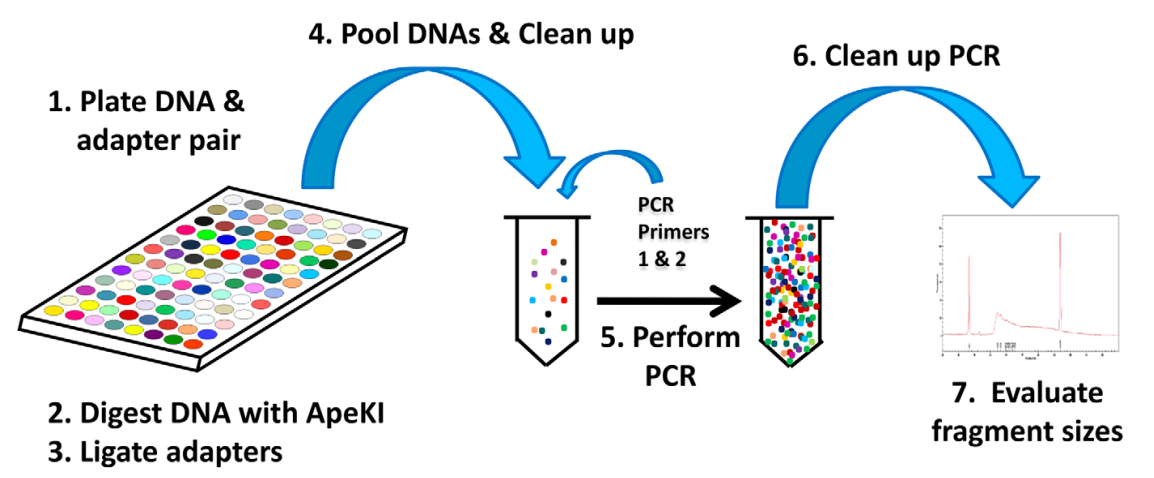
1 Pipeline of experiments
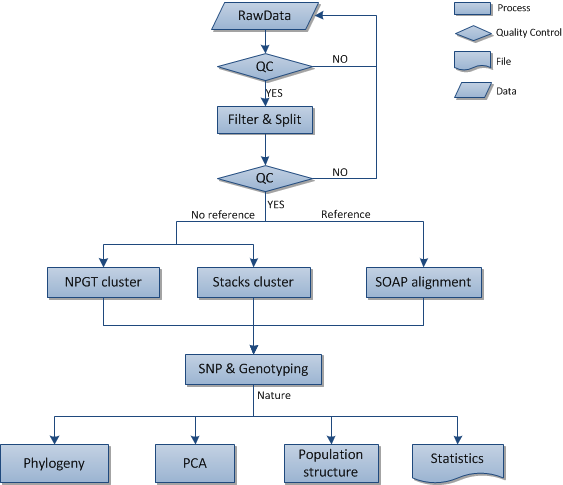
2 Bioinformatics analysis
2.1 Data statistics
52.61 Gb clean data was generated in this project. Data statistics of some samples was shown as following (Please read Data.readme_en.txt for definition of each column.):
| Sample name | Read number (M) | Base number (Mb) | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| 1HL_10A | 6.37 | 615.01 | 42.61 | 95.68 | 90.80 |
| 1HL_11A | 7.14 | 688.72 | 42.69 | 95.72 | 90.91 |
| 1HL_12A | 5.05 | 487.29 | 42.57 | 95.61 | 90.70 |
| 1HL_13A | 5.35 | 516.55 | 42.84 | 95.63 | 90.72 |
| 1HL_14A | 4.03 | 387.00 | 42.69 | 95.59 | 90.64 |
| 1HL_15A | 4.39 | 421.69 | 42.50 | 95.59 | 90.62 |
| 1HL_16A | 4.67 | 447.97 | 42.62 | 95.63 | 90.74 |
| 1HL_17A | 7.31 | 701.52 | 42.51 | 95.61 | 90.70 |
| 1HL_19A | 7.24 | 694.65 | 42.76 | 95.64 | 90.73 |
| 1HL_1A | 4.52 | 436.26 | 42.36 | 95.61 | 90.66 |
| ... | ... | ... | ... | ... | ... |
2.2 SNP Detection
The information about SNPs in one sample was shown as following(Please read SNP.readme_en.txt for definition of each column.):
| Sample name | Total | Homo | Hete | Homo rate (%) | Hete rate (%) |
|---|---|---|---|---|---|
| 1HL_10A | 8058 | 6755 | 1303 | 83.83 | 16.17 |
| 1HL_11A | 8392 | 7033 | 1359 | 83.81 | 16.19 |
| 1HL_12A | 7422 | 6405 | 1017 | 86.30 | 13.70 |
| 1HL_13A | 7580 | 6499 | 1081 | 85.74 | 14.26 |
| 1HL_14A | 6374 | 5552 | 822 | 87.10 | 12.90 |
| 1HL_15A | 6683 | 5706 | 977 | 85.38 | 14.62 |
| 1HL_16A | 7041 | 6043 | 998 | 85.83 | 14.17 |
| 1HL_17A | 8419 | 7050 | 1369 | 83.74 | 16.26 |
| 1HL_19A | 8543 | 7141 | 1402 | 83.59 | 16.41 |
| 1HL_1A | 6919 | 5949 | 970 | 85.98 | 14.02 |
| ... | ... | ... | ... | ... | ... |
2.3 Genotyping
The genotyping result was shown as following (Please read genotype.noref.readme_en.txt for definition of each column.):
| ID | Consensus_Seq | pos | 1SN_9A | 1SN_8A | 1SN_7A |
|---|---|---|---|---|---|
| record_385 | CAGCCCTTATTAGGCCACCTGAGTRAACTGATGTGACCTATATTGTAATTAGTTTTCATCCATCATTGTTAAGTCATATGTA | 25 | A | A | A |
| record_795 | CAGCTTCCGATGCGTCGGAGAAGATCAGCACTTYTGGATCTGATACTAAGCTCAGTGATTGTGGAACAAACATCCGTGGAAT | 34 | Y | Y | Y |
| record_1907 | CAGCTGAAGCATCCAGCAAAGCACGAACAAAGCAGAGAGACACCTACAACACCAAAGTGAGARGAGGAACGGTTAAGGCTGG | 63 | G | G | - |
| record_2412 | CAGCCCTTCACCGGTAATGGTGACGTCTCAATATGAATGAAATATTCTCGACGGGWCGAAAAACAACAAATAATCAATCAAT | 56 | - | - | - |
| record_3293 | CAGCACCTCAGGGCTATCCGTTTCTAACACGGGTACGGGCCMGCGGGGGTTCACGGGCAGTGACCTCCCTTGCACGTTACAA | 42 | - | C | - |
| record_4526 | CTGCACTTGACCACAATAAATGTGATCTTCCTTTAGATTTAATATATATCTGTCTGCCTTCATRAACGTTGATTTTTAAAAT | 64 | R | G | R |
| record_4591 | CAGCTGTATCATCTGACCCCAGAAGAAGCCGGYTTGATAGGAAGCTCTCTTGATGTGTGCTTCTCGAGTTGGAGGAATGGCC | 33 | C | C | C |
| record_5127 | CTGCGTTGTCATGCAAGCATTCTTGACTCCCATGCTCACGAGCCATAGTTACAATGCCGGAATCGCRCATTGGTTTATTCGC | 67 | R | G | R |
| record_5132 | CAGCTTAAGATTTTGTGCATTTTTCAGTTGTGYTCTCTCTACTTTCGATTCCTTGATCCGCCCTTGAAGGAGACCTTCATGT | 33 | C | C | - |
| record_5456 | CAGCTTGTTTACGAGGCCCTGCYCGCACTTTCTACATAAGTCTCCCCGCAGAATTCAGACAGCCATATAACTTGTTAATTAC | 23 | Y | Y | Y |
| ... | ... | ... | ... | ... | ... |
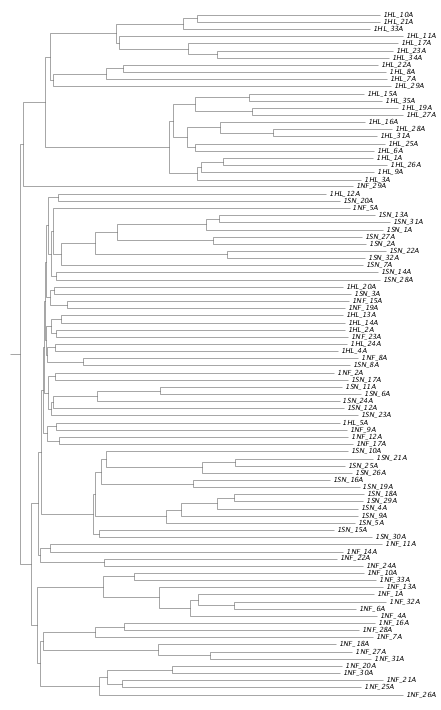
2.4 Phylogeny analysis
According to the SNPs we got, the evolution relationship between groups can be infered and shown in phylogenetic tree as following:
2.5 PCA
PCA (Principal Component Analysis) is a mathematical procedure that uses orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of linearly uncorrelated variables call principal components, which have the largest possible variances. Based on different SNPs between samples, we could separate samples into different subgroups by PCA. The analysis result was shown below:
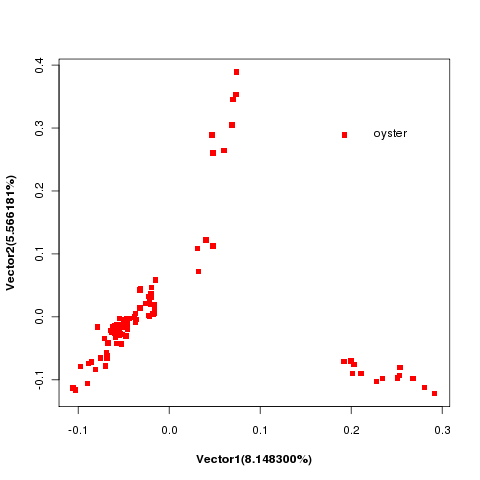
Note:Dots in different color represent in different subgroups.