11/29/2011 -- LAB 9
1. Detail at least 2 reasons why your results turned out the way they did. This should be easy to do if your results are "unexpected", but even expected results can have multiple explanations. Really think about this, the answer "because I messed up in lab" (or any variation thereof) is not acceptable.
Hyperoxic acclimated oyster showed the highest gene expression (mean = 0.12 ± 0.19) , while the controls had lower expression levels (mean = 0.069 ± 0.068) and the low DO oysters the least amount (mean = 0.017 ± 0.029) of HIF-1α mRNA in the system (Fig. 2). However, none of the treatment groups were significantly different (P =0.3967, F-stat = 0.99).
The HIF-1α trends appear to be correlated with the DO acclimation of the oyster, a possible indication that Pacific oysters are capable of altering HIF expression to match the changing oxygen environment. It is also possible that increases in mRNA is not a reasonable proxy for the actual active HIF protein. Never the less, the non-significance of the results is most likely due to the high amount of variance; the high variability could be a natural artifact of individual variation in response to hypoxia and/or a result of the fluctuation in DO during acclimation. A large sample size would help increase the precision of our results.
2. What are two obstacles that you encountered during your lab work and experimental design? Did these obstacles affect your results? Why?
One obstacle was the fluctuations in DO for all acclimation groups due to the experimental equipment available at the time. The Nitrogen and Oxygen tank valves were very sensitive and as a result the DO tended to change +/- 2mg/l everyday. These fluctuation could have impacted the response to the stress, and possibly increased viability among samples and between the groups. We also had issues with mixed up labeling, which we believe we corrected as soon as it was realized and thus did not effect the results.
3. Explain at least one aspect of your research and its results that have a greater impact outside of your own personal learning experience. What would you tell a non-scientist who challenged the importance of your research?
Oysters, and possibly other bivalves, are capable of adjusting their energetic costs and needs essential for survival. This is particularly important considering aquatic species not only have hypoxia to counter, but other changing environmental stressors such as changing global temperature. From a management perspective these findings are particularly important in a natural and aquaculture setting in terms of persistence of the species.
4. What part of your research and analysis has completely stumped you? Is there anything you can do to find the answer or will it always remain a mystery?
Not really stumped, but the high variance might need more explanation. This could be done by increasing our sample size.
5. In about 3 sentences each, summarize 2 papers that you are going to cite in your own paper that give insight into the results that you found.
de Jong (2005) investigated whether phenotypic plasticity causes evolution or is a major evolutionary mechanism. Based on extensions of several preexisting models of evolution, de Jong found that modifiable regulatory mechanism can be advantageous and evolve if the adjusted phenotype increases overall fitness.
Vogel et al. (2010) used large-scale absolute protein expression measurements (APEX) and applied those values to estimate the relative contributions of mRNA gene expression in yeast and E. coli. The researchers found that significant percentages of the variance of yeast (73%) and E.coli (47%) protein could be explained by mRNA expression, and is thus a reasonable proxy for active protein expression in Eukaryotes and Prokaryotes.
11/22/2010 -- LAB 8
Summary:
The purpose of lab 8 was to run qPCR on (1) all remaining C. gigas gill tissue cDNA from dissolve oxygen acclimation groups and (2) house keeping genes for comparison.
Materials & Methods:
qPCR
1) Prepared master mix for 13 remaining templates, duplicates, two NTCs, and +1 for pipetting error
For a 25μl reaction volume:
| Component |
Volume |
Final Conc. |
Multiplier |
Duplicate |
Total volume |
| Master Mix, 2X (Immomix) |
12.5µL |
1x |
x16 |
x2 |
400ul |
| Syto-13 dye (50uM) |
1µL |
2µM |
x16 |
x2 |
32ul |
| upstream primer, 10μM |
1.25μl |
2.5μM |
x16 |
x2 |
40ul |
| downstream primer, 10μM |
1.25μl |
2.5μM |
x16 |
x2 |
40ul |
| Ultra Pure Water |
7uL |
NA |
x16 |
x2 |
224ul |
2. Added mastermix to wells of a white PCR plate
3. Thawed cDNA sample: Hyperoxic 1
4. Added 2uL cDNA template to each reaction
5. Added 2uL of ultra pure water (0.1% DEPC) to the negative control wells
6. Capped the wells securely
7. Spun down strips to collect volume in the bottom of the wells
8. Ensured the lids were clean and place strips on ice
9. Load the plate, verify the PCR conditions and start the run
PCR conditions:
95°C for 10 minutes
95°C for 15s
55 °C for 15 s
72°C for 30 s (+ plate read)
Returned to step 2 39 more times
95°C for 10s
Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
**This procedure was performed again except with Elongation factor-1a (EF-1a) primer pairs; a house keeping gene for relative gene expression comparison.
Results: Will be confirmed after Thanksgiving break.
Conclusion: N/A
Reflection: When I was trying to coax the lids onto the wells of one of the white PCR plates, the caps popped off on one side ejecting some of the solution from the samples. After all that master mix this might have compromised some of the EF-1a samples. Ugh.
11/15/2010 -- LAB 7
Summary:
The purpose of lab 7 was to (1) perform a dot blot on C.gigas DNA and (2) run qPCR on C. gigas gill tissue cDNA from all three dissolve oxygen acclimation groups.
Materials & Methods:
Dot blot: DNA Dilutions
1) Collected sample 55 (tissue and treatment unknown): concentration = 107 ng/ul
2) An initial 50ng/ul dilution was made, total volume = 40ul
C1V1 = C2V2
(107)V1 = (50)(40)
V1 = 18.7ul of DNA
40 - V1 = volume of H20 = 21.3ul
3) The five dilutions below were prepared using the new 50ng/ul DNA sample (1.5ml SNAP CAP tubes - necessary for denaturing in warm water)
| Dilution |
TARGET amount |
ul of H20 |
ul of 20X SSC |
ul of 50ng/ul DNA sample |
| 1 |
800 ng |
124 |
60 |
16 |
| 2 |
400 ng |
132 |
60 |
8 |
| 3 |
200 ng |
136 |
60 |
4 |
| 4 |
100 ng |
138 |
60 |
2 |
| 5 |
50 ng |
139 |
60 |
1 |
Dot blot
1) Nylon membrane for 72 wells of manifold was cut and soaked in 6X SSC for 10 in top of tip box
2) Filter paper was cut to size of membrane and wet in 6X SSC
3) Manifold was assembled: membrane on top of filter paper
4) DNA samples denatured in boiling water for 10min
5) 500ul of 6X SCC was applied to each well and the solution was allowed to filter through the membrane (vacuum speed adjusted so filtration took ~2min)
6) DNA samples were spun down
7) Entire volume was transfer into each well
8) Samples were filtered through
9) While samples were filtering through the membrane, filter paper was soaked in denaturation buffer fro 10min
10) Filter was then neutralization-soaked for 5min
11) Manifold was dismantled and membrane was transferred (dot side up) on to the treated filter paper for 5min
12) Membrane was then placed on dry filter paper and left to dry
13) Dried blot was wrapped in plastic wrap and placed DNA-side-down on UV transluminator for 2min at 120kJ (immobilizes DNA)
WesternBreeze® CHROMOGENIC IMMUNODETECTION
1) 20 mL of Blocking Solution was prepared
Ultra filtered Water = 14 ml
Blocker/Diluent (Part A) = 4 m
Blocker/Diluent (Part B) = 2 m
Total Volume = 20 ml
2) Membrane was placed in 10 ml of Blocking Solution in a covered, plastic dish.
3) Incubated for 30 minutes on a rotary shaker set at 1 revolution/sec.
4) Blocking Solution was decanted
5) Membrane was rinsed with 20 ml of water for 5 minutes, then decanted and repeated
6) 10 mL of Primary Antibody Solution (1:5000 dilution) was prepared
Blocking Solution = 10 ml
5-MeC antibody = 2 µl
Total Volume = 10 ml
7) Membrane was incubated with 10 ml of Primary Antibody Solution for 1 hour
8) Primary antibody was decanted and membrane was washed for 5 mins with 20 ml of 1x TBS-T. Decanted and repeated three more times
9) Membrane was incubated in 10 ml of secondary antibody solution for 30 mins. Decanted
10) Membrane was washed for 5 mins with 20 ml of TBS-T. Decanted and repeated three more times
11) Membrane was rinsed with 20 ml of water for 2 minutes, then decanted. Repeated twice.
12) Incubated the membrane in 5 ml of Chromogenic Substrate until color began to develop (1-60 mins)
13) Rinsed the membrane with 20 ml of water for 2 minutes, then decanted. Repeated twice.
14) Membrane was finally dried on a clean piece of filter paper
qPCR
1) Prepared master mix for template, duplicate, two NTCs, and +1 for pipetting error (= 5 rxns)
For a 25μl reaction volume:
| Component |
Volume |
Final Conc. |
Multiplier |
Total volume |
| Master Mix, 2X (Immomix) |
12.5µL |
1x |
x5 |
62.5ul |
| Syto-13 dye (50uM) |
1µL |
2µM |
x5 |
5ul |
| upstream primer, 10μM |
1.25μl |
2.5μM |
x5 |
6.25ul |
| downstream primer, 10μM |
1.25μl |
2.5μM |
x5 |
6.25ul |
| Ultra Pure Water |
7uL |
NA |
x5 |
30ul |
2. Added mastermix to wells of a white PCR plate
3. Thawed cDNA sample: Hyperoxic 1
4. Added 2uL cDNA template to each reaction
5. Added 2uL of ultra pure water (0.1% DEPC) to the negative control wells
6. Capped the wells securely
7. Spun down strips to collect volume in the bottom of the wells
8. Ensured the lids were clean and place strips on ice
9. Load the plate, verify the PCR conditions and start the run
PCR conditions:
95°C for 10 minutes
95°C for 15s
55 °C for 15 s
72°C for 30 s (+ plate read)
Returned to step 2 39 more times
95°C for 10s
Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Results:
(1) Dot blot: Positive detection of DNA methylation.
 |
| Dot blot |
(2) qPCR: results will be discussed 11/22/2011
Conclusion: Based on the dot blot results, there appears to be DNA methylation for most of the tissue samples. This is not surprising considering it's known that oysters have Mosaic-like methylation. However, there does appear to be more methylation (darker blots) in the last five columns; potentially the more stressed individuals. In addition, the "threshold" concentration appears to be the third dilution (200ng/ul; for the most part dots start to diminish after this dilution).
Reflection: It's fascinating to not only evaluate the genomic expression variation as a result of a stressor, but also the epigenome. I wonder if one is more telling of the physiological consequences of a stressor than the other (depending on the stressor)?
11/9/2010 -- LAB 6
Materials & Methods (continuation of 11/8/2011 lab):
RNA Quantification
RNA samples were always kept on ice
1) 2ul of 0.1% DEPC-H2O was pipetted onto the Nanodrop pedestal and the arm was lowered
2) The Nanodrop spectrophotometer was zero by clicking "Blank"
3) 2ul of RNA was placed on the Nanodrop pedestal and the arm was lowered
4) By clicking "Measure" the RNA concentration and 260/280 ratio was quantified (and recorded)
5) Between each measurement the top and bottom of the pedestal was wiped with a KimWipe
6) Samples were returned to -80oC
Reverse Transcription (RT)
1. Once our stock RNA thawed, we immediately inverted the tube to mix our sample.
2. We labeled a 0.5ml PCR tube with our initials and "cDNA".
3. Pipetted 5µl of our stock RNA into the PCR tube
4. Added 1µl of oligo dT
5. Added 4µl of nuclease free H2O (i.e., DEPC)
6. In a thermocycler, we incubated the mix at 70C for 5min
7. After incubation, we place the mix on ice for 2min
8. Spun the sample down
9. Added 5µl of M-MLV 5x Reaction buffer
10. Added 5µl of dNTPs
11. Added 5µl of M-MLV Reverse Transcriptase (RT)
12. Added 4µl more of M-MLV nuclease free H2O
13. Vortexed the mix for several seconds
14. Spun it down
15. Placed back in the thermocycler, the mix was incubated for 60min at 42C, then heat inactivate at 70C for 3min.
16. Mix spun down and stored at -20C
Result:
Average RNA concentrations & 260/280 ratios for each acclimation group:
Control: 391.5 ng/ul ; 1.83
Low DO: 424.6 ng/ul ; 1.82
High DO: 252.15 ng/ul; 1.82
Conclusions:
Based on the concentration and 260/280 ratios, it appears we have sufficient RNA to proceed to the next steps. We also completed RT and now have our (template) cDNA for qPCR.
Reflections:**
It took an excessive amount of time to complete these processes. I hope in the future I can cut that time in half (if not more)! However, the results appear to be promising....so maybe slow and steady can still win the race....eventually.
11/8/2010
Summary:
The purpose of lab 6 was to (1) isolate and extracting total RNA from C. gigas gill tissue from all three dissolve oxygen acclimation groups and (2) produce cDNA.
Materials & Methods:
RNA Extraction
1) ~0.002-0.003g of gill tissue from each osyter (N=15) was homogenized with a sterile pestle in 500ul of TriReagent in a 1.5ml microfuge tube
2) Shortly vortex
3) Additional 500ul of TriReagent was then added to the test tube
4) Vortexed for 15s
5) Homogenized tissues were incubated at RT for 5min
6) In the fume hood 200ul of chloroform was added to each sample
7) Each sample was then vortex for 30s or until solution becomes "milky"
8) The samples were then incubated again at RT for 5min
9) The samples were spun in a refrigerated (4oC) microfuge for 15min at max speed
10) The tubes were gently removed
11) The aqueous phase (clear top layer) ONLY was carefully transferred into separate 1.5ml microfuge tubes
12) 500ul of isopropanol was then added to each new tube containing the aqueous phase
13) The solution was mixed well by inverting the tubes several times (when solution no longer appeared viscous or lumpy, particularly near the edges)
14) The samples were then incubated at RT for 10min
15) Spun down at max speed for 8min with the TUBE HINGE away from the center
16) A small white pellet should have formed (if not visible, just continue)
17) The supernatant was removed
18) 1ml of 75% Ethanol (EtOH) was added to each pellet
19) Each pellet was vortex to dislodge pellet
20) Samples were spun in refrigerated microfuge (4oC) at 7500g for 5min
21) Supernatant was carefully removed, without removing the pellet
22) 15s of spinning to pool any remaining EtOH
23) Residual EtOH removed with small pipette tip (P10 or P20) and blotted with KimWipes (if necessary)
24) Tubes left open to dry at RT (in the hood) for no more than 5min
25) Due to the larger size of the pellets, 200ul of 0.1% DEPC-H2O (instead of 100ul) was added to each tube to resuspend the pellets
26) The samples were then incubated in 55oC water bath for 5min to help solubilize the pellet
27) Each test tube was flicked a few time to mix. If flicking didn't work, the solution was pipetted.
28) The samples were then stored at -80oC
11/01/11 -- LAB 5
Summary:
The purpose of the lab 5 was (1) protein extraction from experimental oyster tissues, (2) electrophoresis (on agarose gel) of PCR samples from lab 4, (3) SDS-PAGE of extracted oyster protein, and (4) Western Blot Immnodetection of the SDS-PAGE gel.
Materials & Methods:
Protein extraction
Same methods were conducted from Lab 1. However, the microfuge was not refrigerated.
Agarose Gel Electrophoresis
1. Gel was placed in 1x TAE buffer filled gel box
2. 5uL of 100bp ladder was loaded in the far left lane
3. 25uL of each PCR sample (from lab 4--bright green) was loaded into the wells of the gel
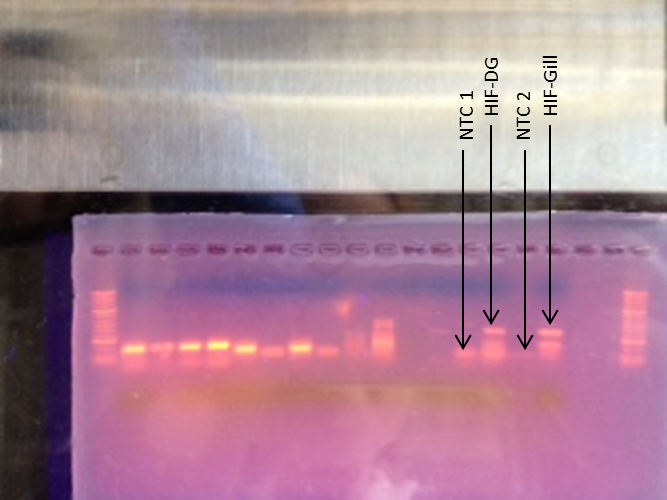
-Our samples were loaded into lanes 14,15,16, and 17 containing NTC 1, HIF digestive gland, NTC 2, and HIF gill, respectively
4. Gel was ran at 100V for ~60min
5. Gel was visualized with UV transilluminator.
SDS-PAGE
1. 15ul of our extracted protein (from this lab) and 15ul of 2X Reducing Sample Buffer to a 1.5mL SCREW CAP.
2. The sample was flicked (mixed) and centrifuged for 10s
3. The samples were boiled for 5min
4. Gel was placed into gel box and wells were rinsed thoroughly with a pipette (creating eddies within each well)
5. The samples centrifuged for 1min after boiling
6. The entire sample (30uL) was then loaded into one of the wells (lane 6,8, and 9)
7. Gel box was hooked up to power supply and ran at 150V for ~20min (the gel ran fast so it didn't take the proposed 45min)
8. Power was turned off and unhooked
9. Lid removed
10, Tension wedge disengaged and gel was removed
11. The cassette containing the gel was carefully cracked open (using reverse end of forceps)
12. Wells were trimmed off
13. The right corner of gel was notched to properly orient gel
14. Next step = Western Blot
Western Blot
1. 4 pieces of filter paper, a membrane and the gel (SDS-PAGE) were soaked in Tri-Glycine Transfer Buffer for 15min
2. The blotting sandwich was assembled in the semi-dry blotting apparatus as follows:
- - - - - - - - - - - - - Cathode
2 pieces of filter paper (Top)
Gel
Membrane
2 pieces of filter paper (Bottom)
+ + + + + + + + + Anode
Air bubbles were rolled out of the first layer of filter paper and the full sandwich (with Falcon tube). Excess water was wiped off.3. Blot transferred for 30min at 20V
4. Gel was removed from the sandwich and rinsed adhering pieces of gel were washed with transfer buffer
5. Membrane washed twice, 5min each, with 20mL of pure water
6. Membrane was then placed in a plastic box and 10mL of Blocking Solution was added. The plastic box was (suppose) to be covered and the membrane was incubated over night a rotary shaker set at 1 revolution/sec
7. Next morning, liquid was decanted
8. Membrane rinsed with 20ml of water for 5min, twice, decanted
9. Membrane incubated in 10ml of Primary Antibody Solution for 30min, then decanted (antibodies bind to HSP70 protein)
10. Membrane rinsed with 20ml of Antibody Wash for 5min x3, decant each time
11. Membrane incubated in 10ml of Secondary Antibody Solution for 30min, then decanted (antibodies bind to mouse protein)
12. Membrane washed for 5min with 20ml of Antibody wash x3, decant each time
13. Membrane rinsed with 20mL of pure water for 2 minutes x2
14. Membrane incubated in 5mL of Chromogenic Substrate until purple bands appeared; this took approximately 40min.
15. Membrane was dried on a clean piece of filter paper in the open air
Results:
Agarose Gel
NTC 1 was contaminated and some primer-dimers occurred in NTC 2. Even worse, we had two product bands for both samples.
SDS-PAGE and Western Blot
None of the oyster protein transferred.
Conclusion:
Our agarose gel didn't work, but we will be using different (and smaller) primers when evaluating the actual experimental (fresher) samples. The Western Blot also didn't work, potentially because the antibodies were not specific enough to bind to the HSP70 protein (Primary Antibody Solution) in oysters. Hopefully we are more successful with our hypoxia treated samples.
Reflection:
After running the slew of gels, it's apparent it's easy to make mistakes. Some where along the line our cDNA was compromised and thus the assay didn't work. I think for the next round concerning our actual treatment samples it will go a lot smoother (and successful), now that one person is responsible for thier portion of the study.
10/25/2011-- LAB 4
Summary:
The purpose of lab 4 was to (1) conduct PCR on the mock-oyster tissue samples using the "first-try" primer pairs of HIF-1a we ordered and (2) dissect gill and mantel samples form C. gigas for the group experiment.
Materials & Methods:
Primer Rehydration
1. Spun down the dry primer for 2-3min
2. Made 100uM primer stock (for each primer) by adding 363ul TE buffer (pH 8.5) to the forward primer (36.3 nm) and 280ul to the reverse primer (28.0 nm).
3. Incubated stocks at 35oC for 10min.
4. Vortexed for several seconds
5. Spun down stocks for several seconds
6. Then prepared a 10uM working stock (for PCR) by adding 90ul of nuclease free water (DEPC) and 10ul of of the 100uM stock into a new 1.5ml test tube.
Master Mix & PCR
7. We then prepared the following master mix (MM) for a 50ul PCR reaction:

** Quantities of 2xGoTag, 10uM primer (i.e., working stock prepared above), and H2O were all multiplied by 5 for 2 rxn (DG and gill cDNA), 2 non-template controls (NTC), and 1 to account for pipetting error.
8. Four 0.5ml PCR tubes were labled and 48ul of MM were pipetted into each one.
9. 2ul of the DG template and 2ul of the gill template were pipetted into separate PCR tubes.
10. 2ul of nuclease free water was added to each NTC PCR tube
11. All four tubes were spun down and placed in the thermocycler to go through the following cycling profile:
| Step |
Temperature |
Time |
Cycles |
| Denaturation |
95C |
5 min |
1 |
| Denaturation |
95C |
30 sec |
40 |
| Annealing |
55C |
30 sec |
|
| Extension |
72C |
90 sec |
|
| Final extension |
72C |
3 min |
1 |
| Hold |
4C |
∞ |
1 |
Tissue Extraction
We collected a total of 30 tissue samples from our 15 oysters: 15 mantle and 15 gill.
1. We dissected one oyster at a time, while the other oysters remain submerged.
2. Oyster were shucked
3. With sterile forceps and scissors (for each organ) the mantel and gill tissue were placed into separate, pre-labeled tubes. All interments were sterilized by dunking them in bleach then ethanol between each tissue extraction.
4. All tissue samples were immediately placed on dry ice, then transferred to a -80oC freezer.
Results:
Amplified segments of the HIF-1a gene in C.gigas.
Reflection:
The purpose of Lab 4 was to finially isolate a gene of interest (HIF-1a) and amplify the target gene with PCR so there is enough to measure (e.g., agarose gel). We now have tissue samples from hypoxia treated oysters and will begin RNA isolation and extraction in Lab 5.
10/18/2011-- LAB 3
Summary:
The purpose of lab 3 was to (1) reverse transcribe the RNA (i.e., stock RNA) extracted from C. gigas (digestive gland), (2) set up the group experiment (3) and develop primers for the lab group study.
Materials & Methods:
Reverse Transcription
1. Once our stock RNA thawed, we immediately inverted the tube to mix our sample.
2. We labeled a 0.5ml PCR tube with our initials and "cDNA".
3. Pipetted 5µl of our stock RNA into the PCR tube
4. Added 1µl of oligo dT
5. Added 4µl of nuclease free H2O (i.e., DEPC)
6. In a thermocycler, we incubated the mix at 70C for 5min
7. After incubation, we place the mix on ice for 2min
8. Spun the sample down
9. Added 5µl of M-MLV 5x Reaction buffer
10. Added 5µl of dNTPs
11. Added 5µl of M-MLV Reverse Transcriptase (RT)
12. Added 4µl more of M-MLV nuclease free H2O
13. Vortexed the mix for several seconds
14. Spun it down
15. Placed back in the thermocycler, the mix was incubated for 60min at 42C, then heat inactivate at 70C for 3min.
16. Mix spun down and stored at -20C
Experimental set up
Three 5 gallon buckets (6L of water each) were set up for the different dissolved oxygen (DO) acclimation levels: high DO (9+ mg/l), normoxia (6-8mg/l), and low DO (3-4mg/l) at 13± 1oC. The high DO saltwater was prepared by pumping O2 gas into the water so DO was > 9mg/l. The normoxic tank was fixed with an air pump, while the low DO acclimation was prepared by bubbling nitrogen gas until DO ~ 4mg/l; the hypoxic treatment tank will be prepared in the same manner, but the DO will be reduced to < 2 mg/l. Each acclimation bucket will be checked three times a day to monitor DO, temperature, and overall condition of the oysters
Primers
See Hypoxia wiki page.
Results
Team work and hopefully cDNA!
Reflection:
The purpose of Lab 3 was to familiarize ourselves with the step-by-step techniques for reverse transcription of an RNA sample, which is an essential step in developing a more stable template (i.e., cDNA) that can later be measured. Again, every step was clear, well organized, and easy to follow. In addition, we completed the set up for our group experiment. The experiment itself will be completed by Monday so tissue dissection can occur Tuesday in lab 4.
10/18/2011-- LAB 2
In the field. Constructed the HYPOXIA experimental design and wiki page.
10/4/2011 -- LAB 1
Summary:
The purpose of lab 1 was to (1) begin extracting RNA from C. gigas -
Materials & Methods:
RNA Extraction
1) 0.0016g of DG was homogenized (as much as possible) with a sterile pestle in 500ul of TriReagent.
2) Shortly vortex
3) Additional 500ul of TriReagent was then added to the test tube
4) Vortexed for 15s
5) Then the sample was stored at -80C
Protein Extraction & Analysis
1) 0.013g of gill was homogenized with a sterile pestle in CellLytic MT solution.
2) Tube was then inverted several times
3) Spun in a refrigerated microfuge for 10min - what temperature? -
4) The spernatant (protein sample) was then transferred into a new tube and stored on ice
5) 15ul of the portein sample was diluted with 15ul of DI in a new test tube
6) A control test tube was filled with 30µl of DI water -
7) 1.5ml of Bradford reagent (grey solution) was added to both test tubes
8) 1000ul from each tube was mixed (w/ pipette) and transferred into disposable cuvettes
9) Control cuvette zeroed spectrometer (wavelength = 595nm) - absorbance was what you measured -
10) Protein sample measured twice (mixing occurred between measurements)
11) Protein sample stored at -20C
Results:
Absorbance 1: 0.122nm
Absorbance 2: 0.120nm
AVERAGE: 0.121nm
Standard curve concentration vs. absorbance:
y = 1013.9x
y = 1013.9*0.121*2 <--- Multiplied by 2 to account for 1:2 dilution
y = 245.36ug/mL
Conclusions:
Based on the standard curve, the measured absorbance, and thus protein concentration, are reasonable results. The next step is to complete the RNA extraction and analysis.
Reflection:
The purpose of Lab 1 was to familiarize ourselves with the step-by-step techniques for RNA and protein extraction. These procedures are the essential first steps in the isolation and quantification of specific genes of interest. Every step was clear, well organized, and easy to follow. Bring on Lab 2!