that I was on had purple bands, which makes sense because some of those organisms were heat stressed, so they should have been expressing heat shock protein.
-
11/3/09
Lab 5: Quantitative PCR
Summary: On this day we prepared our samples to be run in quantitative PCR, prepared sample of cDNA, RNA and negative controls. Running RNA will tell me if I have genomic DNA in my sample, I will know this my RNA sample gets amplified because PCR is not suppose to work on RNA, if it does then I most likely have carry over. And if anything gets amplified in my negative control then that means something was wrong with my water.
Realtime PCR
Have duplicates of cDNA, RNA and negative controls. Label wells or write down the order that the samples were loaded.
1. Prepare Master Mix: prepare enough to run templates 7 times.
Component
Master Mix, 2x (Immomix)- 175 uL
Syto-13 dye (50uM)- 14 uL
upstream primer- 7 uL
downstream primer- 7 uL
Ultra Pure Water- 133 uL
2. Add 48 uL of mastermix to white wells of PCR plate.
3. Add 2 uL of thawed out cDNA sample to two of the wells.
4. Add 2 uL of ultra pure water to negative control wells.
5. Add 2uL of diluted RNA (1 uL RNA + 3 uL of H2O3) to RNA wells.
6. Cap securely, and if needed spin strips to collect volume at bottom of the wells.
Results:

The PCR results looking at C(t) values on the class spreedsheet, tells me that I have no amplification in the cDNA wells, 1 and 2, and my negative controls, 5 and 6, the results for those are N/A. My C(t) values for well 3 is 36.43 and well 4 is 35.06, because they are less then 40 this means that there was amplification in the wells with RNA. The values for the duplicates are also close to each other so that means the precision is close.
The melt curve on from column 10 has protein melting from the 3rd and 4th well, which is my RNA. This most likely the point where my RNA broke apart. The two wells also showed a similar melting point so I did amplify one protein and it was expressed in both of my RNA wells.
Note: I did not dilute my RNA when I loaded them into my wells. So the RNA are straight, so I might get a huge amplification of protein that is does not relate to cDNA.
Conclusion: Because I had no amplification in my cDNA wells that means that there was probably no DNA in those wells to amplify protein. It probably means that something must have gone wrong during reverse transcription. To not be able to get amplification with PCR in the cDNA wells. But however there was genomic DNA in my RNA wells because they were the only ones that amplified proteins.
Lab schedule
11/10 TriReagent and RNA isolation
11/17 Run PCR -
11/24 Quantitate
11/30 Analyze data
11/8 have presentation ready or extra lab day
11/10/09
Lab 6: Cell Sample and RNA isolation
Summary: This day we started on our own projects. I wanted to see how sea stars' immune system would react to a toxin, Roundup, in the sea water. The Monday night before lab the sea stars were put in a tank with 100 ml of sea water with 20 ml of Roundup added in.There was also a control tank with sea stars in normal sea water. Unfortunately the sea stars were overexposed and died the next morning. So in lab we subjected more sea stars to Roundup in the same conditions, only for 30 minutes. Then I harvested the cells I was interested in, hemolymph, from the sea stars in the control and experiment there were 3 controls and 3 experiments.
Obtaining Sample Cell
Procedure:
1. Obtain scissors and tubes to drain hemolymph in. Label tubes with controls and samples. Label as C1 and S1 with initials SV
2. Cut a tip of one arm off the sea star and drain hemolymph into clean tube, it helps to squeeze some of the liquid out, as much as will come out
3. In most of the tubes drained more then enough to pippette 1 ml of the sea star liquid into a 1.5 ml microfuge tube, execpt for Sample 1 which was less then 1 ml
4. Sample 1 pippette 25 ul of liquid into 1.5 ml microfuge tube
5. Since most of the samples I had were more then 2 ml I did duplictes of 1 ml in 1.5 microfuge tubes
6. Sample 1 has duplicate of less then 25 ul
7. Then microfuge the samples in refrigerated microfuge at the lowest speed for 3 minutes to not destroy the cells and release RNA. Check to see if the cells have gathered to the bottom of the microfuge. If not repeat microfuge again (I microfuge my samples 3 times)
8. Remove the clear liquid from the microfuge tubes, be CAREFUL to not distrube the cells at the bottom
RNA Isolation
1. Pippette 500 ul of microfuge into the tube with the cells
2. With disposal pestle smash the cells into solution to homogenize it
3. Add 500ul of TriReagent to tube and close tube
4. Vortex for 15 seconds
5. Add 200uL of chloroform to the sample in the fume hood carefully and quickly and close the tube tightly. Have tube ready for pipetting.
6. Invert tube to mix and vortex for 30s, til it becomes a milky white mixture.
7. Incubate sample at RT for 5 minutes.
8. Microfuge in for 15 minutes in the refrigerated microfuge at max speed.
9. Remove tube from microfuge, do not disturb the sample.
10. Carefully transfer most of the clear aqueous phase to a new microfuge tube. Do not transfer the white interphase, the cell debris and organic phase.
11. Close the tube with the organic phase and interphase. Dispose of the liquid in the liquid waste jar in the fumehood. And tube in the solid waste jar.
12. Add 500uL isopropanol to the new tube sample of RNA and close tube.
13. Mix the solution in tube by inverting till solution appears to be uniform. Solution should not appear viscous/lumpy.
14. Incubate at RT for 10 minutes.
15. Spin in refrigerated microfuge for 8 minutes.
16. Should have a small white pellet (RNA and salts), though the pellet could be too small to see.
17. Remove the supernatant, the clear liquid. Go slowly to not remove the pellet.
18. Add 1ml of 75% EtOH to pellet. Close tube and vortex to loosen the pellet form side of the tube.
19. Spin in microfuge at 7500g for 5 minutes.
20. Carefully remove supernatant. Again go slow because pellet might be loose.
21. Spin quickly ~15s to pool EtOH.
22. Using the P20 or P200 pippet remove the rest of the EtOH.
23. Air dry the tube at RT for 5 minutes ( in the fumehood dries the pellet faster).
24. Resuspend pellet with 100uL of 0.1% DEPC-H2O. Pippet up and down till pellet is dissolved
26. Flick to mix samples and then place on ice.
27. Quantitate RNA yield next lab.
Notes: When I drained the sea stars of fluid it contained sea water and other things besides hemolymph, in order to seperate the liquid from cells I microfuge the liquid at a low speed for a short amount of time to not destroy the cells and release RNA. Since I'm working with hemolymph it is hard to collect enough coelomytes cells then it would be with regular tissue. So this means that I did not get a visable white pellet when if first microfuge with the Triagent, in fact I did not get a white pellet at all during RNA isolation. So when i was pippetting EtOH out of the microfuge tube i left more then necessary to not remove any RNA that was at the bottom of my microfuge tube.
11/17/09
Lab 7: DNAase Protocol
Summary: To remove DNA from RNA samples before going on to make cDNA. (keep all of this on ice!!!!)
1. In new tubes add 2.5 uL of DNAse buffer for all the samples that you have (I had 12 samples).
2. Add 1 uL of Turbo DNAse to those tubes to the also.
3. 20.5 uL of sample RNA.
4. Incubate the samples at 37 degrees C for 30 minutes.
5. After incubation add 1 uL of Turbo DNAse, then incubate for another 37 degrees C for another 30 minutes.
6. After incubation add 2.5 uL inactivation Reagent, incubate at RT for 2 minutes and mix.
7. Spin in 10,000 rpm for 1.5 minutes.
8. Spec
Stock Primers~ 100 uM of Primer solution
1. Take the number of the nm of primer and multiply by 10.
2. The resulting number is the volume of water you add to the RNA tube. This is the 100 uM solution.
ex. P-glycoprotein Primer set 1
Foward primer- 40.6 nm * 10 = 406 uL of water
Reverse primer - 33.6 nm * 10 = 336 uL of water
Stock primer ~ 10 uM
1. Use C1V1=C2V2
2. Take 10 ul of 100 uM of stock primer add to new tube.
3. Add 90 uL of water to dilute the 100 uM of stock primer.
Notes: It took longer then I expected to do the DNase procedure so I fell behind on my time line for labs. I also made a huge mix of the DNase solution of buffer, this might affect all of my samples if I did the procedure wrong.
11/24/09
Lab 8: Spec RNA and Reverse Transcript -
Summary: Spec RNA sample to find out the concentration of RNA is in the stock samples. And then with after the RNA spec do reverse transcription and make cDNA.
Procedure: RNA spec
1. Open the Nanodrop program and use "nucleotide" option.
2. Run blank, pippette 2 uL of 0.1% DEPC-H2O onto Nanodrop pedistal and lower the arm. Click "Blank" to zero instrument.
3. Run own sample, change the name of the sample everytime run new sample.
4. Pippette 2 uL of RNA sample onto Nanodrop pedistal and carefully lower arm.
5. Click "Measure", the absorbance of A260, RNA concentration (ng/uL), A260/280 ration and A260/320 ration will be recorded on a table in the program.
6. I repeated this for all my 12 samples
Procedure: Reverse transcription
Master Mix
5x Buffer (AMV RT Buffer)- 52 uL
dNTPs (10 mM total)- 104 uL
AMV-RTranscriptase- 13 uL
Oligo dT Primer- 13 uL
RNase free water- 13 uL
1. Mix RNA sample stock, inverting a couple of times.
2. Into a .25 uL of PCR tube transfer 5 uL of RNA.
3. Incubate tube at 75 C degrees for 5 minutes in thermal cycler.
4. After incubation immediately transfer to ice.
5. Add 15 uL of Master Mix to RNA so that total volume is 20 uL.
6. Vortex and spot spin.
7. Incubate at RT for 10 minutes.
8. Then incubate at 37 C degrees for 1 hr in thermocycler.
9. Heat activate @ 95 C for 3 minutes.
10. Store cDNA at ~20 C.
Results: The spec on my RNA showed some very mixed results of the absorbance. I expected my RNA concentration to be small because I expected to extract smaller amounts of RNA from my cells, then other peoples' samples. Some where really high and some were really low. It wasn't until Mac suggested looking at the absorbance curve, it showed that the light was absorbing at 230 and 270. Not the absorbation levels of nucleotides (260) so this meant that I had some carry over. the carry over seemed to match the absorbance levels of of the deactivant not being completely removed from the sample. Mac suggested microfuging the samples and testing them again. It should that i still had the same problems. but we decided to move on and make cDNA from the stock RNA because it seemed that I had some RNA in the sample. And hopefully when we spec them for DNA I won't get the same problem again.
Conclusion: The carry over could have happened during DNase. And if my cDNA samples still show contaminant then I will use Chris' RNA and make new cDNA from that and run my primers using Chris' RNA. I don't know how that would work considering that we're looking at different systems in starfish. Chris is looking at the reproductive system and I am looking at immune response so I don't know if my primers will find anything in Chris' RNA.
12/1/09
Lab: 9 Spec and Reverse transcription again
Summary: While quantifying my cDNA, it still showed that I had the same contaminants. So I made cDNA using Chris' samples.
Procedure: RNA spec
1. Open the Nanodrop program and use "nucleotide" option.
2. Run blank, pippette 2 uL of 0.1% DEPC-H2O onto Nanodrop pedistal and lower the arm. Click "Blank" to zero instrument.
3. Run own sample, change the name of the sample everytime run new sample.
4. Pippette 2 uL of RNA sample onto Nanodrop pedistal and carefully lower arm.
5. Click "Measure", the absorbance of A260, RNA concentration (ng/uL), A260/280 ration and A260/320 ration will be recorded on a table in the program.
6. I repeated this for all my 12 samples
Again they show contaminants. So I made cDNA with Chris' samples.
Procedure: Reverse transcription
Master Mix
5x Buffer (AMV RT Buffer)- 52 uL
dNTPs (10 mM total)- 104 uL
AMV-RTranscriptase- 13 uL
Oligo dT Primer- 13 uL
RNase free water- 13 uL
1. Mix RNA sample stock, inverting a couple of times.
2. Into a .25 uL of PCR tube transfer 5 uL of RNA.
3. Incubate tube at 75 C degrees for 5 minutes in thermal cycler.
4. After incubation immediately transfer to ice.
5. Add 15 uL of Master Mix to RNA so that total volume is 20 uL.
6. Vortex and spot spin.
7. Incubate at RT for 10 minutes.
8. Then incubate at 37 C degrees for 1 hr in thermocycler.
9. Heat activate @ 95 C for 3 minutes.
10. Store cDNA at ~20 C.
Results: cDNA quantification. It shows two florescence peaks and neither of them are at the 260, so what is shown is not DNA.


Conclusion: Doing reverse transcription did not get rid of the contaminants, and the contaminant could have happened during anytime of my procedures. So now I'm going to use Chris' samples to do the rest of my experiment. I will use my primers on Chris' samples and see if I can amplify anything.
12/2/09
Lab 10: qPCR
Summary: Run qPCR with Chris' samples and my primers. This is a test run I'm going to run the a couple of my samples with my four sets of primers. This will tell me which ones work for what I'm hoping is going to be expressed.
Procedure:
Master Mix
62.5 uL- 2x (Immomix)
5 uL- Syto-13 dye
2.5 uL- Primer Forward
2.5 uL- Primer Reverse
47.5 uL- PCR H2O
Total = 24 uL
- Make four master mixes for the four different sets of primers.
ATP binding cassette 1
ATP binging cassette 2
P-glycoprotein 1
P-glycoprotein 2
Load the wells in this order
ABC 1- H2O, H2O, S-L, C-L
ABC 2- H2O, H2O, S-M, C-M
P-Gl 1- H2O, H2O, S-S, C-S
P-Gl 2- H2O, H2O, S-L, C-L
Procedure to load wells:
1. Add 24 uL of master mix to qPCR wells.
2. Add 1 uL of cDNA to the wells.
3. Cap the wells.
4. Place in fridge until ready for qPCR.
Results: The fluorescence and melt curves of my qPCR results were not very good. The ABC primers did not work though the primers for P-glycoproteins did show some promise. This the melt curve of P-glyprotein 2 shows some thing was amplified in my samples, but unfortunately there were also amplification in my negative controls also. And the melt curves showed that there could be two things in the samples because there are two melt points.


Conclusion: There could be something in those samples and those primers could work. But to make sure we're going to do more runs with the P-glycoprotein primers. This time I'm going to run all my cDNA samples with P-glycoprotein 2 primers. If this run with the P-glycoprotein works then I will do a real run with all the cDNA and have multiples. And I will also create a new 10 uM stock of primer incase it is the primers that are contaminated.
12/3/09
Lab 11: qPCR with P-glycoprotein
Summary: Do another run for with P-glycoprotein with all my samples on cDNA.
Stock primer ~ 10 uM
1. Use C1V1=C2V2
2. Take 10 ul of 100 uM of stock primer add to new tube.
3. Add 90 uL of water to dilute the 100 uM of stock primer.
Procedure:
Master Mix
112.5 uL- 2x (Immomix)
9 uL- Syto-13 dye
4.5 uL- Primer Forward
4.5 uL- Primer Reverse
85.5 uL- PCR H2O
Total = 24 uL
- add P-glycoprotein 2 primers
Load qPCR wells in this order
P-Gl 2- H2O, H2O, S-L, S-M, S-S, C-L, C-M, C-S
Procedure to load qPCR wells:
1. Add 24 uL of master mix to qPCR wells.
2. Add 1 uL of cDNA to the wells.
3. Cap the wells.
4. Place in fridge until ready for qPCR.
Results: Again my waters showed amplification but it seemed that the cDNA also had some amplification.
Conclusion: Since there was also amplification in my waters I can't say that the amplification of my cDNA means anything because everything could be contaminated. Luckily Mac made some more cDNA from Chris' RNA and ran her own qPCR with my primers. And she had amplicfication in the P-glycoprotein 1 and had clean waters. So next I'm going to run more qPCR with Mac's cDNA. And also we want to know what is being amplified in the samples, so I'm going to do a tradtion PCR and see if the gel will give me any insights on what is being amplified.
12/4/09
Lab 12: PCR gel and qPCR
Summary: Since I had more ampilification in my qPCR results we're going to run a gel and see what is being amplified using gel PCR. I am also trying qPCR one more time with Mac's cDNA.
Tradition PCR
Procedure to make agarose gel:
1. Weigh 2 mg of agarose and mix with 150 mL TAE in a flask
2. Microwave for 3 minutes or until it boils.
3. Cool solution then add 12 uL of ethidium bromide carefully.
4. Mix by swirling then pour into gel tray.
5. Add combs.
6. Wrap gel in plastic and store in fridge.
Procedure prepare qPCR samples:
1. To the qPCR wells add 2.5 uL of (what kind) dye
2. Mix samples in wells with dye and spin.
Run PCR products on gel
1. Put gel in gel box filled with 1x TAE buffer (completely cover wells)
2. Remove combs from wells
3. Pippette in 20 uL 100bp to furthest left lane (ladder)
4. Load 25uL of qPCR sample into gel (refrigerate remaining PCR samples at 20 C)
5. Run gel at about 100 for about an hour
6. Look at gel on UV transilluminator
qPCP
Procedure:
Master Mix
112.5 uL- 2x (Immomix)
9 uL- Syto-13 dye
4.5 uL- Primer Forward
4.5 uL- Primer Reverse
85.5 uL- PCR H2O
Total = 24 uL
- add P-glycoprotein 1 and P-glycoprotein 2 primer sets
Load qPCR wells in this order:
P-Gl 2- H2O, H2O, S-M, S-S, C-L, C-M, C-S, H2O
P-Gl 1- H2O, H2O, S-M, S-S, C-L, C-M, C-S, H2O
Note: Mac cDNA only had 5 samples because the S-L had some organic matter in it, so she did not use S-L. And also I had made an extra amount of master mix, so I decided to put in a extra H2O at the end of the wells.
Results for traditional PCR: Does not show any bands besides the very intense "primer dimer". The primers could have been what accounted for the florescence. My qPCR samples are in the bottom lanes, the furthest left is the ladder and the rest are my samples loaded in the way I loaded my qPCR samples.

ladder H2O H2O S-L S-M S-S C-L C-M C-S
Results qPCR: I did not get any amplificication for P-glycoprotein 2. But was able to get something in P-glycoprotein 1, I was able to get 2 of my waters to be negative, no amplification in them, however one of them had amplification we're going to consider that just that one well is contaminated. Now I have something to compare my amplification with. Also one of my samples came out with unclear melt curve, that was sample C-S, so I'm going to discount that sample and not include it to my results. Using a t-test there seems to be no significant difference in the level of expression of P-glycoprotien between samples and control.
Conclusion: I believe that some of the problems I ran into were due to the choice of tissue I focused on. It was hard to know if I had enough tissue to extract RNA, and even if I was able to extract RNA that amount would have been very small. And of course there was the contamination all I can think to do about that is to be cleaner in my methods. And some of my primers didn't work most likely because my primers were not species specific. I designed my primers based off sea urchin and to be very broad to encompass the whole echinoderm phylum, so it must have not been specific enough to find the genes I was looking for. Next time I would like to choose a different tissue, and design more specific primers for that species, or I might choose a different organism all together, one that was more studied then sea stars.